Isolation, Purification, and Application of Protoplasts and Transient Expression Systems in Plants
Abstract
:1. Introduction
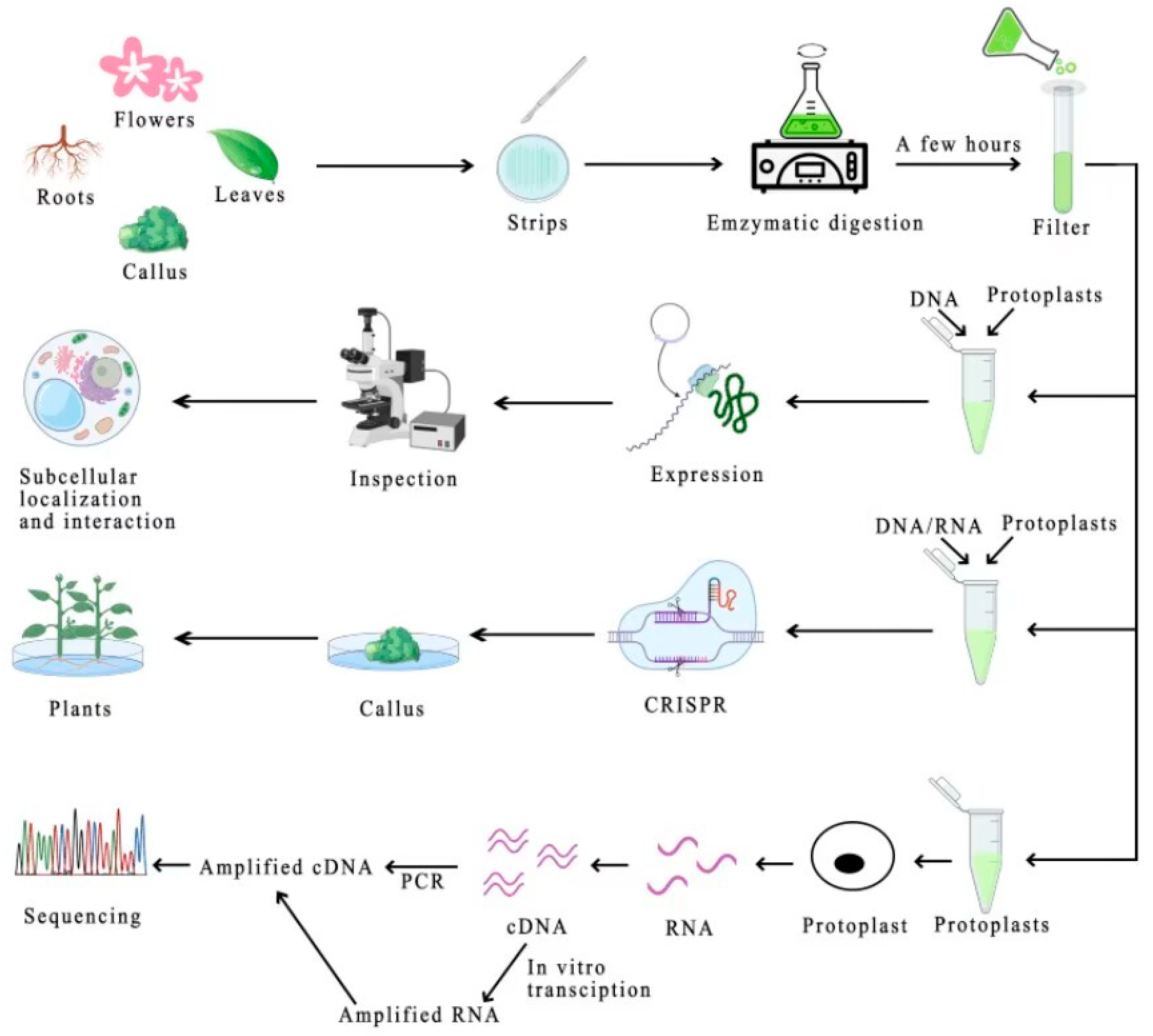
2. Protoplast Isolation
2.1. Plant Tissues for Protoplast Isolation
2.2. Methods of Protoplast Separation and Isolation
2.3. Pretreatment of Enzymatically Isolated Plant Material
2.4. Factors Influencing the Preparation of Protoplasts
3. Purification of Protoplasts
4. Protoplast Yield and Viability Assay
5. Protoplasmic Transient Gene Expression System
6. Applications of Protoplasts and Their Transient Transformation Systems
6.1. Protein Subcellular Localization and Interaction Analysis
6.2. Gene Expression Regulation and Functional Assays
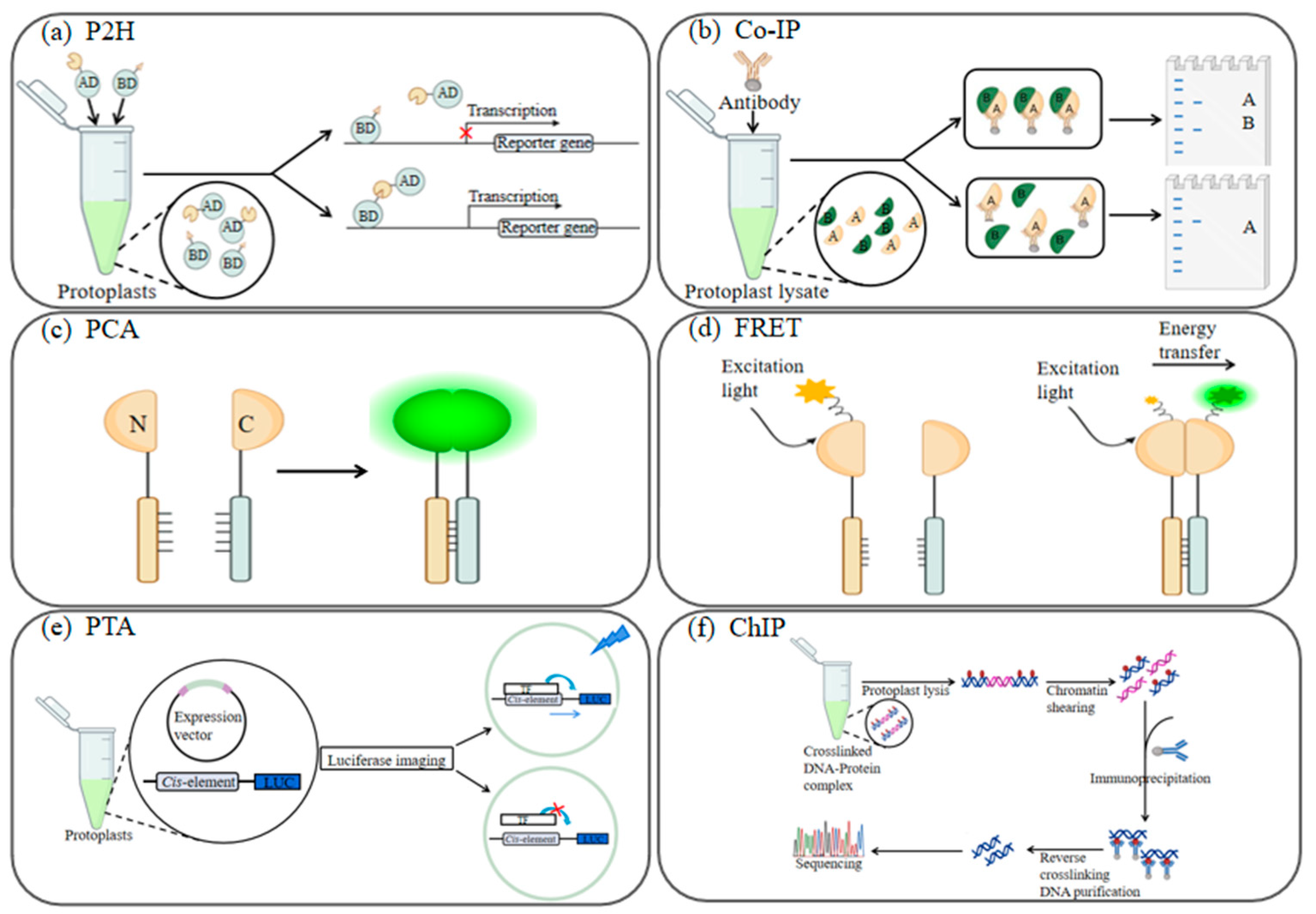
6.3. Gene Editing Advancements in Protoplasts
6.4. Protoplasts Empowering Single-Cell RNA Sequencing
7. Summary Remarks and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xu, Y.; Li, R.; Luo, H.; Wang, Z.; Li, M.-W.; Lam, H.-M.; Huang, C. Protoplasts: Small Cells with Big Roles in Plant Biology. Trends Plant Sci. 2022, 27, 828–829. [Google Scholar] [CrossRef] [PubMed]
- Reyna-Llorens, I.; Ferro-Costa, M.; Burgess, S.J. Plant Protoplasts in the Age of Synthetic Biology. J. Exp. Bot. 2023, 74, 3821–3832. [Google Scholar] [CrossRef] [PubMed]
- Klimek-Chodacka, M.; Kadluczka, D.; Lukasiewicz, A.; Malec-Pala, A.; Baranski, R.; Grzebelus, E. Effective Callus Induction and Plant Regeneration in Callus and Protoplast Cultures of Nigella damascena L. Plant Cell Tissue Organ Cult. 2020, 143, 693–707. [Google Scholar] [CrossRef]
- Sakai, K.; Charlot, F.; Le Saux, T.; Bonhomme, S.; Nogué, F.; Palauqui, J.-C.; Fattaccioli, J. Design of a Comprehensive Microfluidic and Microscopic Toolbox for the Ultra-Wide Spatio-Temporal Study of Plant Protoplasts Development and Physiology. Plant Methods 2019, 15, 79. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Liu, H.; Gou, B.; Hu, W.; Qin, L.; Shen, W.; Wang, A.; Cui, H.; Dai, Z. Direct Leaf-Peeling Method for Areca Protoplasts: A Simple and Efficient System for Protoplast Isolation and Transformation in Areca Palm (Areca catechu). BMC Plant Biol. 2023, 23, 56. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.-J.; Hung, Y.-L.; Chen, T.-Y.; Shih, Y.-A.; Lin, Y.-C.J.; Wang, C.-N. Development of a Petal Protoplast Transfection System for Sinningia speciosa. Appl. Plant Sci. 2022, 10, e11476. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, P.; Cheng, S.; Zhao, Z.; Liu, Y.; Wei, Y.; Lu, Q.; Han, J.; Cai, X.; Zhou, Z.; et al. Protoplast Dissociation and Transcriptome Analysis Provides Insights to Salt Stress Response in Cotton. Int. J. Mol. Sci. 2022, 23, 2845. [Google Scholar] [CrossRef]
- Bertini, E.; Tornielli, G.B.; Pezzotti, M.; Zenoni, S. Regeneration of Plants from Embryogenic Callus-Derived Protoplasts of Garganega and Sangiovese Grapevine (Vitis vinifera L.) Cultivars. Plant Cell Tissue Organ Cult. 2019, 138, 239–246. [Google Scholar] [CrossRef]
- Masani, M.Y.A.; Parveez, G.K.A.; Noll, G.; Fizree, M.D.P.M.A.A.; Sambanthamurthi, R.; Pruefer, D. Protoplast Isolation and Transformation in Oil Palm. Methods Mol. Biol. 2022, 2464, 187–202. [Google Scholar] [CrossRef]
- Kang, H.H.; Naing, A.H.; Kim, C.K. Protoplast Isolation and Shoot Regeneration from Protoplast-Derived Callus of Petunia Hybrida cv. Mirage Rose. Biology 2020, 9, 228. [Google Scholar] [CrossRef]
- Ren, R.; Gao, J.; Lu, C.; Wei, Y.; Jin, J.; Wong, S.-M.; Zhu, G.; Yang, F. Highly Efficient Protoplast Isolation and Transient Expression System for Functional Characterization of Flowering Related Genes in Cymbidium Orchids. Int. J. Mol. Sci. 2020, 21, 2264. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-H.; Xue, H.-W. Plant Regeneration from Cultured Protoplasts. In Morphogenesis in Plant Tissue Cultures; Soh, W.-Y., Bhojwani, S.S., Eds.; Springer: Dordrecht, The Netherlands, 1999; pp. 37–70. ISBN 978-94-015-9253-6. [Google Scholar]
