The Competing Endogenous RNAs Regulatory Genes Network Mediates Leaf Shape Variation and Main Effector Gene Function in Mulberry Plant (Morus alba)
Abstract
1. Introduction
2. Results
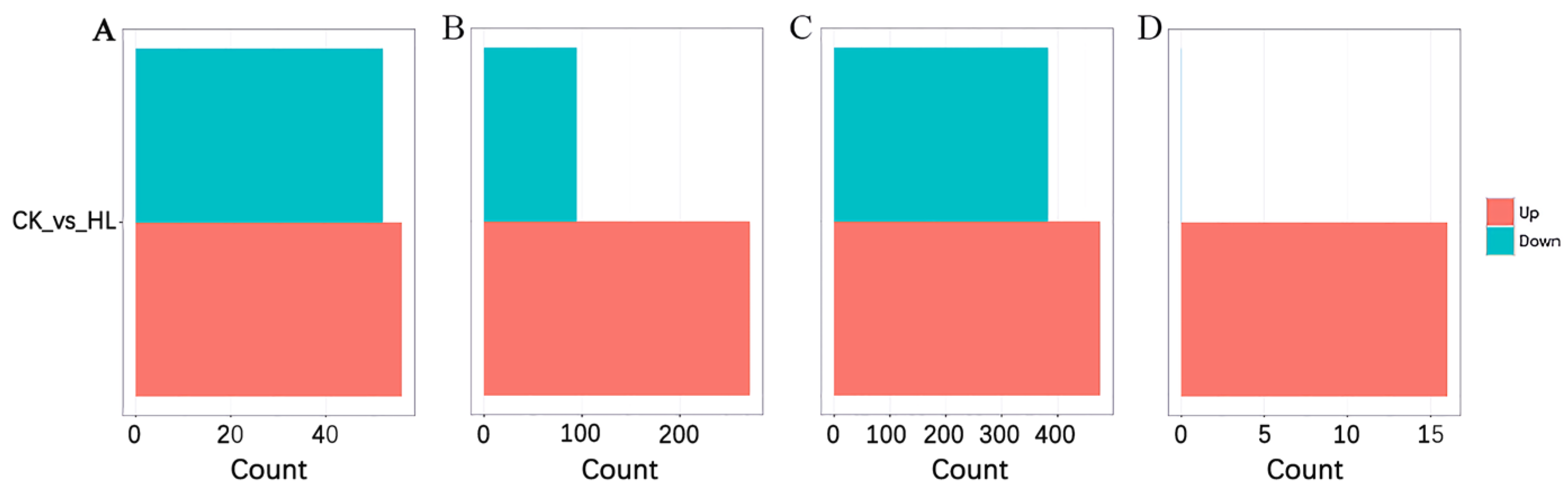
2.1. Transcriptome Profile of ceRNAs and DEGs Analysis
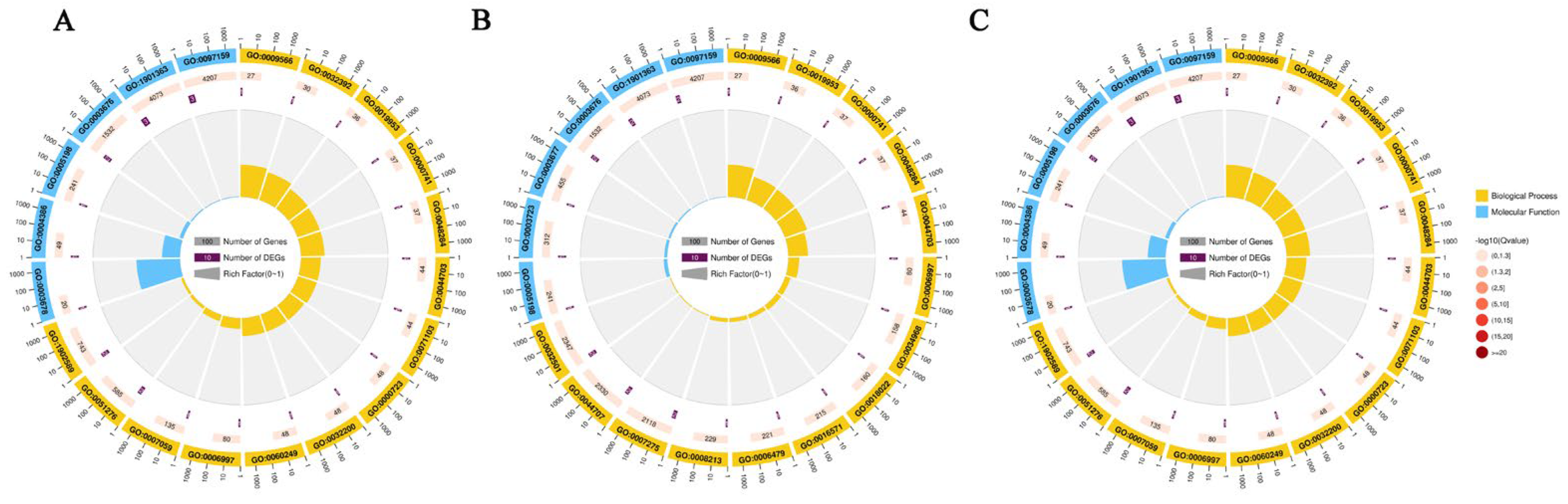
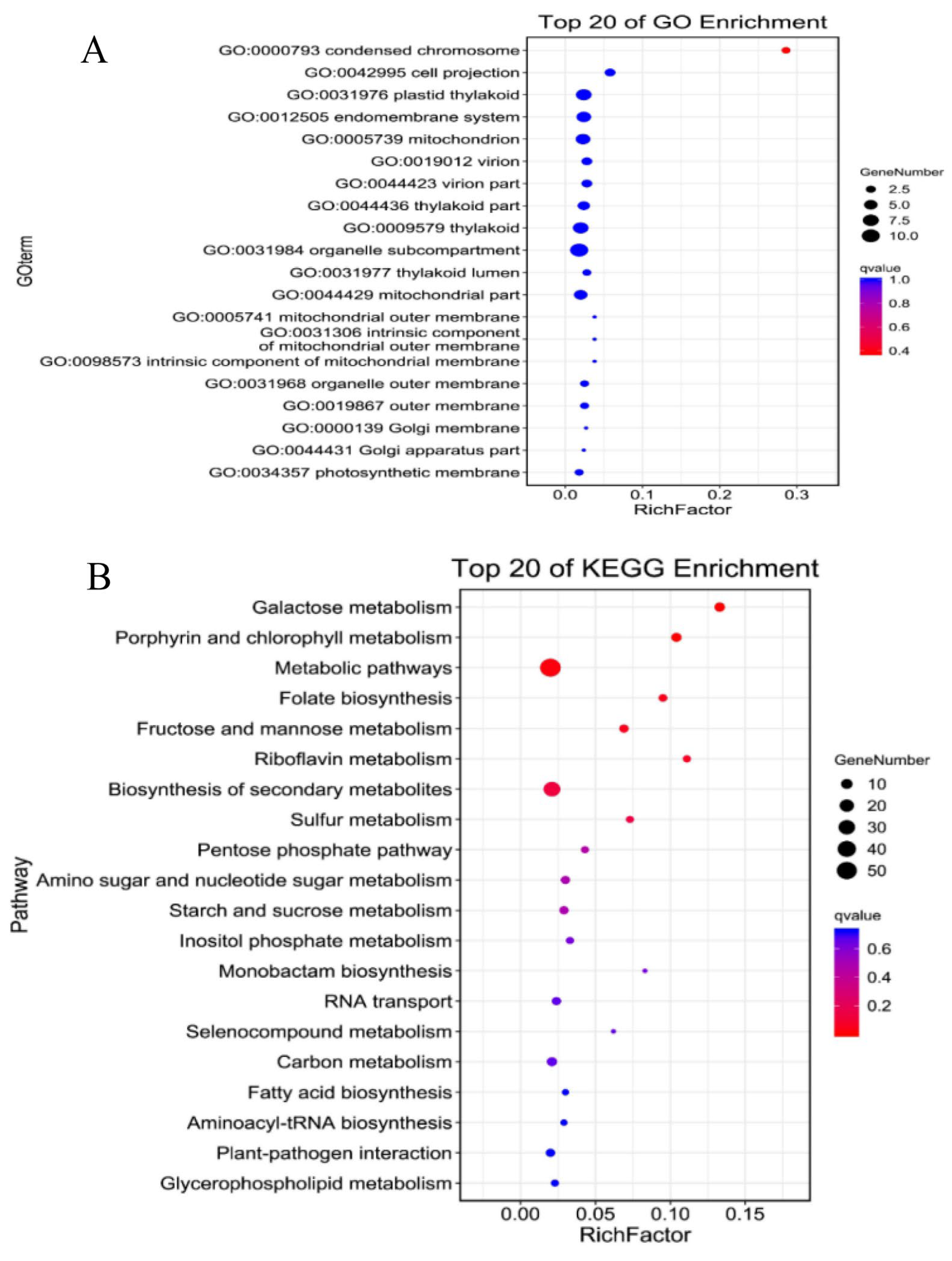
2.2. Functional Enrichment Analysis
2.3. ceRNA Regulatory Network
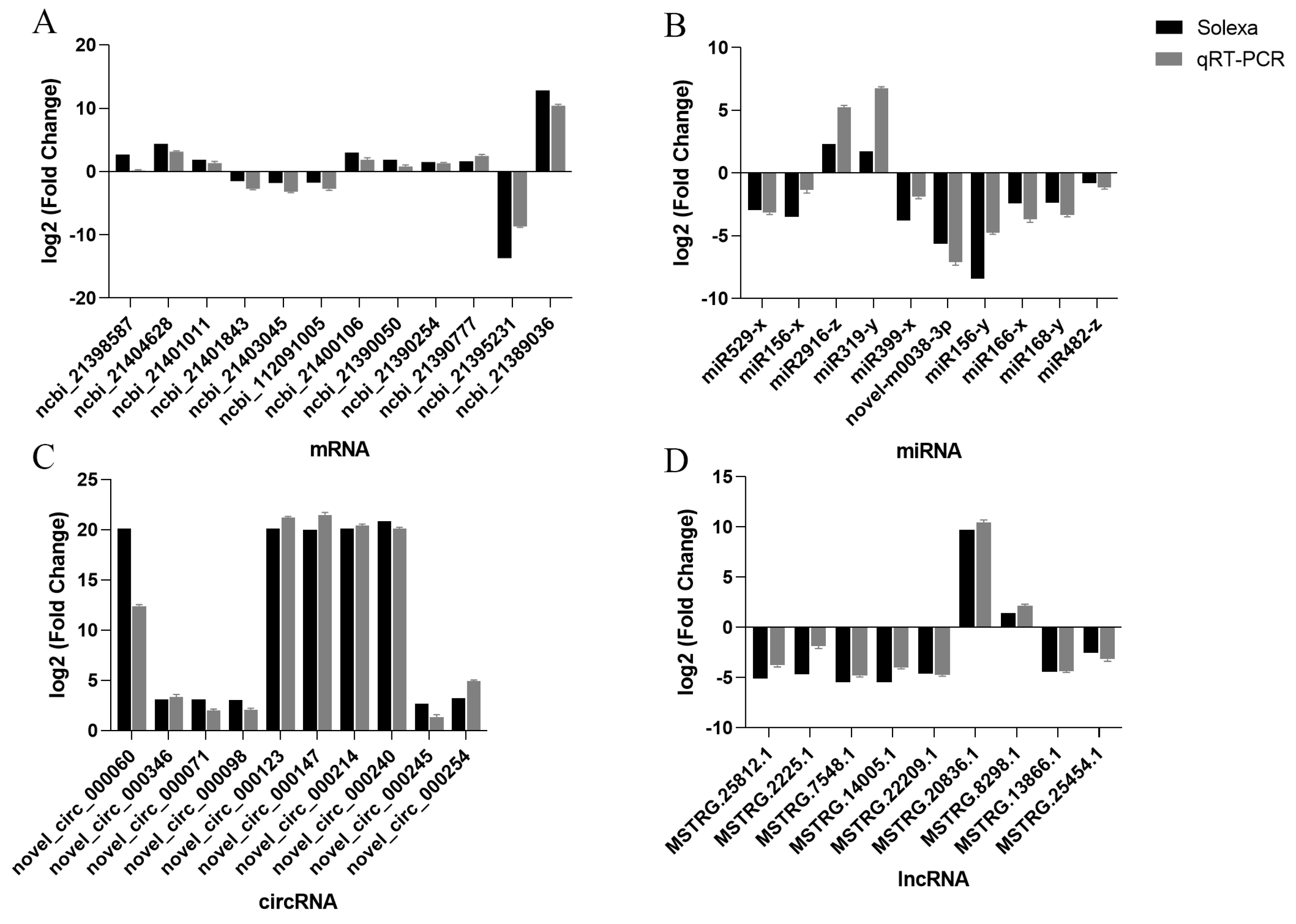
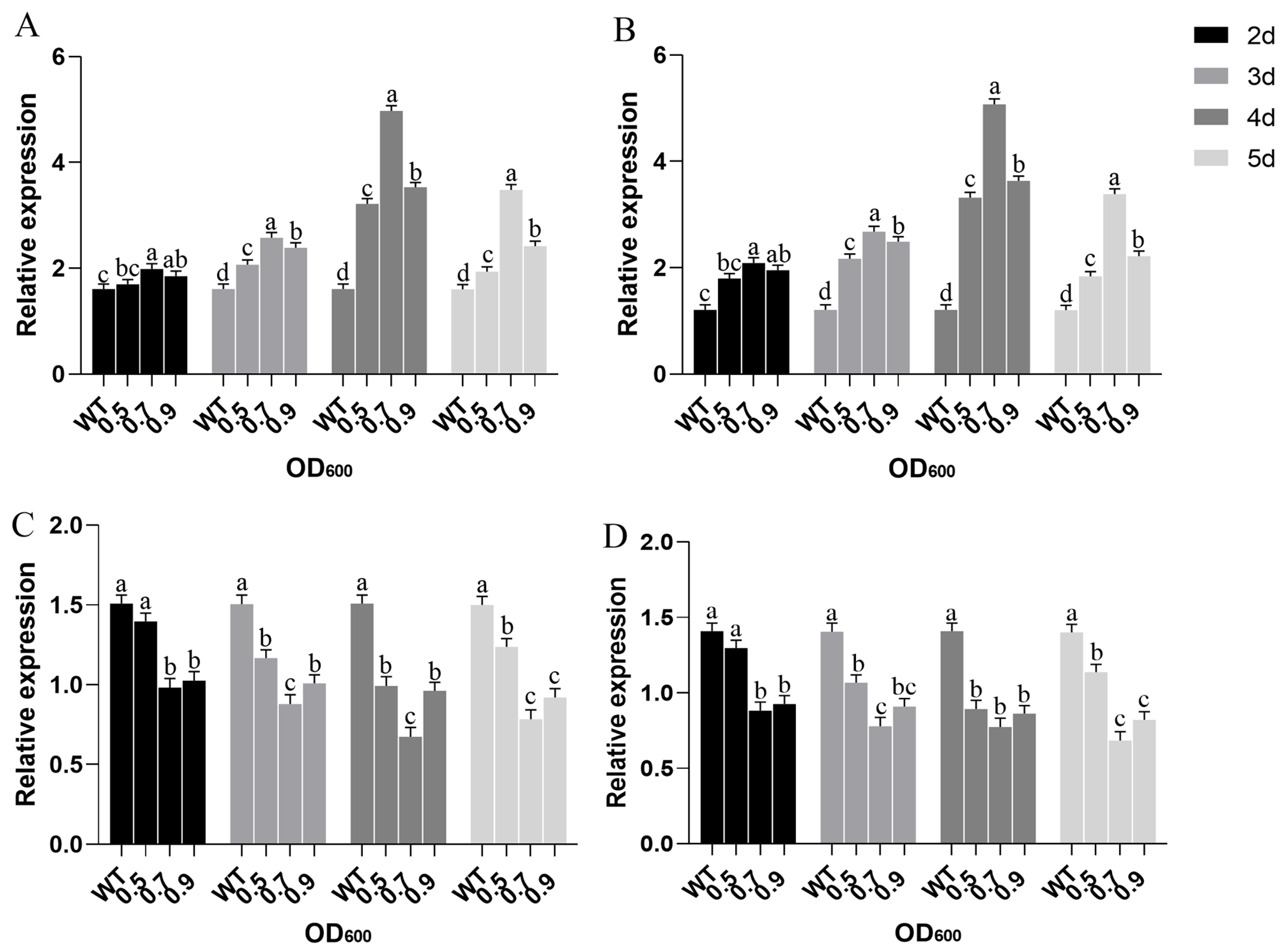
2.4. The DEG Validation by Real-Time RT-PCR Analysis
2.5. Secondary Structure Analysis of miR156x
2.6. miR156x Overexpression Analysis
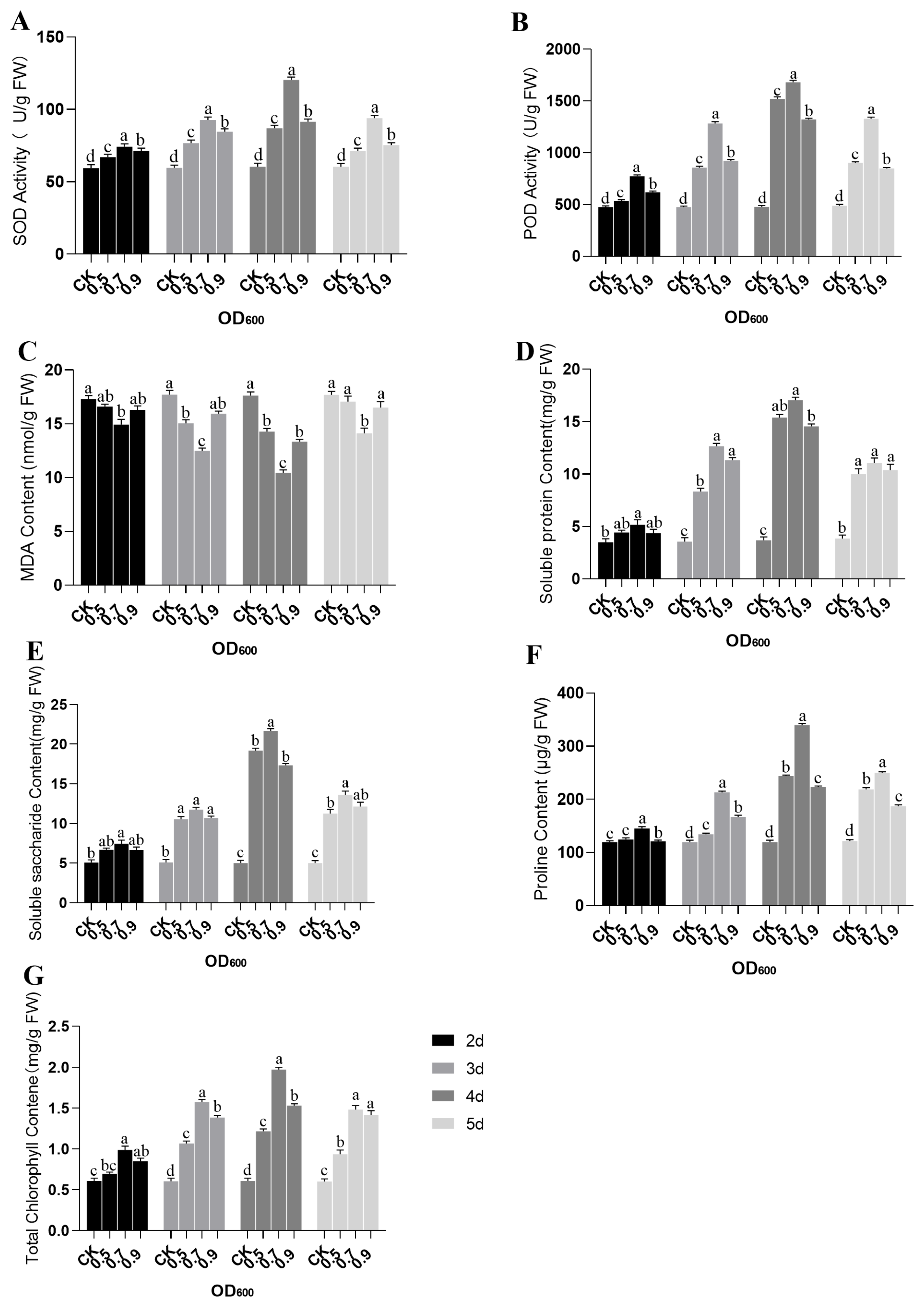
2.7. The Impact of miR156x Overexpression in Mulberry Plant on Physiological and Biochemical Indicators Related to Leaf Morphology Regulation
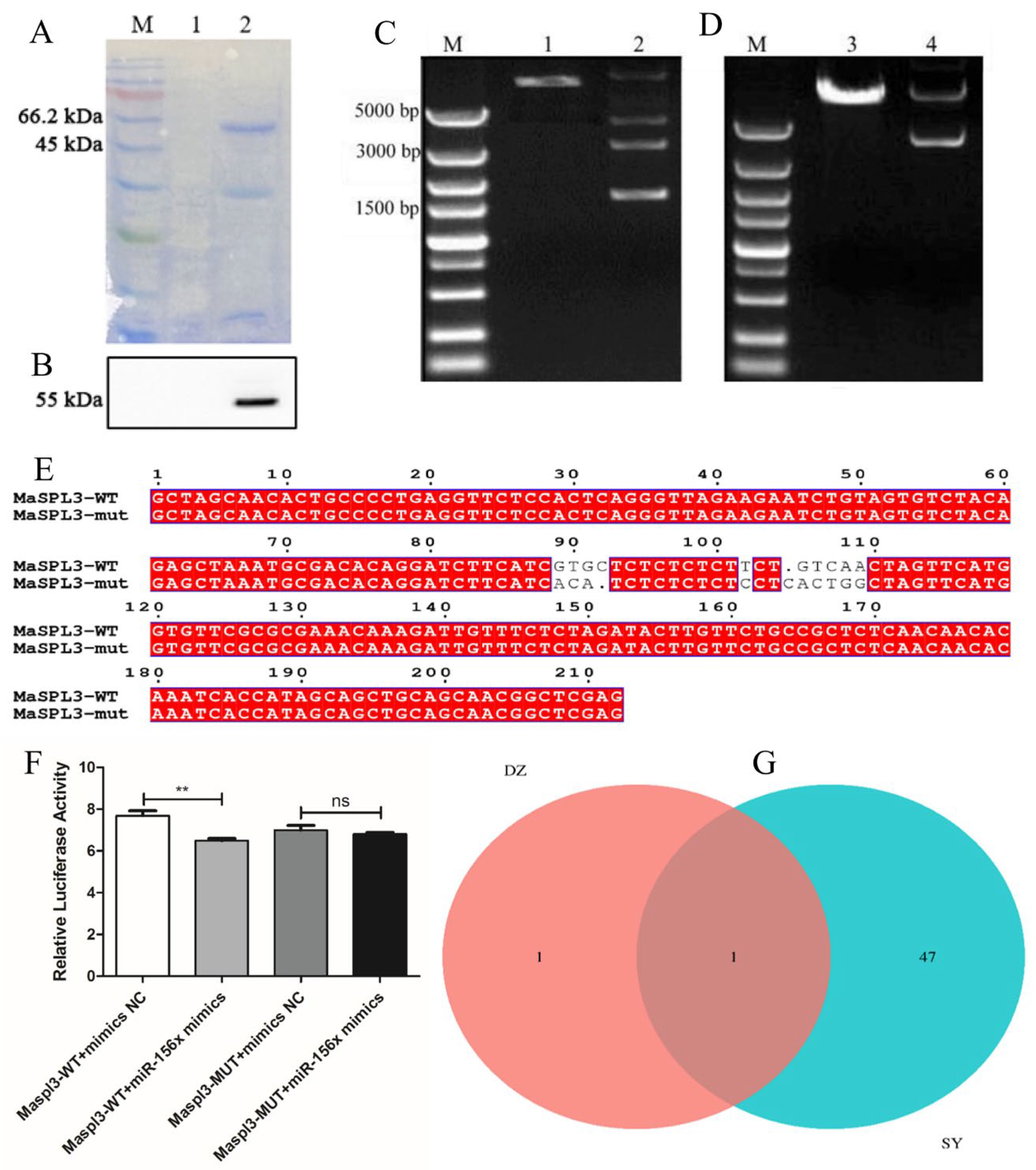
2.8. Cloning and Verification of the MaSPL3 Gene in Mulberry
2.9. Dual Luciferase Validation of the miR156x and Maspl3 Targeting Relationship
2.10. GST-Pull Down Analysis of the Interacting Proteins of MaSPL3 in Mulberry Plants
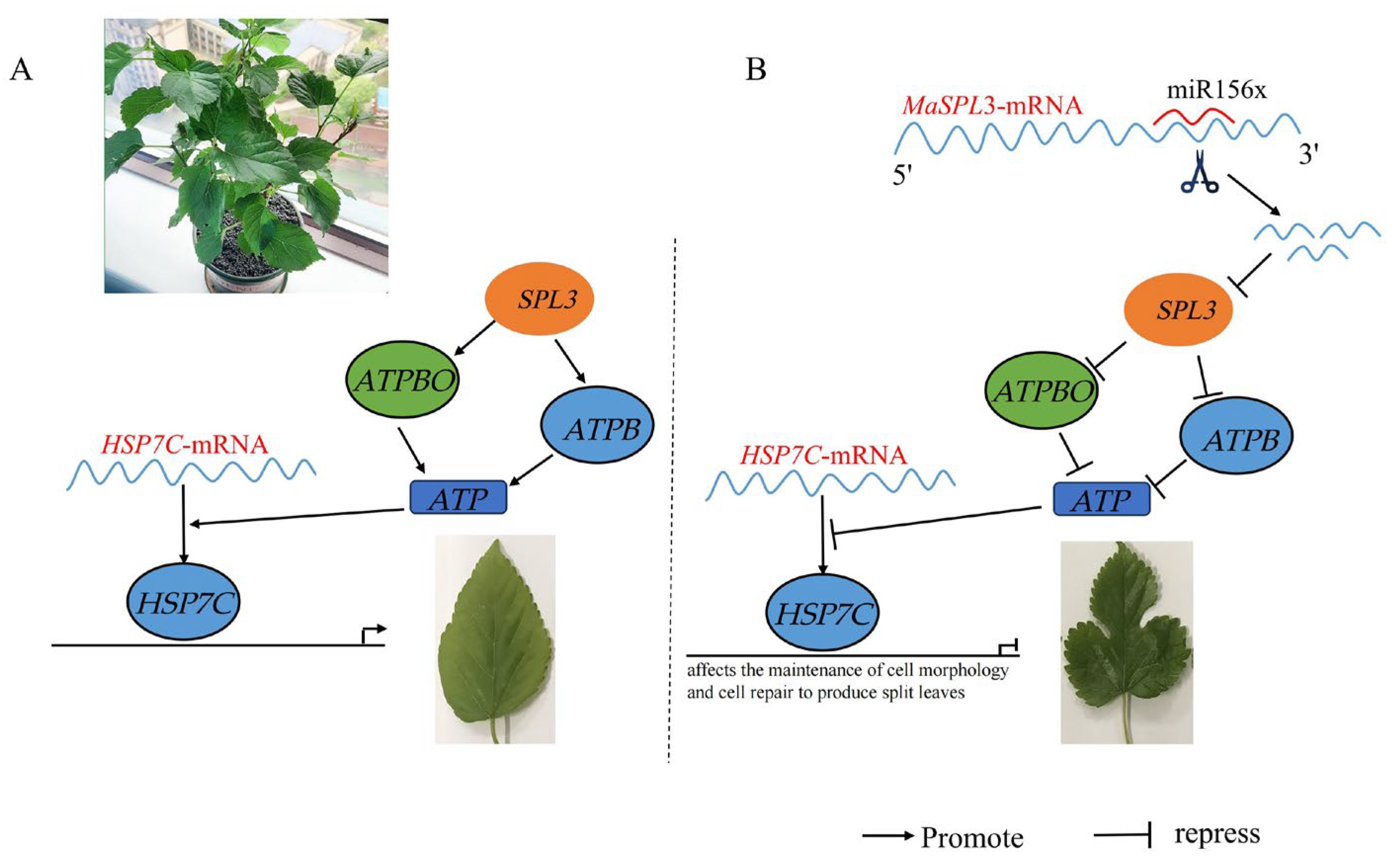
3. Discussion
4. Materials and Methods
4.1. Plant Material, Growth Conditions, and Treatments
4.2. RNA Isolation, Library Construction, and Sequencing
4.3. Sequence Data Analysis and Assembly
4.4. ceRNAs Network Construction and Functional Enrichment Analysis
4.5. Functional Enrichment Analysis
4.6. Quantitative Real-Time PCR (qRT-PCR) Verification of DEGs
4.7. PCR Amplification and Prokaryotic Expression Vector Construction
4.8. Sequence Analysis of the miR156x Precursor
4.9. Analysis of miR156x Overexpression Patterns
4.10. Detection of Physiological and Biochemical Indicators
4.11. Luciferase Reporter Assay
4.12. GST (Glutathione S-Transferase) Pull-Down Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qin, C.; Li, Y.; Niu, W.; Ding, Y.; Zhang, R.; Shang, X. Analysis and characterisation of anthocyanins in mulberry fruit. Czech J. Food Sci. 2010, 28, 117–126. [Google Scholar] [CrossRef]
