Genetic and Epigenetic Factors in Gestational Diabetes Mellitus Pathology
Abstract
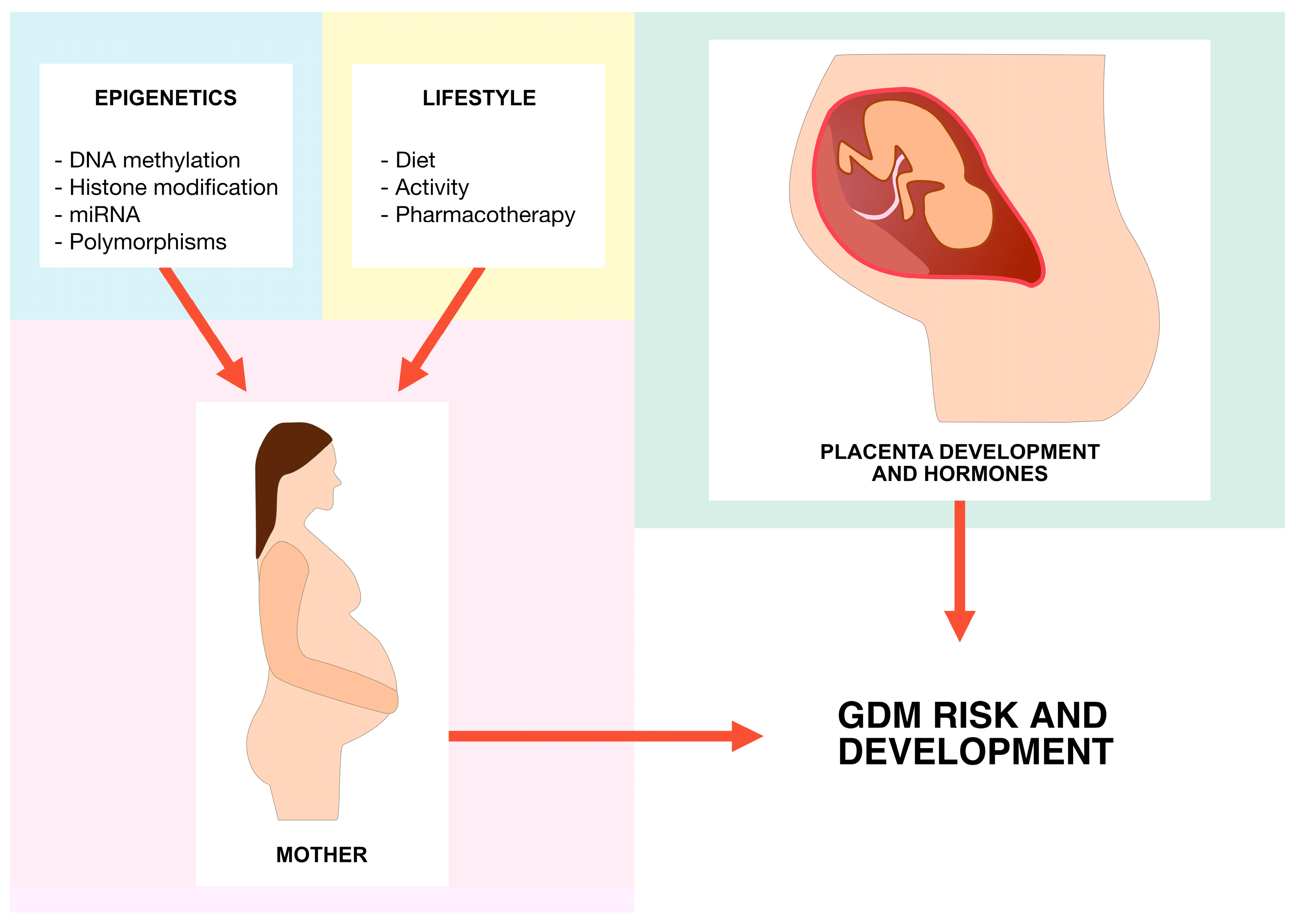
:1. Introduction
2. MODY
- MODY 3: hepatocyte nuclear factor 1 alpha (HNF1A)
- MODY 1: hepatocyte nuclear factor 4 alpha (HNF4A)
- MODY 2: glucokinase (GCK)
- MODY 5: hepatocyte nuclear factor 1 beta (HNF1B)
2.1. MODY 1
2.2. MODY 2
2.3. MODY 3
2.4. MODY 4
2.5. MODY 5
2.6. MODY 12
2.7. MODY 13
3. Adiponectin S, Leptin, and Interleukins
4. The β3-Adrenergic Receptor (ADRB3)
5. Insulin Receptor
5.1. Insulin Receptor Substrate 1
5.2. Insulin-Sensitive Glucose Transporter Protein 4/Solute Carrier Family 2 Member 4
6. Plasma Cell Membrane Glycoprotein 1
7. Calpain 10
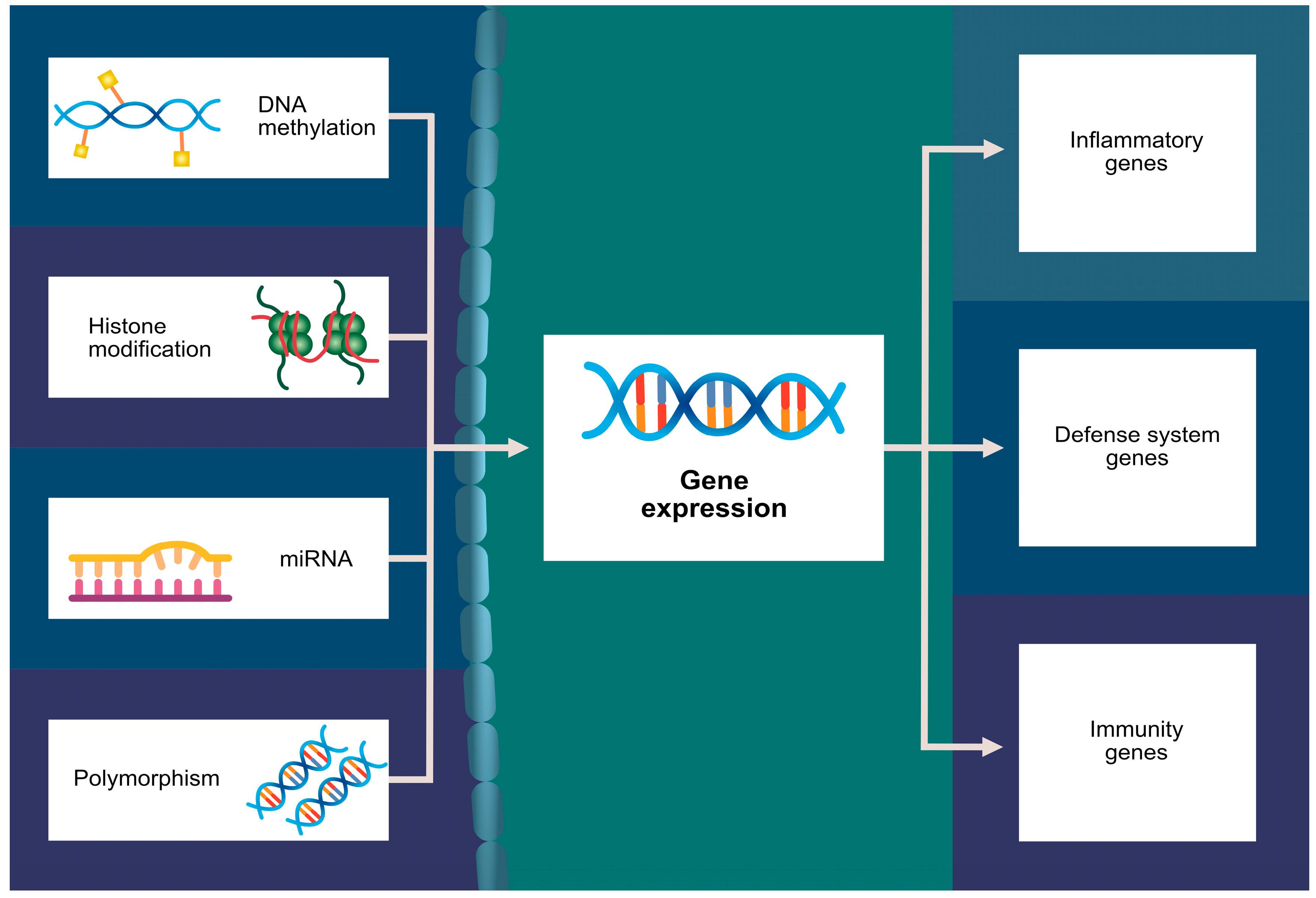
8. Histone Modification
9. miRNA
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- American Diabetes Association. 2. Classification and Diagnosis of Diabetes. Diabetes Care 2020, 43 (Suppl. S1), S14–S31. [Google Scholar] [CrossRef] [PubMed]
- Rosik, J.; Szostak, B.; Machaj, F.; Pawlik, A. The role of genetics and epigenetics in the pathogenesis of gestational diabetes mellitus. Ann. Hum. Genet. 2020, 84, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Sweeting, A.; Wong, J.; Murphy, H.R.; Ross, G.P. A Clinical Update on Gestational Diabetes Mellitus. Endocr. Rev. 2022, 43, 763–793. [Google Scholar] [CrossRef] [PubMed]
- Kharroubi, A.T.; Darwish, H.M. Diabetes mellitus: The epidemic of the century. World J. Diabetes 2015, 6, 850–867. [Google Scholar] [CrossRef] [PubMed]
- Metzger, B.E.; Buchanan, T.A.; Coustan, D.R.; de Leiva, A.; Dunger, D.B.; Hadden, D.R.; Hod, M.; Kitzmiller, J.L.; Kjos, S.L.; Oats, J.N.; et al. Summary and Recommendations of the Fifth International Workshop-Conference on Gestational Diabetes Mellitus. Diabetes Care 2007, 30 (Suppl. S2), S251–S260. [Google Scholar] [CrossRef]
- Bellamy, L.; Casas, J.-P.; Hingorani, A.D.; Williams, D. Type 2 diabetes mellitus after gestational diabetes: A systematic review and meta-analysis. Lancet 2009, 373, 1773–1779. [Google Scholar] [CrossRef]
- Galtier, F. Definition, epidemiology, risk factors. Diabetes Metab. 2010, 36, 628–651. [Google Scholar] [CrossRef]
- Kim, C. Gestational diabetes: Risks, management, and treatment options. Int. J. Womens Health 2010, 2, 339–351. [Google Scholar] [CrossRef]
- Tarnowski, M.; Malinowski, D.; Pawlak, K.; Dziedziejko, V.; Safranow, K.; Pawlik, A. GCK, GCKR, FADS1, DGKB/TMEM195 and CDKAL1 Gene Polymorphisms in Women with Gestational Diabetes. Can. J. Diabetes 2017, 41, 372–379. [Google Scholar] [CrossRef]
- Franzago, M.; Fraticelli, F.; Stuppia, L.; Vitacolonna, E. Nutrigenetics, epigenetics and gestational diabetes: Consequences in mother and child. Epigenetics 2019, 14, 215–235. [Google Scholar] [CrossRef]
- Dalfrà, M.G.; Burlina, S.; Del Vescovo, G.G.; Lapolla, A. Genetics and Epigenetics: New Insight on Gestational Diabetes Mellitus. Front. Endocrinol. 2020, 11, 602477. [Google Scholar] [CrossRef] [PubMed]
- Lambrinoudaki, I.; Vlachou, S.A.; Creatsas, G. Genetics in gestational diabetes mellitus: Association with incidence, severity, pregnancy outcome and response to treatment. Curr. Diabetes Rev. 2010, 6, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, C.-M.; Hu, X.-Q.; Zhao, Y. Relationship between melatonin receptor 1B and insulin receptor substrate 1 polymorphisms with gestational diabetes mellitus: A systematic review and meta-analysis. Sci. Rep. 2014, 4, 6113. [Google Scholar] [CrossRef] [PubMed]
- Urakami, T. Maturity-onset diabetes of the young (MODY): Current perspectives on diagnosis and treatment. Diabetes Metab. Syndr. Obesity Targets Ther. 2019, 12, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Majewska, A.; Stanirowski, P.; Wielgoś, M.; Bomba-Opoń, D. Maturity-onset Diabetes of the Young (MODY) in Pregnancy: A Review. Curr. Diabetes Rev. 2023, 19, e280122200657. [Google Scholar] [CrossRef]