- Rahmani, M.-S.; Pijut, P.M.; Shabanian, N. Protoplast Isolation and Genetically True-to-Type Plant Regeneration from Leaf- and Callus-Derived Protoplasts of Albizia julibrissin. Plant Cell Tissue Organ Cult. 2016, 127, 475–488. [Google Scholar] [CrossRef]
- Priyadarshani, S.V.G.N.; Hu, B.; Li, W.; Ali, H.; Jia, H.; Zhao, L.; Ojolo, S.P.; Azam, S.M.; Xiong, J.; Yan, M.; et al. Simple Protoplast Isolation System for Gene Expression and Protein Interaction Studies in Pineapple (Ananas comosus L.). Plant Methods 2018, 14, 95. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Jung, H.-I.; Vatamaniuk, O.K. Isolation of Protoplasts from Tissues of 14-Day-Old Seedlings of Arabidopsis Thaliana. J. Vis. Exp. 2009, 30, 1149. [Google Scholar] [CrossRef]
- Sun, B.; Yuan, Q.; Zheng, H.; Liang, S.; Jiang, M.; Wang, M.-M.; Chen, Q.; Li, M.-Y.; Zhang, Y.; Luo, Y.; et al. An Efficient and Economical Protocol for Isolating, Purifying and PEG-Mediated Transient Gene Expression of Chinese Kale Hypocotyl Protoplasts. Plants 2019, 8, 385. [Google Scholar] [CrossRef]
- Ma, W.; Yi, F.; Xiao, Y.; Yang, G.; Chen, F.; Wang, J. Isolation of Leaf Mesophyll Protoplasts Optimized by Orthogonal Design for Transient Gene Expression in Catalpa bungei. Sci. Hortic. 2020, 274, 109684. [Google Scholar] [CrossRef]
- Meyer, L.; Serek, M.; Winkelmann, T. Protoplast Isolation and Plant Regeneration of Different Genotypes of Petunia and Calibrachoa. Plant Cell Tissue Organ Cult. 2009, 99, 27–34. [Google Scholar] [CrossRef]
- Lin, Z.; Huang, L.-J.; Yu, P.; Chen, J.; Du, S.; Qin, G.; Zhang, L.; Li, N.; Yuan, D. Development of a Protoplast Isolation System for Functional Gene Expression and Characterization Using Petals of Camellia Oleifera. Plant Physiol. Biochem. 2023, 201, 107885. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhao, R.; Ye, T.; Guan, R.; Xu, L.; Ma, X.; Zhang, J.; Xiao, S.; Yuan, D. Isolation, Purification and PEG-Mediated Transient Expression of Mesophyll Protoplasts in Camellia Oleifera. Plant Methods 2022, 18, 141. [Google Scholar] [CrossRef]
- Kim, A.L.; Yun, Y.J.; Choi, H.W.; Hong, C.-H.; Shim, H.J.; Lee, J.H.; Kim, Y.-C. Establishment of Efficient Cannabis (Cannabis sativa L.) Protoplast Isolation and Transient Expression Condition. Plant Biotechnol. Rep. 2022, 16, 613–619. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Lü, T.; Yang, X.; Liu, J.; Dong, Y.; Wang, Y. An Efficient and Universal Protoplast Isolation Protocol Suitable for Transient Gene Expression Analysis and Single-Cell RNA Sequencing. Int. J. Mol. Sci. 2022, 23, 3419. [Google Scholar] [CrossRef] [PubMed]
- Adedeji, O.S.; Naing, A.H.; Kim, C.K. Protoplast Isolation and Shoot Regeneration from Protoplast-Derived Calli of Chrysanthemum cv. White ND. Plant Cell Tissue Organ Cult. 2020, 141, 571–581. [Google Scholar] [CrossRef]
- Huang, H.; Wang, Z.; Cheng, J.; Zhao, W.; Li, X.; Wang, H.; Zhang, Z.; Sui, X. An Efficient Cucumber (Cucumis sativus L.) Protoplast Isolation and Transient Expression System. Sci. Hortic. 2013, 150, 206–212. [Google Scholar] [CrossRef]
- Ren, R.; Gao, J.; Yin, D.; Li, K.; Lu, C.; Ahmad, S.; Wei, Y.; Jin, J.; Zhu, G.; Yang, F. Highly Efficient Leaf Base Protoplast Isolation and Transient Expression Systems for Orchids and Other Important Monocot Crops. Front. Plant Sci. 2021, 12, 626015. [Google Scholar] [CrossRef] [PubMed]
- Liqing, Z.; Bochu, W.; Jing, Z.; Lingxi, C.; Chuanyun, D.; Chuanren, D. Protoplast Isolation of Callus in Echinacea augustifolia. Colloids Surf. B Biointerfaces 2005, 44, 1–5. [Google Scholar] [CrossRef]
- Lai, Q.; Wang, Y.; Zhou, Q.; Zhao, Z. Isolation and Purification of Mesophyll Protoplasts from Ginkgo biloba L. Cytologia 2020, 85, 27–32. [Google Scholar] [CrossRef]
- Han, X.; Rong, H.; Feng, Y.; Xin, Y.; Luan, X.; Zhou, Q.; Xu, M.; Xu, L.-A. Protoplast Isolation and Transient Transformation System for Ginkgo biloba L. Front. Plant Sci. 2023, 14, 1145754. [Google Scholar] [CrossRef]
- Wang, P.; Pu, Y.; Abid, M.A.; Kang, L.; Ye, Y.; Zhang, M.; Liang, C.; Wei, Y.; Zhang, R.; Meng, Z. A Rapid and Efficient Method for Isolation and Transformation of Cotton Callus Protoplast. Int. J. Mol. Sci. 2022, 23, 8368. [Google Scholar] [CrossRef]