- Ackah, M.; Shi, Y.; Wu, M.; Wang, L.; Guo, P.; Guo, L.; Jin, X.; Li, S.; Zhang, Q.; Qiu, C. Metabolomics response to drought stress in Morus alba L. variety Yu-711. Plants 2021, 10, 1636. [Google Scholar] [CrossRef] [PubMed]
- Karanikas, C.; Scaltsoyiannes, A.; Iliev, I.; Tsoulpha, P.; Tsaktsira, M. Micropropagation of mature Morus alba “pendula” trees for ornamental use. Epistem. Epeterida Tou Tm. Dasologies Kai Fus. Periballontos Aristot. Panepistemio Thessalon. 2000, 43, 59–69. [Google Scholar]
- Jokar, M.; Farahani, T.S.; Ramezanzadeh, B. Electrochemical and surface characterizations of Morus alba pendula leaves extract (MAPLE) as a green corrosion inhibitor for steel in 1 M HCl. J. Taiwan Inst. Chem. Eng. 2016, 63, 436–452. [Google Scholar] [CrossRef]
- Zhang, Q.; Ackah, M.; Wang, M.; Amoako, F.K.; Shi, Y.; Wang, L.; Dari, L.; Li, J.; Jin, X.; Jiang, Z. The impact of boron nutrient supply in mulberry (Morus alba) response to metabolomics, enzyme activities, and physiological parameters. Plant Physiol. Biochem. 2023, 200, 107649. [Google Scholar] [CrossRef]
- Jin, X.; Ackah, M.; Wang, L.; Amoako, F.K.; Shi, Y.; Essoh, L.G.; Li, J.; Zhang, Q.; Li, H.; Zhao, W. Magnesium Nutrient Application Induces Metabolomics and Physiological Responses in Mulberry (Morus alba) Plants. Int. J. Mol. Sci. 2023, 24, 9650. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Y.; Wang, J.; Jin, X.; Zhang, Q.; Ackah, M.; Wang, Y.; Xu, D.; Zhao, W. ATAC-seq exposes differences in chromatin accessibility leading to distinct leaf shapes in mulberry. Plant Direct. 2022, 6, e464. [Google Scholar] [CrossRef]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Ackah, M.; Jin, X.; Zhang, Q.; Amoako, F.K.; Wang, L.; Attaribo, T.; Zhao, M.; Yuan, F.; Herman, R.A.; Qiu, C. Long noncoding RNA transcriptome analysis reveals novel lncRNAs in Morus alba ‘Yu-711’ response to drought stress. Plant Genome 2022, e20273. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, J.; Chen, J.; Wang, J.; Xu, J. Construction and analysis of an aberrant lncRNA-miRNA-mRNA network associated with papillary thyroid cancer. Medicine 2020, 99, e22705. [Google Scholar] [CrossRef]