- Yorifuji, T.; Fujimaru, R.; Hosokawa, Y.; Tamagawa, N.; Shiozaki, M.; Aizu, K.; Jinno, K.; Maruo, Y.; Nagasaka, H.; Tajima, T.; et al. Comprehensive molecular analysis of Japanese patients with pediatric-onset MODY-type diabetes mellitus. Pediatr. Diabetes 2012, 13, 26–32. [Google Scholar] [CrossRef]
- Yahaya, T.O.; Ufuoma, S.B. Genetics and Pathophysiology of Maturity-onset Diabetes of the Young (MODY): A Review of Current Trends. Oman Med. J. 2020, 35, e126. [Google Scholar] [CrossRef]
- Yau, T.T.L.; Yu, S.C.Y.; Cheng, J.Y.; Kwok, J.S.S.; Ma, R.C.W. GCK-MODY in pregnancy: A pregnant woman with diabetes and a small-for-gestational-age fetus. Clin. Case Rep. 2022, 10, e6629. [Google Scholar] [CrossRef]
- Dickens, L.T.; Naylor, R.N. Clinical Management of Women with Monogenic Diabetes During Pregnancy. Curr. Diabetes Rep. 2018, 18, 12. [Google Scholar] [CrossRef]
- Shepherd, M.; Brook, A.J.; Chakera, A.J.; Hattersley, A.T. Management of sulfonylurea-treated monogenic diabetes in pregnancy: Implications of placental glibenclamide transfer. Diabet. Med. 2017, 34, 1332–1339. [Google Scholar] [CrossRef]
- Pearson, E.R.; Boj, S.F.; Steele, A.M.; Barrett, T.; Stals, K.; Shield, J.P.; Ellard, S.; Ferrer, J.; Hattersley, A.T. Macrosomia and Hyperinsulinaemic Hypoglycaemia in Patients with Heterozygous Mutations in the HNF4A Gene. PLoS Med. 2007, 4, e118. [Google Scholar] [CrossRef]
- Matschinsky, F.M.; Wilson, D.F. The Central Role of Glucokinase in Glucose Homeostasis: A Perspective 50 Years After Demonstrating the Presence of the Enzyme in Islets of Langerhans. Front. Physiol. 2019, 10, 148. [Google Scholar] [CrossRef] [PubMed]
- Kirzhner, A.; Barak, O.; Vaisbuch, E.; Zornitzki, T.; Schiller, T. The Challenges of Treating Glucokinase MODY during Pregnancy: A Review of Maternal and Fetal Outcomes. Int. J. Environ. Res. Public Health 2022, 19, 5980. [Google Scholar] [CrossRef] [PubMed]
- Angueira, A.R.; Ludvik, A.E.; Reddy, T.E.; Wicksteed, B.; Lowe, W.L.; Layden, B.T. New Insights Into Gestational Glucose Metabolism: Lessons Learned From 21st Century Approaches. Diabetes 2015, 64, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Chakera, A.J.; Steele, A.M.; Gloyn, A.L.; Shepherd, M.H.; Shields, B.; Ellard, S.; Hattersley, A.T. Recognition and Management of Individuals With Hyperglycemia Because of a Heterozygous Glucokinase Mutation. Diabetes Care 2015, 38, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Ye, H.; Hong, Q.; Wang, L.; Wang, Q.; Wang, H.; Xu, L.; Bu, S.; Zhang, L.; Cheng, J.; et al. Elevated CpG island methylation of GCK gene predicts the risk of type 2 diabetes in Chinese males. Gene 2014, 547, 329–333. [Google Scholar] [CrossRef]
- Steele, A.M.; Shields, B.M.; Wensley, K.J.; Colclough, K.; Ellard, S.; Hattersley, A.T. Prevalence of Vascular Complications Among Patients With Glucokinase Mutations and Prolonged, Mild Hyperglycemia. JAMA 2014, 311, 279–286. [Google Scholar] [CrossRef]
- Chen, B.; Du, Y.-R.; Zhu, H.; Sun, M.-L.; Wang, C.; Cheng, Y.; Pang, H.; Ding, G.; Gao, J.; Tan, Y.; et al. Maternal inheritance of glucose intolerance via oocyte TET3 insufficiency. Nature 2022, 605, 761–766. [Google Scholar] [CrossRef]
- Stride, A.; Shields, B.; Gill-Carey, O.; Chakera, A.J.; Colclough, K.; Ellard, S.; Hattersley, A.T. Cross-sectional and longitudinal studies suggest pharmacological treatment used in patients with glucokinase mutations does not alter glycaemia. Diabetologia 2014, 57, 54–56. [Google Scholar] [CrossRef]
- Bellanné-Chantelot, C.; Lévy, D.J.; Carette, C.; Saint-Martin, C.; Riveline, J.-P.; Larger, E.; Valéro, R.; Gautier, J.-F.; Reznik, Y.; Sola, A.; et al. Clinical Characteristics and Diagnostic Criteria of Maturity-Onset Diabetes Of The Young (MODY) due to Molecular Anomalies of the HNF1A Gene. J. Clin. Endocrinol. Metab. 2011, 96, E1346–E1351. [Google Scholar] [CrossRef]
- Stride, A.; Ellard, S.; Clark, P.; Shakespeare, L.; Salzmann, M.; Shepherd, M.; Salzman, M. Beta-cell dysfunction, insulin sensitivity, and glycosuria precede diabetes in hepatocyte nuclear factor-1alpha mutation carriers. Diabetes Care 2005, 28, 1751–1756. [Google Scholar] [CrossRef] [PubMed]