- Li, N.-N.; Ding, L.-Y.; Zhang, Z.-Y.; Guo, W.-Z. Isolation of Mesophyll Protoplast and Establishment of Gene Transient Expression System in Cotton. Acta Agron. Sin. 2014, 40, 231. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, S.; Fu, Y.; Wang, Z.; Yang, X.; Li, W.; Zhang, C.; Zhang, D.; Li, J. Establishment of an Efficient Cotton Root Protoplast Isolation Protocol Suitable for Single-Cell RNA Sequencing and Transient Gene Expression Analysis. Plant Methods 2023, 19, 5. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; He, C.; Luo, H. An Efficient Transient Mesophyll Protoplast System for Investigation of the Innate Immunity Responses in the Rubber Tree (Hevea brasiliensis). Plant Cell Tissue Organ Cult. 2016, 126, 281–290. [Google Scholar] [CrossRef]
- Ahmed, M.A.A.; Miao, M.; Pratsinakis, E.D.; Zhang, H.; Wang, W.; Yuan, Y.; Lyu, M.; Iftikhar, J.; Yousef, A.F.; Madesis, P.; et al. Protoplast Isolation, Fusion, Culture and Transformation in the Woody Plant Jasminum spp. Agriculture 2021, 11, 699. [Google Scholar] [CrossRef]
- Huo, A.; Chen, Z.; Wang, P.; Yang, L.; Wang, G.; Wang, D.; Liao, S.; Cheng, T.; Chen, J.; Shi, J. Establishment of Transient Gene Expression Systems in Protoplasts from Liriodendron Hybrid Mesophyll Cells. PLoS ONE 2017, 12, e0172475. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-Z.; Liu, Q.; Geng, X.-S.; Li, K.-M.; Luo, L.-J.; Liu, J.-P. Highly Efficient Mesophyll Protoplast Isolation and PEG-Mediated Transient Gene Expression for Rapid and Large-Scale Gene Characterization in Cassava (Manihot esculenta Crantz). BMC Biotechnol. 2017, 17, 29. [Google Scholar] [CrossRef] [PubMed]
- Jia, N.; Zhu, Y.; Xie, F. An Efficient Protocol for Model Legume Root Protoplast Isolation and Transformation. Front. Plant Sci. 2018, 9, 670. [Google Scholar] [CrossRef]
- Hsu, C.-T.; Lee, W.-C.; Cheng, Y.-J.; Yuan, Y.-H.; Wu, F.-H.; Lin, C.-S. Genome Editing and Protoplast Regeneration to Study Plant-Pathogen Interactions in the Model Plant Nicotiana benthamiana. Front. Genome 2020, 2, 627803. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, J.; Duan, S.; Ao, Y.; Dai, J.; Liu, J.; Wang, P.; Li, Y.; Liu, B.; Feng, D.; et al. A Highly Efficient Rice Green Tissue Protoplast System for Transient Gene Expression and Studying Light/Chloroplast-Related Processes. Plant Methods 2011, 7, 30. [Google Scholar] [CrossRef]
- Lin, C.-S.; Hsu, C.-T.; Yang, L.-H.; Lee, L.-Y.; Fu, J.-Y.; Cheng, Q.-W.; Wu, F.-H.; Hsiao, H.C.-W.; Zhang, Y.; Zhang, R.; et al. Application of Protoplast Technology to CRISPR/Cas9 Mutagenesis: From Single-Cell Mutation Detection to Mutant Plant Regeneration. Plant Biotechnol. J. 2018, 16, 1295–1310. [Google Scholar] [CrossRef]
- Li, J.; Liao, X.; Zhou, S.; Liu, S.; Jiang, L.; Wang, G. Efficient Protoplast Isolation and Transient Gene Expression System for Phalaenopsis hybrid Cultivar ‘Ruili Beauty’. Vitr. Cell. Dev. Biol. Plant 2018, 54, 87–93. [Google Scholar] [CrossRef]
- Torres, K.C. Overview of Protoplast Isolation and Culture. In Tissue Culture Techniques for Horticultural Crops; Torres, K.C., Ed.; Springer: Boston, MA, USA, 1989; pp. 189–199. ISBN 978-1-4615-9756-8. [Google Scholar]
- Nanjareddy, K.; Arthikala, M.-K.; Blanco, L.; Arellano, E.S.; Lara, M. Protoplast Isolation, Transient Transformation of Leaf Mesophyll Protoplasts and Improved Agrobacterium-Mediated Leaf Disc Infiltration of Phaseolus vulgaris: Tools for Rapid Gene Expression Analysis. BMC Biotechnol. 2016, 16, 53. [Google Scholar] [CrossRef]
- Tan, B.; Xu, M.; Chen, Y.; Huang, M. Transient Expression for Functional Gene Analysis Using Populus protoplasts. Plant Cell Tissue Organ Cult. 2013, 114, 11–18. [Google Scholar] [CrossRef]
- Guo, J.; Morrell-Falvey, J.L.; Labbé, J.L.; Muchero, W.; Kalluri, U.C.; Tuskan, G.A.; Chen, J.-G. Highly Efficient Isolation of Populus Mesophyll Protoplasts and Its Application in Transient Expression Assays. PLoS ONE 2012, 7, e44908. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Liao, X.; Gan, Z.; Peng, X.; Wang, P.; Li, S.; Li, T. Protoplast Isolation and Development of a Transient Expression System for Sweet Cherry (Prunus avium L.). Sci. Hortic. 2016, 209, 14–21. [Google Scholar] [CrossRef]