- He, H.; Shen, Z.; Gu, Q.; Xie, R. Construction of ceRNA network and identification of two differentially expressed circRNAs in hepatocellular carcinoma by bioinformatic analysis. Int. J. Clin. Exp. Pathol. 2020, 13, 2727. [Google Scholar]
- Deng, R.; Cui, X.; Dong, Y.; Tang, Y.; Tao, X.; Wang, S.; Wang, J.; Chen, L. Construction of circRNA-based ceRNA network to reveal the role of circRNAs in the progression and prognosis of hepatocellular carcinoma. Front. Genet. 2021, 12, 626764. [Google Scholar] [CrossRef]
- Pawełkowicz, M.E.; Skarzyńska, A.; Koter, M.D.; Turek, S.; Pląder, W. miRNA Profiling and Its Role in Multi-Omics Regulatory Networks Connected with Somaclonal Variation in Cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2022, 23, 4317. [Google Scholar] [CrossRef]
- Li, M.; Sun, X.; Cai, H.; Sun, Y.; Plath, M.; Li, C.; Lan, X.; Lei, C.; Lin, F.; Bai, Y. Long non-coding RNA ADNCR suppresses adipogenic differentiation by targeting miR-204. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 2016, 1859, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Song, Y.; Liu, L.; Zhang, M.; Li, Z.; Ji, C.; Xu, W.; Liu, T.; Xu, B.; Wang, X. Long-term safety and efficacy of orelabrutinib monotherapy in Chinese patients with relapsed or refractory mantle cell lymphoma: A multicenter, open-label, phase II study. Blood 2020, 136, 1. [Google Scholar] [CrossRef]
- Feng, J.; Bian, Q.; He, X.; Zhang, H.; He, J. A LncRNA-miRNA-mRNA ceRNA regulatory network based tuberculosis prediction model. Microb. Pathog. 2021, 158, 105069. [Google Scholar] [CrossRef]
- Wu, X.; Sui, Z.; Zhang, H.; Wang, Y.; Yu, Z. Integrated Analysis of lncRNA–Mediated ceRNA Network in lung adenocarcinoma. Front. Oncol. 2020, 10, 554759. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, H.; Wu, C.; Yan, M.; Wu, H.; Wang, J.; Yang, X.; Shao, Q. Construction and investigation of lncRNA-associated ceRNA regulatory network in papillary thyroid cancer. Oncol. Rep. 2018, 39, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Lei, K.J.; Lin, Y.M.; Ren, J.; Bai, L.; Miao, Y.C.; An, G.Y.; Song, C.P. Modulation of the phosphate-deficient responses by microRNA156 and its targeted SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 in Arabidopsis. Plant Cell Physiol. 2016, 57, 192–203. [Google Scholar] [CrossRef]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef]