- Ebrahim, N.; Shakirova, K.; Dashinimaev, E. PDX1 is the cornerstone of pancreatic β-cell functions and identity. Front. Mol. Biosci. 2022, 9, 1091757. [Google Scholar] [CrossRef] [PubMed]
- Baumel-Alterzon, S.; Scott, D.K. Regulation of Pdx1 by oxidative stress and Nrf2 in pancreatic beta-cells. Front. Endocrinol. 2022, 13, 1011187. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fan, H.; Zhou, L.; Wu, Y.; Lu, H.; Luo, J. Altered expression of PGC-1 α and PDX1 and their methylation status are associated with fetal glucose metabolism in gestational diabetes mellitus. Biochem. Biophys. Res. Commun. 2018, 501, 300–306. [Google Scholar] [CrossRef]
- Kaimala, S.; Kumar, C.A.; Allouh, M.Z.; Ansari, S.A.; Emerald, B.S. Epigenetic modifications in pancreas development, diabetes, and therapeutics. Med. Res. Rev. 2022, 42, 1343–1371. [Google Scholar] [CrossRef]
- Chang, H.; Wang, D.; Xia, W.; Pan, X.; Huo, W.; Xu, S.; Li, Y. Epigenetic disruption and glucose homeostasis changes following low-dose maternal bisphenol A exposure. Toxicol. Res. 2016, 5, 1400–1409. [Google Scholar] [CrossRef]
- Dubois-Laforgue, D.; Cornu, E.; Saint-Martin, C.; Coste, J.; Bellanné-Chantelot, C.; Timsit, J. Diabetes, Associated Clinical Spectrum, Long-term Prognosis, and Genotype/Phenotype Correlations in 201 Adult Patients With Hepatocyte Nuclear Factor 1B. Diabetes Care 2017, 40, 1436–1443. [Google Scholar] [CrossRef]
- Pearson, E.R.; Badman, M.K.; Lockwood, C.R.; Clark, P.M.; Ellard, S.; Bingham, C.; Hattersley, A.T. Contrasting diabetes phenotypes associated with hepatocyte nuclear factor-1alpha and -1beta mutations. Diabetes Care 2004, 27, 1102–1107. [Google Scholar] [CrossRef]
- Edghill, E.L.; Bingham, C.; Slingerland, A.S.; Minton, J.A.; Noordam, C.; Ellard, S.; Hattersley, A.T. Hepatocyte nuclear factor-1 beta mutations cause neonatal diabetes and intrauterine growth retardation: Support for a critical role of HNF-1β in human pancreatic development. Diabet. Med. 2006, 23, 1301–1306. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, B.; Cheng, Y.; Zhou, Y.; Yan, Y.-S.; Luo, Q.; Jiang, Y.; Sheng, J.-Z.; Ding, G.-L.; Huang, H.-F. Insulin Therapy for Gestational Diabetes Mellitus Does Not Fully Protect Offspring From Diet-Induced Metabolic Disorders. Diabetes 2019, 68, 696–708. [Google Scholar] [CrossRef]
- Zhu, H.; Ding, G.; Liu, X.; Huang, H. Developmental origins of diabetes mellitus: Environmental epigenomics and emerging patterns. J. Diabetes 2023, 15, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Bowman, P.; Flanagan, S.E.; Edghill, E.L.; Damhuis, A.; Shepherd, M.H.; Paisey, R.; Hattersley, A.T.; Ellard, S. Heterozygous ABCC8 mutations are a cause of MODY. Diabetologia 2012, 55, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Haghvirdizadeh, P.; Mohamed, Z.; Abdullah, N.A.; Haghvirdizadeh, P.; Haerian, M.S.; Haerian, B.S. KCNJ11: Genetic Polymorphisms and Risk of Diabetes Mellitus. J. Diabetes Res. 2015, 2015, 908152. [Google Scholar] [CrossRef] [PubMed]
- Kocova, M. Genetic spectrum of neonatal diabetes. Balk. J. Med. Genet. 2020, 23, 5–15. [Google Scholar] [CrossRef]
- Madani, H.A.; Fawzy, N.; Afif, A.; Abdelghaffar, S.; Gohar, N. Study of kcnj11 gene mutations in association with monogenic diabetes of infancy and response to sulfonylurea treatment in a cohort study in Egypt. Acta Endocrinol. 2016, 12, 157–160. [Google Scholar] [CrossRef] [PubMed]
- van Otterdijk, S.D.; Binder, A.M.; Szic, K.S.V.; Schwald, J.; Michels, K.B. DNA methylation of candidate genes in peripheral blood from patients with type 2 diabetes or the metabolic syndrome. PLoS ONE 2017, 12, e0180955. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.A.H.; Ansari, S.A.; Mensah-Brown, E.P.K.; Emerald, B.S. The role of DNA methylation in the pathogenesis of type 2 diabetes mellitus. Clin. Epigenet. 2020, 12, 104. [Google Scholar] [CrossRef]