- Bai, L.; Cheng, Y.; She, J.; He, Z.; Liu, H.; Zhang, G.; Cao, R.; Chen, Y. Development of an Efficient Protoplast Isolation and Transfection System for Castor Bean (Ricinus communis L.). Plant Cell Tissue Organ Cult. 2020, 143, 457–464. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, G.; Chen, Z.; Han, J.; Hu, Y.; Wang, K. Optimization of Protoplast Isolation, Transformation and Its Application in Sugarcane (Saccharum spontaneum L). Crop J. 2021, 9, 133–142. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Dong, Y.; Li, D.; Shi, S.; Li, S.; Li, L.; He, Y.; Li, J.; Chen, H.; et al. A Highly Efficient Mesophyll Protoplast Isolation and PEG-Mediated Transient Expression System in Eggplant. Sci. Hortic. 2022, 304, 111303. [Google Scholar] [CrossRef]
- Zhang, L.; Yung, W.-S.; Wang, Z.; Li, M.-W.; Huang, M. Optimization of an Efficient Protoplast Transformation System for Transient Expression Analysis Using Leaves of Torenia fournieri. Plants 2022, 11, 2106. [Google Scholar] [CrossRef]
- Jia, X.; Zhang, X.; Qu, J.; Han, R. Optimization Conditions of Wheat Mesophyll Protoplast Isolation. Agric. Sci. 2016, 7, 850–858. [Google Scholar] [CrossRef]
- Shao, Y.; Mu, D.; Pan, L.; Wilson, I.W.; Zheng, Y.; Zhu, L.; Lu, Z.; Wan, L.; Fu, J.; Wei, S.; et al. Optimization of Isolation and Transformation of Protoplasts from Uncaria rhynchophylla and Its Application to Transient Gene Expression Analysis. Int. J. Mol. Sci. 2023, 24, 3633. [Google Scholar] [CrossRef]
- Gu, Z.; Yang, R.; Chen, H. The Establishment of Isolation and Transient Transformation Methods of Protoplasts of Vernicia fordii Mesophyll Cells. Linye Kexue Sci. Silvae Sin. 2018, 54, 46–53. [Google Scholar]
- Wang, H.; Wang, W.; Zhan, J.; Huang, W.; Xu, H. An Efficient PEG-Mediated Transient Gene Expression System in Grape Protoplasts and Its Application in Subcellular Localization Studies of Flavonoids Biosynthesis Enzymes. Sci. Hortic. 2015, 191, 82–89. [Google Scholar] [CrossRef]
- Zhao, F.; Li, Y.-J.; Hu, Y.; Gao, Y.-R.; Zang, X.-W.; Ding, Q.; Wang, Y.-J.; Wen, Y. A Highly Efficient Grapevine Mesophyll Protoplast System for Transient Gene Expression and the Study of Disease Resistance Proteins. Plant Cell Tissue Organ Cult. 2016, 125, 43–57. [Google Scholar] [CrossRef]
- Coy, M.R.; Abbitt, S.E.; Frank, M.J. Protoplast Isolation and Transfection in Maize. Methods Mol. Biol. 2022, 2464, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Yao, D.; Lin, F.; Jiang, M. PEG-Mediated Transient Gene Expression and Silencing System in Maize Mesophyll Protoplasts: A Valuable Tool for Signal Transduction Study in Maize. Acta Physiol. Plant. 2014, 36, 1271–1281. [Google Scholar] [CrossRef]
- Hu, Y.; Song, D.; Gao, L.; Ajayo, B.S.; Wang, Y.; Huang, H.; Zhang, J.; Liu, H.; Liu, Y.; Yu, G.; et al. Optimization of Isolation and Transfection Conditions of Maize Endosperm Protoplasts. Plant Methods 2020, 16, 96. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, R.; Han, S.; Li, Z.; Xiao, J.; Li, Y.; Wang, L.; Li, S. Transcriptome Analysis of Sugarcane Young Leaves and Protoplasts after Enzymatic Digestion. Life 2022, 12, 1210. [Google Scholar] [CrossRef]
- Avila-Peltroche, J.; Won, B.Y.; Cho, T.O. Protoplast Isolation from Dictyopteris Pacifica and Scytosiphon Lomentaria, Using a Simple Commercial Enzyme Preparation. J. Genet. Eng. Biotechnol. 2021, 19, 135. [Google Scholar] [CrossRef]
- Zhou, Y.; Deng, R.; Xu, X.; Yang, Z.; Zhou, Y.; Deng, R.; Xu, X.; Yang, Z. Isolation of Mesophyll Protoplasts from Tea (Camellia sinensis) and Localization Analysis of Enzymes Involved in the Biosynthesis of Specialized Metabolites. Beverage Plant Res. 2021, 1, 2. [Google Scholar] [CrossRef]
- Shrestha, S.; Rahman, M.S.; Qin, W. New Insights in Pectinase Production Development and Industrial Applications. Appl. Microbiol. Biotechnol. 2021, 105, 9069–9087. [Google Scholar] [CrossRef]