- Wyrzykowska, A.; Bielewicz, D.; Plewka, P.; Sołtys-Kalina, D.; Wasilewicz-Flis, I.; Marczewski, W.; Jarmolowski, A.; Szweykowska-Kulinska, Z. The MYB33, MYB65, and MYB101 transcription factors affect Arabidopsis and potato responses to drought by regulating the ABA signaling pathway. Physiol. Plant. 2022, 174, e13775. [Google Scholar] [CrossRef] [PubMed]
- Kanazawa, A.; Inaba, J.I.; Kasai, M.; Shimura, H.; Masuta, C. RNA-mediated epigenetic modifications of an endogenous gene targeted by a viral vector: A potent gene silencing system to produce a plant that does not carry a transgene but has altered traits. Plant Signal. Behav. 2011, 6, 1090–1093. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Buxdorf, K.; Hendelman, A.; Stav, R.; Lapidot, M.; Ori, N.; Arazi, T. Identification and characterization of a novel miR159 target not related to MYB in tomato. Planta 2010, 232, 1009–1022. [Google Scholar] [CrossRef] [PubMed]
- Bologna, N.G.; Mateos, J.L.; Bresso, E.G.; Palatnik, J.F. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 2009, 28, 3646–3656. [Google Scholar] [CrossRef]
- Lal, S.; Pacis, L.B.; Smith, H.M. Regulation of the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE genes/microRNA156 module by the homeodomain proteins PENNYWISE and POUND-FOOLISH in Arabidopsis. Mol. Plant 2011, 4, 1123–1132. [Google Scholar] [CrossRef]
- Preston, J.C.; Hileman, L.C. Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 2013, 4, 80. [Google Scholar] [CrossRef]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA promoter binding protein–like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef]
- Kirihara, T.; Ogawa, M.; Inoue, H.; Kodera, H.; Ishida, K. Lossless and low-crosstalk characteristics in an InP-based 4/spl times/4 optical switch with integrated single-stage optical amplifiers. IEEE Photonics Technol. Lett. 1994, 6, 218–221. [Google Scholar] [CrossRef]