- García-Cardona, M.C.; Huang, F.; García-Vivas, J.M.; López-Camarillo, C.; del Rion Navarro, B.E.; Olivos, E.N.; Hong-Chong, E.; Bolaños-Jiménez, F.; A Marchat, L. DNA methylation of leptin and adiponectin promoters in children is reduced by the combined presence of obesity and insulin resistance. Int. J. Obes. 2014, 38, 1457–1465. [Google Scholar] [CrossRef]
- Xu, P.; Dong, S.; Wu, L.; Bai, Y.; Bi, X.; Li, Y.; Shu, C. Maternal and Placental DNA Methylation Changes Associated with the Pathogenesis of Gestational Diabetes Mellitus. Nutrients 2023, 15, 70. [Google Scholar] [CrossRef]
- Bouchard, L.; Hivert, M.-F.; Guay, S.-P.; St-Pierre, J.; Perron, P.; Brisson, D. Placental Adiponectin Gene DNA Methylation Levels Are Associated With Mothers’ Blood Glucose Concentration. Diabetes 2012, 61, 1272–1280. [Google Scholar] [CrossRef]
- Lesseur, C.; Armstrong, D.A.; Paquette, A.G.; Li, Z.; Padbury, J.F.; Marsit, C.J. Maternal obesity and gestational diabetes are associated with placental leptin DNA methylation. Am. J. Obstet. Gynecol. 2014, 211, 654.e1–654.e9. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, L.; Thibault, S.; Guay, S.-P.; Santure, M.; Monpetit, A.; St-Pierre, J.; Perron, P.; Brisson, D. Leptin Gene Epigenetic Adaptation to Impaired Glucose Metabolism During Pregnancy. Diabetes Care 2010, 33, 2436–2441. [Google Scholar] [CrossRef] [PubMed]
- Dłuski, D.F.; Wolińska, E.; Skrzypczak, M. Epigenetic Changes in Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2021, 22, 7649. [Google Scholar] [CrossRef] [PubMed]
- Halvatsiotis, P.; Tsokaki, T.; Chrelias, C.; Kassanos, D.; Domali, E.; Gazouli, M.; Dimitriadis, G.; Kalantaridou, S. Methylation profile of genes involved in inflammation, in the blood from pregnancies with maternal preeclampsia due to untreated gestational diabetes mellitus. Hormones 2019, 18, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Lee, C.-N.; Li, H.-Y.; Hsu, K.-H.; Wang, S.-H.; Lin, S.-Y. Association of Interleukin-10 Methylation Levels With Gestational Diabetes in a Taiwanese Population. Front. Genet. 2018, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, L.; Liu, B.; Li, Q.; Wang, Z.; Fan, S.; Wang, H.; Wang, L. Functional Defects of Regulatory T Cell Through Interleukin 10 Mediated Mechanism in the Induction of Gestational Diabetes Mellitus. DNA Cell Biol. 2018, 37, 278–285. [Google Scholar] [CrossRef]
- Qiu, H.; Liu, X.; Yao, S.; Zhou, J.; Zhang, X.; Du, J. Regulation and Mechanism of miR-518d through the PPARα-Mediated NF-κB Pathway in the Development of Gestational Diabetes Mellitus. J. Diabetes Res. 2020, 2020, 7019597. [Google Scholar] [CrossRef]
- Valencia-Ortega, J.; Saucedo, R.; Sánchez-Rodríguez, M.A.; Cruz-Durán, J.G.; Martínez, E.G.R. Epigenetic Alterations Related to Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2021, 22, 9462. [Google Scholar] [CrossRef]
- Evans, B.A.; Merlin, J.; Bengtsson, T.; Hutchinson, D.S. Adrenoceptors in white, brown, and brite adipocytes. Br. J. Pharmacol. 2019, 176, 2416–2432. [Google Scholar] [CrossRef]
- Chamberlain, P.D.; Jennings, K.H.; Paul, F.; Cordell, J.; Berry, A.; Holmes, S.D.; Park, J.; Chambers, J.; Sennit, M.V.; Stock, M.J.; et al. The tissue distribution of the human beta3-adrenoceptor studied using a monoclonal antibody: Direct evidence of the beta3-adrenoceptor in human adipose tissue, atrium and skeletal muscle. Int. J. Obes. Relat. Metab. Disord. 1999, 23, 1057–1065. [Google Scholar] [CrossRef]
- Strosberg, A.D.; Pietri-Rouxel, F. Function and regulation of the beta 3-adrenoceptor. Trends Pharmacol. Sci. 1996, 17, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Parikh, H.; Groop, L. Candidate Genes for Type 2 Diabetes. Rev. Endocr. Metab. Disord. 2004, 5, 151–176. [Google Scholar] [CrossRef] [PubMed]
- Perfetti, R.; Hui, H.; Chamie, K.; Binder, S.; Seibert, M.; McLenithan, J.; Silver, K.; Walston, J. Pancreatic beta-cells expressing the Arg64 variant of the beta(3)-adrenergic receptor exhibit abnormal insulin secretory activity. J. Mol. Endocrinol. 2001, 27, 133–144. [Google Scholar] [CrossRef]