- Wu, S.C.; Kuniyuki, A.H. Isolation and Culture of Almond Protoplasts. Plant Sci. 1985, 41, 55–60. [Google Scholar] [CrossRef]
- Augustynowicz, J.; Lekka, M.; Burda, K.; Gabryś, H. Correlation between Chloroplast Motility and Elastic Properties of Tobacco Mesophyll Protoplasts. Acta Physiol. Plant 2001, 23, 291–302. [Google Scholar] [CrossRef]
- Du, J.; Zhang, H.; Li, W.; Li, X.; Wang, Z.; Zhang, Y.; Xiong, A.; Li, M. Optimization of Protoplast Preparation System from Leaves and Establishment of a Transient Transformation System in Apium Graveolens. Agronomy 2023, 13, 2154. [Google Scholar] [CrossRef]
- de Souza, T.S.P.; Kawaguti, H.Y. Cellulases, Hemicellulases, and Pectinases: Applications in the Food and Beverage Industry. Food Bioprocess. Technol. 2021, 14, 1446–1477. [Google Scholar] [CrossRef]
- Qiu, L.; Wang, Y.; Qu, H. Loading Calcium Fluorescent Probes into Protoplasts to Detect Calcium in the Flesh Tissue Cells of Malus domestica. Hortic. Res. 2020, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.Y.; Lee, H.-Y.; Kim, S.W.; Noh, Y.-S.; Seo, P.J. Optimization of Protoplast Regeneration in the Model Plant Arabidopsis Thaliana. Plant Methods 2021, 17, 21. [Google Scholar] [CrossRef] [PubMed]
- Matchett-Oates, L.; Mohamaden, E.; Spangenberg, G.C.; Cogan, N.O.I. Development of a Robust Transient Expression Screening System in Protoplasts of Cannabis. Vitr. Cell. Dev. Biol. Plant 2021, 57, 1040–1050. [Google Scholar] [CrossRef]
- Gou, Y.-J.; Li, Y.-L.; Bi, P.-P.; Wang, D.-J.; Ma, Y.-Y.; Hu, Y.; Zhou, H.-C.; Wen, Y.-Q.; Feng, J.-Y. Optimization of the Protoplast Transient Expression System for Gene Functional Studies in Strawberry (Fragaria vesca). Plant Cell Tissue Organ Cult. 2020, 141, 41–53. [Google Scholar] [CrossRef]
- Xia, P.; Hu, W.; Liang, T.; Yang, D.; Liang, Z. An Attempt to Establish an Agrobacterium-Mediated Transient Expression System in Medicinal Plants. Protoplasma 2020, 257, 1497–1505. [Google Scholar] [CrossRef]
- Sharma, R.; Liang, Y.; Lee, M.Y.; Pidatala, V.R.; Mortimer, J.C.; Scheller, H.V. Agrobacterium-Mediated Transient Transformation of Sorghum Leaves for Accelerating Functional Genomics and Genome Editing Studies. BMC Res. Notes 2020, 13, 116. [Google Scholar] [CrossRef]
- Ozyigit, I.I. Gene Transfer to Plants by Electroporation: Methods and Applications. Mol. Biol. Rep. 2020, 47, 3195–3210. [Google Scholar] [CrossRef]
- Lv, Z.; Jiang, R.; Chen, J.; Chen, W. Nanoparticle-Mediated Gene Transformation Strategies for Plant Genetic Engineering. Plant J. 2020, 104, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Tian, X.; Wang, L.; Zhao, J.; Zhou, J.; He, H.; Dai, L.; Qu, S. Characterization of Agrobacterium-Mediated Co-Transformation Events in Rice Using Green and Red Fluorescent Proteins. Mol. Biol. Rep. 2022, 49, 9613–9622. [Google Scholar] [CrossRef] [PubMed]
- Zhi, H.; Zhou, S.; Pan, W.; Shang, Y.; Zeng, Z.; Zhang, H. The Promising Nanovectors for Gene Delivery in Plant Genome Engineering. Int. J. Mol. Sci. 2022, 23, 8501. [Google Scholar] [CrossRef] [PubMed]
- Kausch, A.P.; Wang, K.; Kaeppler, H.F.; Gordon-Kamm, W. Maize Transformation: History, Progress, and Perspectives. Mol. Breed. 2021, 41, 38. [Google Scholar] [CrossRef] [PubMed]
- Malik, W.A.; Wang, X.; Wang, X.; Shu, N.; Cui, R.; Chen, X.; Wang, D.; Lu, X.; Yin, Z.; Wang, J.; et al. Genome-Wide Expression Analysis Suggests Glutaredoxin Genes Response to Various Stresses in Cotton. Int. J. Biol. Macromol. 2020, 153, 470–491. [Google Scholar] [CrossRef]
- Li, C.; Yue, J.; Wu, X.; Xu, C.; Yu, J. An ABA-Responsive DRE-Binding Protein Gene from Setaria italica, SiARDP, the Target Gene of SiAREB, Plays a Critical Role under Drought Stress. J. Exp. Bot. 2014, 65, 5415–5427. [Google Scholar] [CrossRef]
- Hillwig, M.S.; Contento, A.L.; Meyer, A.; Ebany, D.; Bassham, D.C.; Macintosh, G.C. RNS2, a Conserved Member of the RNase T2 Family, Is Necessary for Ribosomal RNA Decay in Plants. Proc. Natl. Acad. Sci. USA 2011, 108, 1093–1098. [Google Scholar] [CrossRef]