- Jung, J.-H.; Seo, P.J.; Kang, S.K.; Park, C.-M. miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Mol. Biol. 2011, 76, 35–45. [Google Scholar] [CrossRef]
- Jung, J.-H.; Lee, H.-J.; Ryu, J.Y.; Park, C.-M. SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol. Plant 2016, 9, 1647–1659. [Google Scholar] [CrossRef]
- Xie, Y.; Zhou, Q.; Zhao, Y.; Li, Q.; Liu, Y.; Ma, M.; Wang, B.; Shen, R.; Zheng, Z.; Wang, H. FHY3 and FAR1 integrate light signals with the miR156-SPL module-mediated aging pathway to regulate Arabidopsis flowering. Mol. Plant 2020, 13, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Luo, Y.; Ma, B.; Hu, J.; Lv, Z.; Wei, W.; Hao, H.; Yuan, J.; He, N. Hierarchical action of mulberry miR156 in the vegetative phase transition. Int. J. Mol. Sci. 2021, 22, 5550. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Snyder, J.A.; Bartel, D. Common functions for diverse small RNAs of land plants. Plant Cell 2007, 19, 1750–1769. [Google Scholar] [CrossRef] [PubMed]
- Dugas, D.V.; Bartel, B. MicroRNA regulation of gene expression in plants. Curr. Opin. Plant Biol. 2004, 7, 512–520. [Google Scholar] [CrossRef]
- Huijser, P.; Schmid, M. The control of developmental phase transitions in plants. Development 2011, 138, 4117–4129. [Google Scholar] [CrossRef]
- Tripathi, R.K.; Bregitzer, P.; Singh, J. Genome-wide analysis of the SPL/miR156 module and its interaction with the AP2/miR172 unit in barley. Sci. Rep. 2018, 8, 7085. [Google Scholar] [CrossRef]
- Guo, C.; Xu, Y.; Shi, M.; Lai, Y.; Wu, X.; Wang, H.; Zhu, Z.; Poethig, R.S.; Wu, G. Repression of miR156 by miR159 regulates the timing of the juvenile-to-adult transition in Arabidopsis. Plant Cell 2017, 29, 1293–1304. [Google Scholar] [CrossRef]
- Li, R.; Fan, T.; Wang, T.; Dominic, K.; Hu, F.; Liu, L.; Zhang, L.; Fang, R.; Pan, G.; Li, L. Characterization and functional analysis of miR166f in drought stress tolerance in mulberry (Morus multicaulis). Mol. Breed. 2018, 38, 1–14. [Google Scholar] [CrossRef]