- Pilch, W.; Piotrowska, A.; Wyrostek, J.; Czerwińska-Ledwig, O.; Ziemann, E.; Antosiewicz, J.; Zasada, M.; Kulesa-Mrowiecka, M.; Żychowska, M. Different Changes in Adipokines, Lipid Profile, and TNF-Alpha Levels between 10 and 20 Whole Body Cryostimulation Sessions in Individuals with I and II Degrees of Obesity. Biomedicines 2022, 10, 269. [Google Scholar] [CrossRef]
- Walston, J.; Silver, K.; Hilfiker, H.; Andersen, R.E.; Seibert, M.; Beamer, B.; Roth, J.; Poehlman, E.; Shuldiner, A.R. Insulin response to glucose is lower in individuals homozygous for the Arg 64 variant of the beta-3-adrenergic receptor. J. Clin. Endocrinol. Metab. 2000, 85, 4019–4022. [Google Scholar] [PubMed]
- Wang, H.-D.; Zhang, C.-S.; Li, M.-W.; Lin, Q.; Zhang, Q.; Liu, D.-F.; Ma, Z.-Y.; Dong, J. The Association of Trp64Arg Polymorphism in the Beta-Adrenergic Receptor With Insulin Resistance: Meta-Analysis. Front. Endocrinol. 2021, 12, 708139. [Google Scholar] [CrossRef] [PubMed]
- Krugluger, W.; Festa, A.; Shnawa, N.; Bucher, J.; Boltz-Nitulescu, G.; Schernthaner, G.; Hopmeier, P. A serine/alanine polymorphism in the nucleotide-binding fold-2 of the sulphonylurea receptor-1 (S1369A) is associated with enhanced glucose-induced insulin secretion during pregnancy. J. Inherit. Metab. Dis. 2000, 23, 705–712. [Google Scholar] [CrossRef]
- Festa, A.; Krugluger, W.; Shnawa, N.; Hopmeier, P.; Haffner, S.M.; Schernthaner, G. Trp64Arg polymorphism of the beta3-adrenergic receptor gene in pregnancy: Association with mild gestational diabetes mellitus. J. Clin. Endocrinol. Metab. 1999, 84, 1695–1699. [Google Scholar]
- Alevizaki, M.; Thalassinou, L.; Grigorakis, S.I.; Philippou, G.; Lili, K.; Souvatzoglou, A.; Anastasiou, E. Study of the Trp64Arg polymorphism of the beta3-adrenergic receptor in Greek women with gestational diabetes. Diabetes Care 2000, 23, 1079–1083. [Google Scholar] [CrossRef]
- Zhang, C.; Bao, W.; Rong, Y.; Yang, H.; Bowers, K.; Yeung, E.; Kiely, M. Genetic variants and the risk of gestational diabetes mellitus: A systematic review. Hum. Reprod. Update 2013, 19, 376–390. [Google Scholar] [CrossRef]
- Lee, J.; Pilch, P.F. The insulin receptor: Structure, function, and signaling. Am. J. Physiol. 1994, 266, C319–C334. [Google Scholar] [CrossRef] [PubMed]
- Shaat, N.; Groop, L. Genetics of Gestational Diabetes Mellitus. Curr. Med. Chem. 2007, 14, 569–583. [Google Scholar] [CrossRef] [PubMed]
- Ott, R.; Melchior, K.; Stupin, J.H.; Ziska, T.; Schellong, K.; Henrich, W.; Rancourt, R.C.; Plagemann, A. Reduced Insulin Receptor Expression and Altered DNA Methylation in Fat Tissues and Blood of Women With GDM and Offspring. J. Clin. Endocrinol. Metab. 2019, 104, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Lowe, W.L. Genetics and Epigenetics: Implications for the Life Course of Gestational Diabetes. Int. J. Mol. Sci. 2023, 24, 6047. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Chen, L.; Song, B.; Cui, Z.; Chen, G.; Yu, Z. Insulin-like Growth Factor-2 (IGF-2) in Fibrosis. Biomolecules 2022, 12, 1557. [Google Scholar] [CrossRef]
- Ober, C.; Xiang, K.-S.; Thisted, R.A.; Indovina, K.A.; Wason, C.J.; Dooley, S.; Rao, D.C. Increased risk for gestational diabetes mellitus associated with insulin receptor and insulin-like growth factor II restriction fragment length polymorphisms. Genet. Epidemiology 1989, 6, 559–569. [Google Scholar] [CrossRef]
- Copps, K.D.; White, M.F. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia 2012, 55, 2565–2582. [Google Scholar] [CrossRef]
- Karami, M.; Mousavi, S.H.; Rafiee, M.; Heidari, R.; Shahrokhi, S.Z. Biochemical and molecular biomarkers: Unraveling their role in gestational diabetes mellitus. Diabetol. Metab. Syndr. 2023, 15, 5. [Google Scholar] [CrossRef]