- Djemal, R.; Mila, I.; Bouzayen, M.; Pirrello, J.; Khoudi, H. Molecular Cloning and Characterization of Novel WIN1/SHN1 Ethylene Responsive Transcription Factor HvSHN1 in Barley (Hordeum vulgare L.). J. Plant Physiol. 2018, 228, 39–46. [Google Scholar] [CrossRef]
- Wang, P.; Ge, M.; Yu, A.; Song, W.; Fang, J.; Leng, X. Effects of Ethylene on Berry Ripening and Anthocyanin Accumulation of “Fujiminori” Grape in Protected Cultivation. J. Sci. Food Agric. 2022, 102, 1124–1136. [Google Scholar] [CrossRef]
- Xu, J.; Yang, X.; Li, B.; Chen, L.; Min, L.; Zhang, X. GhL1L1 Affects Cell Fate Specification by Regulating GhPIN1-Mediated Auxin Distribution. Plant Biotechnol. J. 2019, 17, 63–74. [Google Scholar] [CrossRef]
- Liu, X.; Liu, H.; Liu, W.-C.; Gao, Z. The Nuclear Localized RIN13 Induces Cell Death through Interacting with ARF1. Biochem. Biophys. Res. Commun. 2020, 527, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, M.B.; Collings, D.A.; Rose, R.J.; McCurdy, D.W. ACTIN7 Is Required for Perinuclear Clustering of Chloroplasts during Arabidopsis Protoplast Culture. Plants 2020, 9, 225. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, S.K.; Kanwar, P.; Fernandes, J.L.; Mahiwal, S.; Yadav, A.K.; Samtani, H.; Srivastava, A.K.; Suprasanna, P.; Pandey, G.K. Arabidopsis Mitochondrial Voltage-Dependent Anion Channels Are Involved in Maintaining Reactive Oxygen Species Homeostasis, Oxidative and Salt Stress Tolerance in Yeast. Front. Plant Sci. 2020, 11, 50. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, J.; Chaze, T.; Miranda, K.; Roberson, R.W.; Gorgette, O.; Nimrichter, L.; Matondo, M.; Latgé, J.-P.; Beauvais, A.; Rodrigues, M.L. Characterization of Extracellular Vesicles Produced by Aspergillus Fumigatus Protoplasts. mSphere 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Susila, H.; Jurić, S.; Liu, L.; Gawarecka, K.; Chung, K.S.; Jin, S.; Kim, S.-J.; Nasim, Z.; Youn, G.; Suh, M.C.; et al. Florigen Sequestration in Cellular Membranes Modulates Temperature-Responsive Flowering. Science 2021, 373, 1137–1142. [Google Scholar] [CrossRef]
- Hu, Y.; Huang, Y. Isolation and Transfection of Maize Endosperm Protoplasts. Methods Mol. Biol. 2022, 2464, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Górska, A.M.; Gouveia, P.; Borba, A.R.; Zimmermann, A.; Serra, T.S.; Carvalho, P.; Lourenço, T.F.; Oliveira, M.M.; Peterhänsel, C.; Saibo, N.J.M. ZmOrphan94 Transcription Factor Downregulates ZmPEPC1 Gene Expression in Maize Bundle Sheath Cells. Front. Plant Sci. 2021, 12, 559967. [Google Scholar] [CrossRef]
- Kim, C.-Y.; Vo, K.T.X.; Nguyen, C.D.; Jeong, D.-H.; Lee, S.-K.; Kumar, M.; Kim, S.-R.; Park, S.-H.; Kim, J.-K.; Jeon, J.-S. Functional Analysis of a Cold-Responsive Rice WRKY Gene, OsWRKY71. Plant Biotechnol. Rep. 2016, 10, 13–23. [Google Scholar] [CrossRef]
- Nakashima, K.; Tran, L.-S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional Analysis of a NAC-Type Transcription Factor OsNAC6 Involved in Abiotic and Biotic Stress-Responsive Gene Expression in Rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Chen, J.; Inoue, Y.; Kumakura, N.; Mise, K.; Shirasu, K.; Takano, Y. Comparative Transient Expression Analyses on Two Conserved Effectors of Colletotrichum Orbiculare Reveal Their Distinct Cell Death-Inducing Activities between Nicotiana benthamiana and Melon. Mol. Plant Pathol. 2021, 22, 1006–1013. [Google Scholar] [CrossRef]
- Sakuma, Y.; Maruyama, K.; Osakabe, Y.; Qin, F.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional Analysis of an Arabidopsis Transcription Factor, DREB2A, Involved in Drought-Responsive Gene Expression. Plant Cell 2006, 18, 1292–1309. [Google Scholar] [CrossRef]
- Chen, J.; Yi, Q.; Cao, Y.; Wei, B.; Zheng, L.; Xiao, Q.; Xie, Y.; Gu, Y.; Li, Y.; Huang, H.; et al. ZmbZIP91 Regulates Expression of Starch Synthesis-Related Genes by Binding to ACTCAT Elements in Their Promoters. J. Exp. Bot. 2016, 67, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Uhrig, J.F.; Thurow, C.; Huang, L.-J.; Gatz, C. Reconstitution of the Jasmonate Signaling Pathway in Plant Protoplasts. Cells 2019, 8, 1532. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Cao, L.; Miu, W.; Cao, R.; Peng, M.; Wan, W.; Huang, L.-J. Molecular Rewiring of the Jasmonate Signaling Pathway to Control Auxin-Responsive Gene Expression. Cells 2020, 9, 641. [Google Scholar] [CrossRef] [PubMed]