- Rössner, C.; Lotz, D.; Becker, A. VIGS goes viral: How VIGS transforms our understanding of plant science. Annu. Rev. Plant Biol. 2022, 73, 703–728. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, H.; Wall, M.M.; Yang, J. Roles of transcription factor SQUAMOSA promoter binding protein-like gene family in papaya (Carica papaya) development and ripening. Genomics 2020, 112, 2734–2747. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Z.; Liu, D.; Zhang, K.; Li, A.; Mao, L. SQUAMOSA promoter-binding protein-like transcription factors: Star players for plant growth and development. J. Integr. Plant Biol. 2010, 52, 946–951. [Google Scholar] [CrossRef]
- Kim, J.J.; JLee, H.; Kim, W.; Jung, H.S.; Huijser, P.; Ahn, J.H. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 2012, 159, 461–478. [Google Scholar] [CrossRef]
- Gandikota, M.; Birkenbihl, R.P.; Höhmann, S.; Cardon, G.H.; Saedler, H.; Huijser, P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007, 49, 683–693. [Google Scholar] [CrossRef]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Bellah, S.F.; MensahSedzro, D.; Akbar, H.; Billah, S.S. Structure, enzymatic mechanism of action, applications, advantages, disadvantages and modifications of luciferase enzyme. J. Gene. Engg. Bio. Res. 2019, 1, 1–6. [Google Scholar]
- Wu, N.; Rathnayaka, T.; Kuroda, Y. Bacterial expression and re-engineering of Gaussia princeps luciferase and its use as a reporter protein. Biochim. Biophys. Acta (BBA)–Proteins Proteom. 2015, 1854, 1392–1399. [Google Scholar] [CrossRef]
- Miraglia, L.J.; JKing, F.; Damoiseaux, R. Seeing the light: Luminescent reporter gene assays. Comb. Chem. High Throughput Screen. 2011, 14, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Grentzmann, G.; Ingram, J.A.; Kelly, P.J.; Gestaland, R.F.; Atkins, J.F. A dual-luciferase reporter system for studying recoding signals. Rna 1998, 4, 479–486. [Google Scholar] [PubMed]
- Wang, X.; Chen, P.; Sun, Y.; Chen, Y.; Mao, M.; Jiang, T.; Ouyang, J. Diagnostic value of the dual-luciferase report assay for predicting response to glucocorticoid in children with acute lymphoblastic leukemia. Clin. Transl. Oncol. 2017, 19, 1241–1246. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, X.; Wang, Z.; Jiang, H.; Lv, Z.; Shen, J.; Xia, G.; Wen, K. Universal simultaneous multiplex ELISA of small molecules in milk based on dual luciferases. Anal. Chim. Acta 2018, 1001, 125–133. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, H.; Yu, B.; Xiong, L.; Xia, Y. Proteomic identification of early salicylate-and flg22-responsive redox-sensitive proteins in Arabidopsis. Sci. Rep. 2015, 5, 8625. [Google Scholar] [CrossRef] [PubMed]
- Katam, R.; Lin, C.; Grant, K.; Katam, C.S.; Chen, S. Advances in plant metabolomics and its applications in stress and single-cell biology. Int. J. Mol. Sci. 2022, 23, 6985. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.K.; Pandey, A.; Hamurcu, M.; Avsaroglu, Z.Z.; Ozbek, M.; Omay, A.H.; Elbasan, F.; Omay, M.R.; Gokmen, F.; Topal, A. Variability in physiological traits reveals boron toxicity tolerance in Aegilops species. Front. Plant Sci. 2021, 12, 736614. [Google Scholar] [CrossRef] [PubMed]
- Louche, A.; Salcedo, S.; Bigot, S. Protein–protein interactions: Pull-down assays. Bact. Protein Secret. Syst. Methods Protoc. 2017, 1615, 247–255. [Google Scholar]
- Luo, L.; King, N.P.; Yeo, J.C.; Jones, A.; Stow, J.L. Stow, Single-step protease cleavage elution for identification of protein–protein interactions from GST pull-down and mass spectrometry. Proteomics 2014, 14, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Réthi-Nagy, Z.; Ábrahám, E.; Lipinszki, Z. GST-IVTT pull-down: A fast and versatile in vitro method for validating and mapping protein–protein interactions. FEBS Open Bio. 2022, 12, 1988–1995. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36 (Suppl. S1), D480–D484. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Target_Type | Target_Num | miRNA_Num | Pairs_Num |
|---|---|---|---|
| mRNA | 64 | 31 | 88 |
| lncRNA | 9 | 9 | 11 |
| circRNA | 4 | 4 | 6 |
| Sample | Total Number of Mass Spectra | Number of Identified Spectra | Spectral Interpretation Rate (%) | Number of Identified Peptide Segments | Number of Identified Proteins | Unique-2 |
|---|---|---|---|---|---|---|
| SY | 7096 | 915 | 12.89 | 115 | 48 | 26 |
| DZ | 2487 | 574 | 23.08 | 3 | 2 | 1 |