- Wada, T.; Hori, S.; Sugiyama, M.; Fujisawa, E.; Nakano, T.; Tsuneki, H.; Nagira, K.; Saito, S.; Sasaoka, T. Progesterone inhibits glucose uptake by affecting diverse steps of insulin signaling in 3T3-L1 adipocytes. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E881–E888. [Google Scholar] [CrossRef]
- Barbour, L.A.; Shao, J.; Qiao, L.; Leitner, W.; Anderson, M.; Friedman, J.E.; Draznin, B. Human Placental Growth Hormone Increases Expression of the P85 Regulatory Unit of Phosphatidylinositol 3-Kinase and Triggers Severe Insulin Resistance in Skeletal Muscle. Endocrinology 2004, 145, 1144–1150. [Google Scholar] [CrossRef]
- Barreto-Andrade, J.N.; de Fátima, L.A.; Campello, R.S.; Guedes, J.A.C.; de Freitas, H.S.; Machado, M.M.O.U.F. Estrogen Receptor 1 (ESR1) Enhances. Int. J. Med Sci. 2018, 15, 1320–1328. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhang, L. miR-335-5p aggravates type 2 diabetes by inhibiting SLC2A4 expression. Biochem. Biophys. Res. Commun. 2021, 558, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Corrêa-Giannella, M.L.; Machado, U.F. SLC2A4 gene: A promising target for pharmacogenomics of insulin resistance. Pharmacogenomics 2013, 14, 847–850. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Ma, S.; Li, X.; Tian, Z.; Liang, H.; Yan, J.; Chen, M.; Tan, H. Relationships of SLC2A4, RBP4, PCK1, and PI3K Gene Polymorphisms with Gestational Diabetes Mellitus in a Chinese Population. BioMed Res. Int. 2019, 2019, 7398063. [Google Scholar] [CrossRef]
- Tarquini, F.; Picchiassi, E.; Centra, M.; Pennacchi, L.; Bini, V.; Cappuccini, B.; Torlone, E.; Coata, G.; Di Renzo, G.; Brancorsini, S. Body mass index associated to rs2021966 ENPP1 polymorphism increases the risk for gestational diabetes mellitus. Gynecol. Endocrinol. 2015, 31, 83–86. [Google Scholar] [CrossRef]
- Vasudevan, A.A.J.; Perković, M.; Bulliard, Y.; Cichutek, K.; Trono, D.; Häussinger, D.; Münk, C. Prototype Foamy Virus Bet Impairs the Dimerization and Cytosolic Solubility of Human APOBEC3G. J. Virol. 2013, 87, 9030–9040. [Google Scholar] [CrossRef]
- Branca, D. Calpain-related diseases. Biochem. Biophys. Res. Commun. 2004, 322, 1098–1104. [Google Scholar] [CrossRef]
- Biswas, S.; Harris, F.; Singh, J.; Phoenix, D. Role of calpains in diabetes mellitus-induced cataractogenesis: A mini review. Mol. Cell. Biochem. 2004, 261, 151–159. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, K.K. The calpain family and human disease. Trends Mol. Med. 2001, 7, 355–362. [Google Scholar] [CrossRef]
- Turner, M.D.; Cassell, P.G.; Hitman, G.A. Calpain-10: From genome search to function. Diabetes/Metab. Res. Rev. 2005, 21, 505–514. [Google Scholar] [CrossRef]
- Shang, L.; Huang, J.-F.; Ding, W.; Chen, S.; Xue, L.-X.; Ma, R.-F.; Xiong, K. Calpain: A molecule to induce AIF-mediated necroptosis in RGC-5 following elevated hydrostatic pressure. BMC Neurosci. 2014, 15, 63. [Google Scholar] [CrossRef]
- Laske, C.; Stellos, K.; Kempter, I.; Stransky, E.; Maetzler, W.; Fleming, I.; Randriamboavonjy, V. Increased cerebrospinal fluid calpain activity and microparticle levels in Alzheimer’s disease. Alzheimers Dement. 2015, 11, 465–474. [Google Scholar] [CrossRef]
- Wu, K.; Cai, Y. The SNP43 (G/A) polymorphism in CAPN10 gene confers an increased risk of cognitive impairment in cerebral small vessel disease. J. Clin. Lab. Anal. 2018, 32, e22615. [Google Scholar] [CrossRef] [PubMed]
- Perez-Martinez, P.; Delgado-Lista, J.; Garcia-Rios, A.; Ferguson, J.F.; Gulseth, H.L.; Williams, C.M.; Karlström, B.; Kieć-Wilk, B.; E Blaak, E.; Helal, O.; et al. Calpain-10 interacts with plasma saturated fatty acid concentrations to influence insulin resistance in individuals with the metabolic syndrome. Am. J. Clin. Nutr. 2011, 93, 1136–1141. [Google Scholar] [CrossRef] [PubMed]
- Ono, Y.; Doi, N.; Shindo, M.; Pánico, P.; Salazar, A.M. Cryptic splicing events result in unexpected protein products from calpain-10 (CAPN10) cDNA. Biochim. Biophys. Acta Mol. Cell Res. 2022, 1869, 119188. [Google Scholar] [CrossRef] [PubMed]