- Naing, A.H.; Adedeji, O.S.; Kim, C.K. Protoplast Technology in Ornamental Plants: Current Progress and Potential Applications on Genetic Improvement. Sci. Hortic. 2021, 283, 110043. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, X.; Chen, S.; Chen, S. Overexpression of BpERF1.1 in Betula platyphylla Enhanced Tolerance to Multiple Abiotic Stresses. Physiol. Mol. Biol. Plants 2022, 28, 1159–1172. [Google Scholar] [CrossRef]
- Malnoy, M.; Viola, R.; Jung, M.-H.; Koo, O.-J.; Kim, S.; Kim, J.-S.; Velasco, R.; Kanchiswamy, C.N. DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Andersson, M.; Turesson, H.; Nicolia, A.; Fält, A.-S.; Samuelsson, M.; Hofvander, P. Efficient Targeted Multiallelic Mutagenesis in Tetraploid Potato (Solanum tuberosum) by Transient CRISPR-Cas9 Expression in Protoplasts. Plant Cell Rep. 2017, 36, 117–128. [Google Scholar] [CrossRef]
- Xie, Y.; Jiang, S.; Li, L.; Yu, X.; Wang, Y.; Luo, C.; Cai, Q.; He, W.; Xie, H.; Zheng, Y.; et al. Single-Cell RNA Sequencing Efficiently Predicts Transcription Factor Targets in Plants. Front. Plant Sci. 2020, 11, 603302. [Google Scholar] [CrossRef]
- Li, H.; Dai, X.; Huang, X.; Xu, M.; Wang, Q.; Yan, X.; Sederoff, R.R.; Li, Q. Single-Cell RNA Sequencing Reveals a High-Resolution Cell Atlas of Xylem in Populus. J. Integr. Plant Biol. 2021, 63, 1906–1921. [Google Scholar] [CrossRef]
| Plant Species | Material | Enzymes | Protoplasts Yield | References |
|---|---|---|---|---|
| Albizia julibrissin | Leaf (in vitro) | 1.50% C + 1.00% P | 7.77 × 105/g FW 94% | [13] |
| Albizia julibrissin | Callus | 2.00% C + 1.00% P | 6.92 × 105/g FW 92% | [13] |
| Ananas comosus | Leaf (in vitro) | 1.50% C + 0.50% M | 6.5 × 105/g FW 51.0% | [14] |
| Arabidopsis thaliana | 14-day seedlings | 1.00% C + 1.00% M | >5 × 106/g FW | [15] |
| Brassica oleracea | Leaf | 2.00% C + 0.10% P | 6.00 × 107/g FW 95.0% | [16] |
| Catalpa bungei | Leaf (in vitro) | 3.00% C + 2.00% M | 1 × 106/g FW 90% | [17] |
| Calibrachoa elegans | Leaf (in vitro) | 2.00% C + 0.60% M | 0.4~1.7 × 106/g FW | [18] |
| Camellia Oleifera | Flower petal | 3.00% C + 1.00% M | 1.42 × 107/g FW 88.69% | [19] |
| Camellia Oleifera | Young leaf (in vitro) | 1.50% C + 0.50% M + 0.25% S | 3.50 × 107/g FW 90.90% | [20] |
| Cannabis sativa | Young leaf | 1.50% C + 0.40% M + 1.00% P | 9.7 × 106/g FW | [21] |
| Chirita pumila | Young leaf | 1.00% C + 0.50% M + 0.25% P | 6.83 × 105/g FW 92.97% | [22] |
| Chrysanthemum morifolium | Leaf (in vitro) | 1.50% C + 0.50% M | 6.32 × 105/g FW 91.70% | [23] |
| Cucumis sativus | Leaf (in vitro) | 1.50% C + 0.40% M | 6.0~7.0 × 106/g FW 90.0% | [24] |
| Cymbidium sinense | Flower petal | 1.20% C + 0.60% M | 3.50 × 107/g FW 94.21% | [11] |
| Cymbidium sinense | Flower pedicel | 1.20% C + 0.60% M | 5.3 × 106/g FW 90.3% | [25] |
| Cymbidium sinense | Young leaf | 1.20% C + 0.60% M | 3.3 × 106/g FW 91.3% | [25] |
| Cymbidium sinense | Leaf base | 1.20% C + 0.60% M | 2.5 × 107/g FW 92.1% | [25] |
| Cymbidium sinense | Root tip | 1.20% C + 0.60% M | 7.8 × 105/g FW 89.3% | [25] |
| Dendrobium catenatum | Leaf | 1.20% C + 0.60% M | 8.2 × 106/g FW 91.1% | [25] |
| Echinacea augustifolia | Callus | 2.00% C +1.00% P +0.50% H | 5.0 × 105/g FW | [26] |
| Ginkgo biloba | Leaf | 2.00% C + 0.2% P+ 1.5% M | 5.39 × 106/g FW 80.23% | [27] |
| Ginkgo biloba | Leaf | 2.00% C +0.25% P | 1.0 × 106/g FW 80.0% | [28] |
| Gossypium hirsutum | Callus | 1.50% C + 1.50% P + 1.50%/0.50% M + 0.50% H | 3.3 × 106/g FW 97% | [29] |
| Gossypium hirsutum | Young leaf | 1.50% C + 0.40% M | >1.00 × 106/g FW >90% | [30] |
| Gossypium hirsutum | Taproot | 1.50% C + 0.75% M | 3.55 × 105/g FW 93.3% | [31] |
| Hevea brasiliensis | Leaf | 1.50% C + 0.30% M | 18.6 × 107/g FW 97% | [32] |