| Protein ID | Coverage (%) | Mass (Da) | Unique Peptide |
|---|---|---|---|
| Experimental group (SY) | |||
| sp|C0Z361|CPNB3_ARATH | 17.92 | 63,324.0 | 10 |
| sp|P21238|CPNA1_ARATH | 17.58 | 62,071.3 | 8 |
| sp|P19366|ATPB_ARATH | 16.67 | 53,933.4 | 5 |
| sp|Q9LTX9|HSP7G_ARATH | 9.47 | 76,995.9 | 6 |
| sp|O65719|HSP7C_ARATH | 10.17 | 71,147.0 | 7 |
| sp|Q9LD57|PGKH1_ARATH | 11.43 | 50,111.2 | 3 |
| sp|P53496|ACT11_ARATH | 14.59 | 41,674.3 | 4 |
| sp|Q9C5A9|ATPBO_ARATH | 12.16 | 59,858.6 | 4 |
| sp|O03042|RBL_ARATH | 6.89 | 52,954.7 | 3 |
| tr|F4IGL5|F4IGL5_ARATH | 9.51 | 41,807.3 | 3 |
| Control group (DZ) | |||
| sp|P59259|H4_ARATH | 21.36 | 11,409.3 | 2 |
| sp|P53497|ACT12_ARATH | 4.77 | 41,794.5 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Wang, L.; Ackah, M.; Amoako, F.K.; Jiang, Z.; Shi, Y.; Li, H.; Zhao, W. The Competing Endogenous RNAs Regulatory Genes Network Mediates Leaf Shape Variation and Main Effector Gene Function in Mulberry Plant (Morus alba). Int. J. Mol. Sci. 2023, 24, 16860. https://doi.org/10.3390/ijms242316860
Li J, Wang L, Ackah M, Amoako FK, Jiang Z, Shi Y, Li H, Zhao W. The Competing Endogenous RNAs Regulatory Genes Network Mediates Leaf Shape Variation and Main Effector Gene Function in Mulberry Plant (Morus alba). International Journal of Molecular Sciences. 2023; 24(23):16860. https://doi.org/10.3390/ijms242316860
Chicago/Turabian StyleLi, Jianbin, Lei Wang, Michael Ackah, Frank Kwarteng Amoako, Zijie Jiang, Yisu Shi, Haonan Li, and Weiguo Zhao. 2023. "The Competing Endogenous RNAs Regulatory Genes Network Mediates Leaf Shape Variation and Main Effector Gene Function in Mulberry Plant (Morus alba)" International Journal of Molecular Sciences 24, no. 23: 16860. https://doi.org/10.3390/ijms242316860
APA StyleLi, J., Wang, L., Ackah, M., Amoako, F. K., Jiang, Z., Shi, Y., Li, H., & Zhao, W. (2023). The Competing Endogenous RNAs Regulatory Genes Network Mediates Leaf Shape Variation and Main Effector Gene Function in Mulberry Plant (Morus alba). International Journal of Molecular Sciences, 24(23), 16860. https://doi.org/10.3390/ijms242316860