- Görisch, S.M.; Wachsmuth, M.; Tóth, K.F.; Lichter, P.; Rippe, K. Histone acetylation increases chromatin accessibility. J. Cell Sci. 2005, 118, 5825–5834. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H. Histone modifications for human epigenome analysis. J. Hum. Genet. 2013, 58, 439–445. [Google Scholar] [CrossRef] [PubMed]
- McGee, S.L.; Hargreaves, M. Exercise and skeletal muscle glucose transporter 4 expression: Molecular mechanisms. Clin. Exp. Pharmacol. Physiol. 2006, 33, 395–399. [Google Scholar] [CrossRef]
- Fernández-Morera, J.L.; Rodríguez-Rodero, S.; Menéndez-Torre, E.; Fraga, M.F. The Possible Role of Epigenetics in Gestational Diabetes: Cause, Consequence, or Both. Obstet. Gynecol. Int. 2010, 2010, 605163. [Google Scholar] [CrossRef]
- Michalczyk, A.A.; Dunbar, J.A.; Janus, E.D.; Best, J.D.; Ebeling, P.R.; Ackland, M.J.; Asproloupos, D. Epigenetic markers to predict conversion from gestational diabetes to type 2 diabetes. J. Clin. Endocrinol. Metab. 2016, 101, 2396–2404. [Google Scholar] [CrossRef]
- Hepp, P.; Hutter, S.; Knabl, J.; Hofmann, S.; Kuhn, C.; Mahner, S.; Jeschke, U. Histone H3 lysine 9 acetylation is downregulated in GDM Placentas and Calcitriol supplementation enhanced this effect. Int. J. Mol. Sci. 2018, 19, 4061. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, N. MicroRNAs and Exosomal microRNAs May Be Possible Targets to Investigate in Gestational Diabetes Mellitus. Diabetes Metab. Syndr. Obes. 2022, 15, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Dong, J.; Jiang, T.; Shi, Z.; Yu, B.; Zhu, Y.; Chen, D.; Xu, J.; Huo, R.; Dai, J.; et al. Early Second-Trimester Serum MiRNA Profiling Predicts Gestational Diabetes Mellitus. PLoS ONE 2011, 6, e23925. [Google Scholar] [CrossRef]
- Moen, G.-H.; Sommer, C.; Prasad, R.B.; Sletner, L.; Groop, L.; Qvigstad, E.; Birkeland, K. MECHANISMS IN ENDOCRINOLOGY: Epigenetic modifications and gestational diabetes: A systematic review of published literature. Eur. J. Endocrinol. 2017, 176, R247–R267. [Google Scholar] [CrossRef] [PubMed]
- da Silva, P.H.C.M.; Santos, K.F.; da Silva, L.; da Costa, C.C.P.; Santos, R.D.S.; Reis, A.A.D.S. MicroRNAs Associated with the Pathophysiological Mechanisms of Gestational Diabetes Mellitus: A Systematic Review for Building a Panel of miRNAs. J. Pers. Med. 2023, 13, 1126. [Google Scholar] [CrossRef]
- Kwon, D.-N.; Chang, B.-S.; Kim, J.-H. MicroRNA Dysregulation in Liver and Pancreas of CMP-Neu5Ac Hydroxylase Null Mice Disrupts Insulin/PI3K-AKT Signaling. BioMed Res. Int. 2014, 2014, 236385. [Google Scholar] [CrossRef]
- Pfeiffer, S.; Sánchez-Lechuga, B.; Donovan, P.; Halang, L.; Prehn, J.H.M.; Campos-Caro, A.; Byrne, M.M.; López-Tinoco, C. Circulating miR-330-3p in Late Pregnancy is Associated with Pregnancy Outcomes Among Lean Women with GDM. Sci. Rep. 2020, 10, 908. [Google Scholar] [CrossRef]
- Zhu, Y.; Tian, F.; Li, H.; Zhou, Y.; Lu, J.; Ge, Q. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int. J. Gynaecol. Obstet. 2015, 130, 49–53. [Google Scholar] [CrossRef]
- Pheiffer, C.; Dias, S.; Rheeder, P.; Adam, S. Decreased Expression of Circulating miR-20a-5p in South African Women with Gestational Diabetes Mellitus. Mol. Diagn. Ther. 2018, 22, 345–352. [Google Scholar] [CrossRef]
- Shi, Z.; Zhao, C.; Guo, X.; Ding, H.; Cui, Y.; Shen, R.; Liu, J. Differential Expression of MicroRNAs in Omental Adipose Tissue From Gestational Diabetes Mellitus Subjects Reveals miR-222 as a Regulator of ERα Expression in Estrogen-Induced Insulin Resistance. Endocrinology 2014, 155, 1982–1990. [Google Scholar] [CrossRef]
- Filardi, T.; Catanzaro, G.; Grieco, G.E.; Splendiani, E.; Trocchianesi, S.; Santangelo, C.; Brunelli, R.; Guarino, E.; Sebastiani, G.; Dotta, F.; et al. Identification and Validation of miR-222-3p and miR-409-3p as Plasma Biomarkers in Gestational Diabetes Mellitus Sharing Validated Target Genes Involved in Metabolic Homeostasis. Int. J. Mol. Sci. 2022, 23, 4276. [Google Scholar] [CrossRef] [PubMed]