| Jasminum sambac | Callus | 1.50% C + 0.40% M + 0.80% P | 2.38 × 107/g FW 88% | [33] |
| Liriodendron × sinoamericanum | Leaf (in vitro) | 1.50% C + 0.50% M + 0.10% P | 1.2 × 107/g FW 97.0% | [34] |
| Manihot esculenta | Leaf (in vitro) | 1.60% C + 0.80% M | 4.4 × 107/g FW 92.6% | [35] |
| Medicago sativa | Legumes root | 1.50% C + 2.00% M | 1.0 × 106/g FW >90.0% | [36] |
| Nicotiana benthamiana | Leaf (in vitro) | 1.00% C + 0.50% M | 4~5 × 106/g FW | [37] |
| Oryza sativa | Stem and sheath | 1.50% C + 0.75% M | 1.0 × 107/g FW >95% | [38] |
| Oryza sativa | Leaf base | 1.20% C + 0.60% M | 4.3 × 107/g FW | [25] |
| Oryza sativa | Leaf | 1.20% C + 0.60% M | No viable protoplast | [25] |
| Petunia hybrida | Leaf (in vitro) | 2.00% C + 0.60% M | 0.3~2.0 × 106/g FW | [18] |
| Phalaenopsis aphrodite | Flower petals | 1.00% C + 0.25% M | 1.9 × 105/g FW 90.9% | [39] |
| Phalaenopsis aphrodite | Leaf (in vitro) | 1.00% C + 0.70% M | 5.9 × 106/g FW 57.9% | [40] |
| Phalaenopsis aphrodite | Leaf (in vitro) | 2.00% C + 1.00% M | 1.1 × 106/g FW 83.8% | [41] |
| Phalaenopsis equestris | Leaf base | 1.20% C + 0.60% M | 1.8 × 107/g FW 92.8% | [25] |
| Phaseolus vulgaris | Leaf | 1.50% C + 0.37% M | 3.0 × 105/g FW | [42] |
| Phaseolus vulgaris | Flower petals | 1.50% C + 0.37% M | 2.0 × 105/g FW | [42] |
| Phaseolus vulgaris | Hypocotyl and root | 2.00% C + 0.30% M + 4.00% H | 2.0 × 105/g FW | [42] |
| Phaseolus vulgaris | Nodule | 1.00% C + 0.30% M + 4.00% H | 1.0 × 105/g FW | [42] |
| Populus przewalskii | Leaf (in vitro) | 2.00% C + 0.50% P | 1.0 × 108/g FW >82.0% | [43] |
| Populus przewalskii | Leaf | 3.00% C + 0.80% P | 1.0 × 107/g FW >90.0% | [44] |
| Prunus avium | Cell suspension | 1.00% C + 0.50% P | 4.3 × 106/g FW 84.1% | [45] |
| Ricinus communis | Leaf | 1.50% C + 0.40% M | 6.1 × 106/g FW 85% | [46] |
| Saccharum officinarum | Young leaf base | 2.00% C + 0.50% M | 12.6 × 107/g FW 80.19% | [47] |
| Solanum melongena | Leaf | 1.25% C + 0.40% M | 1.2 × 107/g FW 96.0% | [48] |
| Torenia fournieri | Leaf | 1.50% C + 0.50% M | 6.0~7.0 × 105/g FW | [49] |
| Triticum aestivum | Leaf | 1.00% C + 0.25% M | 7.3 × 106/g FW 95.0% | [50] |
| Uncaria rhynchophylla | Leaf | 1.25% C + 0.6% M | 1.5 × 107/g FW >90% | [51] |
| Vernicia fordii | Mature leaf | 1.50% C + 1.00% M | 7.21 × 106/g FW 93.19% | [52] |
| Vernicia fordii | Young leaf (in vitro) | 2.00% C + 1.00% M | 7.08 × 106/g FW 95.06% | [52] |
| Vitis vinifera | Cell suspension | 2.00% C + 1.00% M | 3~4 × 107/g FW >95% | [53] |
| Vitis vinifera | Leaf (in vitro) | 1.50% C + 0.40% M | 3.3 × 106/g FW 96.0% | [54] |
| Zea mays | Yellow leaf bases | 1.50% C + 0.50% P | 1.0~5.0 × 106/g FW 80~90% | [55] |
| Zea mays | Leaf | 0.10% C + 0.01% M | 1.8~1.9 × 107/g FW 95.0% | [56] |
| Zea mays | Leaf | 1.20% C + 0.60% M | 0.7 × 107/g FW 89.2% | [25] |
| Zea mays | Leaf base | 1.20% C + 0.60% M | 3.2 × 107/g FW 94.3% | [25] |
| Zea mays | Endosperm | 1.00% C + 0.75% M | 2.43 × 106/g FW >80% | [57] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, K.; Chen, J.; Pi, X.; Huang, L.-J.; Li, N. Isolation, Purification, and Application of Protoplasts and Transient Expression Systems in Plants. Int. J. Mol. Sci. 2023, 24, 16892. https://doi.org/10.3390/ijms242316892
Chen K, Chen J, Pi X, Huang L-J, Li N. Isolation, Purification, and Application of Protoplasts and Transient Expression Systems in Plants. International Journal of Molecular Sciences. 2023; 24(23):16892. https://doi.org/10.3390/ijms242316892
Chicago/Turabian StyleChen, Kebin, Jiali Chen, Xin Pi, Li-Jun Huang, and Ning Li. 2023. "Isolation, Purification, and Application of Protoplasts and Transient Expression Systems in Plants" International Journal of Molecular Sciences 24, no. 23: 16892. https://doi.org/10.3390/ijms242316892
APA StyleChen, K., Chen, J., Pi, X., Huang, L.-J., & Li, N. (2023). Isolation, Purification, and Application of Protoplasts and Transient Expression Systems in Plants. International Journal of Molecular Sciences, 24(23), 16892. https://doi.org/10.3390/ijms242316892