- Qian, K.; Hu, L.; Chen, H.; Li, H.; Liu, N.; Li, Y.; Ai, J.; Zhu, G.; Tang, Z.; Zhang, H. Hsa-miR-222 is involved in differentiation of endometrial stromal cells in vitro. Endocrinology 2009, 150, 4734–4743. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.Y.; Li, P.Y.; Wang, Y.F.; Wang, P.; Wu, Y.H.; Gao, S.G.; Li, S. High glucose-increased miR-200c contributes to cellular senescence and DNA damage in neural stem cells. Birth Defects Res. 2023, 115, 1770–1779. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.M.; Wei, J.; Xu, L.L.; Yan, Y.S.; Chen, Y.; Lv, M.; Jiang, Y.; Luo, Q. Altered expression of long noncoding RNA MEG3 in the offspring of gestational diabetes mellitus induces impaired glucose tolerance in adulthood. Acta Diabetol. 2023, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mitra, T.; Gulati, R.; Uppal, A.; Kumari, S.R.; Tripathy, S.; Ranjan, P.; Janardhanan, R. Prospecting of exosomal-miRNA signatures as prognostic marker for gestational diabetes mellitus and other adverse pregnancy outcomes. Front. Endocrinol. 2023, 14, 1097337. [Google Scholar] [CrossRef]
| MODY Type | Gene | Full Name | Mutation Influence on Pathophysiology |
|---|---|---|---|
| Most common mutations accounting for 70–90% of MODY cases | |||
| MODY 3 | HNF1A | Hepatocyte nuclear factor-1 alpha | Gradual beta-cell dysfunction, reduced insulin production, and progressive hyperglycemia |
| MODY 1 | HNF4A | Hepatocyte nuclear factor-4 alpha | Progressive beta-cell dysfunction, fetal macrosomia, and hyperinsulinemic hypoglycemia |
| MODY 2 | GCK | Glucokinase | Disrupted glucose sensing and hyperglycemia |
| MODY 5 | HNF1B | Hepatocyte nuclear factor 1B | Dysfunctional pancreatic development, suppressed cytokine signaling, and formation of kidney cyst |
| MODY mutations of lower prevalence | |||
| MODY 4 | IPFI/PDX1 | Insulin promoter factor/pancreatic duodenal homeobox | Pancreatic agenesis, beta-cell development, and defective insulin secretion |
| MODY 13 | KCNJII | Inward-rectifier potassium channel, subfamily J, member 11 | Congenital hyperinsulinism |
| MODY 12 | ABCC8 | ATP binding cassette subfamily C member 8 | Congenital hyperinsulinism, disrupted biogenesis, and insulin trafficking of KATP channels |
| Other GDM mutations | |||
| CAPN10 | Calpain-10 | Dysfunction of cell metabolism and signal transduction and elevated fasting glucose levels | |
| ADRB3 | β3-adrenergic receptor | Decreased insulin excretion, disrupted thermogenesis, and lipolysis | |
| INSR | Insulin receptor | Disrupted metabolism of β-cell and elevated glucose levels | |
| IRS1 | Insulin receptor substrate 1 | Dysfunction of intracellular signaling and increased insulin resistance | |
| GLUT4/SCLA4 | Insulin-sensitive glucose transporter protein 4/solute carrier family 2, member 4 | Progressively increasing insulin resistance | |
| PC-1 | Plasma cell membrane glycoprotein 1 | Increased insulin resistance | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ustianowski, Ł.; Udzik, J.; Szostak, J.; Gorący, A.; Ustianowska, K.; Pawlik, A. Genetic and Epigenetic Factors in Gestational Diabetes Mellitus Pathology. Int. J. Mol. Sci. 2023, 24, 16619. https://doi.org/10.3390/ijms242316619
Ustianowski Ł, Udzik J, Szostak J, Gorący A, Ustianowska K, Pawlik A. Genetic and Epigenetic Factors in Gestational Diabetes Mellitus Pathology. International Journal of Molecular Sciences. 2023; 24(23):16619. https://doi.org/10.3390/ijms242316619
Chicago/Turabian StyleUstianowski, Łukasz, Jakub Udzik, Joanna Szostak, Anna Gorący, Klaudia Ustianowska, and Andrzej Pawlik. 2023. "Genetic and Epigenetic Factors in Gestational Diabetes Mellitus Pathology" International Journal of Molecular Sciences 24, no. 23: 16619. https://doi.org/10.3390/ijms242316619
APA StyleUstianowski, Ł., Udzik, J., Szostak, J., Gorący, A., Ustianowska, K., & Pawlik, A. (2023). Genetic and Epigenetic Factors in Gestational Diabetes Mellitus Pathology. International Journal of Molecular Sciences, 24(23), 16619. https://doi.org/10.3390/ijms242316619






