Human Serum and Salivary Metabolomes: Diversity and Closeness
Abstract
:1. Introduction
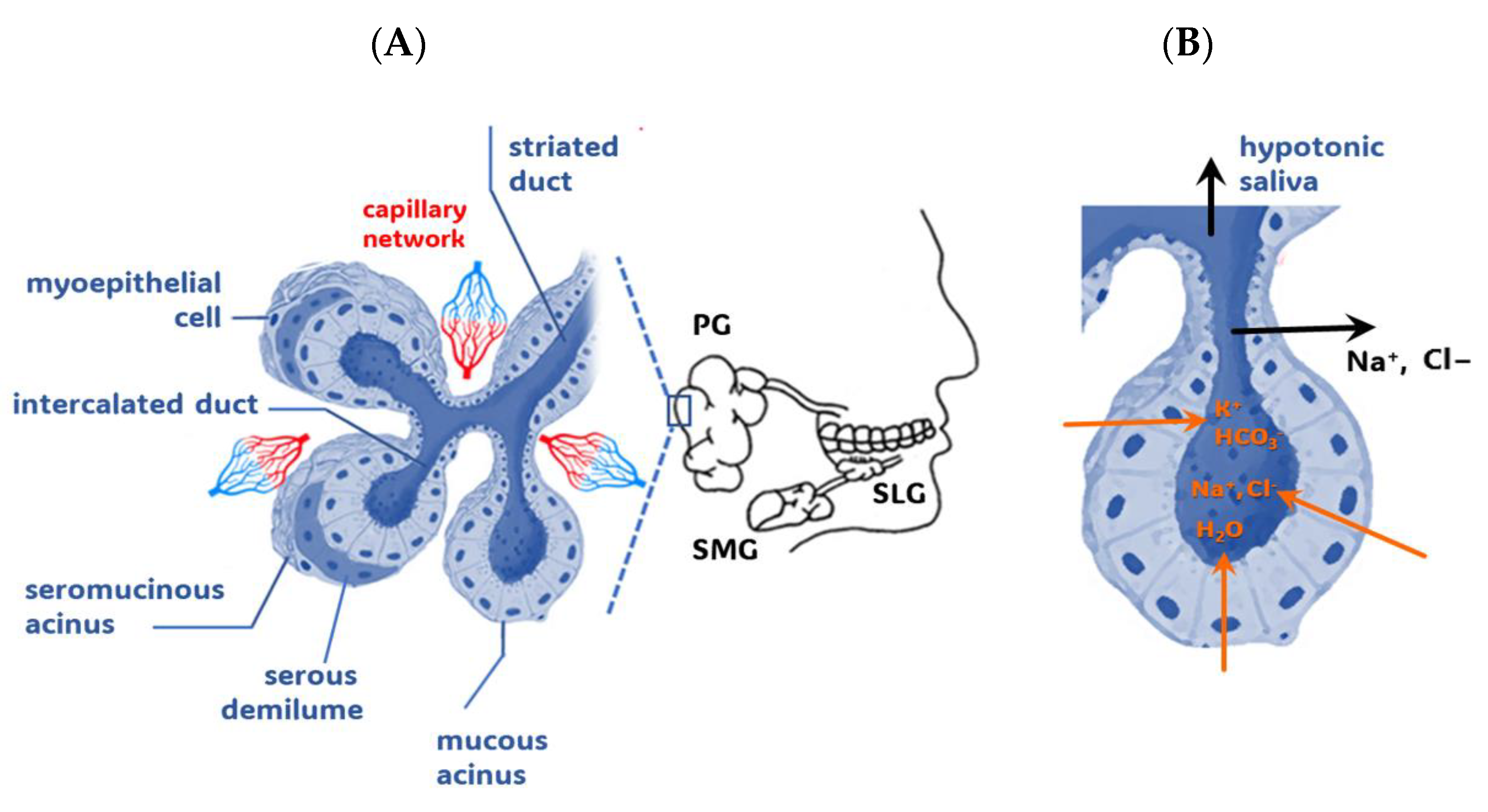
1.1. Salivary Glands
1.2. From Capillary Blood to Whole Saliva
1.3. Salivary Metabolomics
2. Results
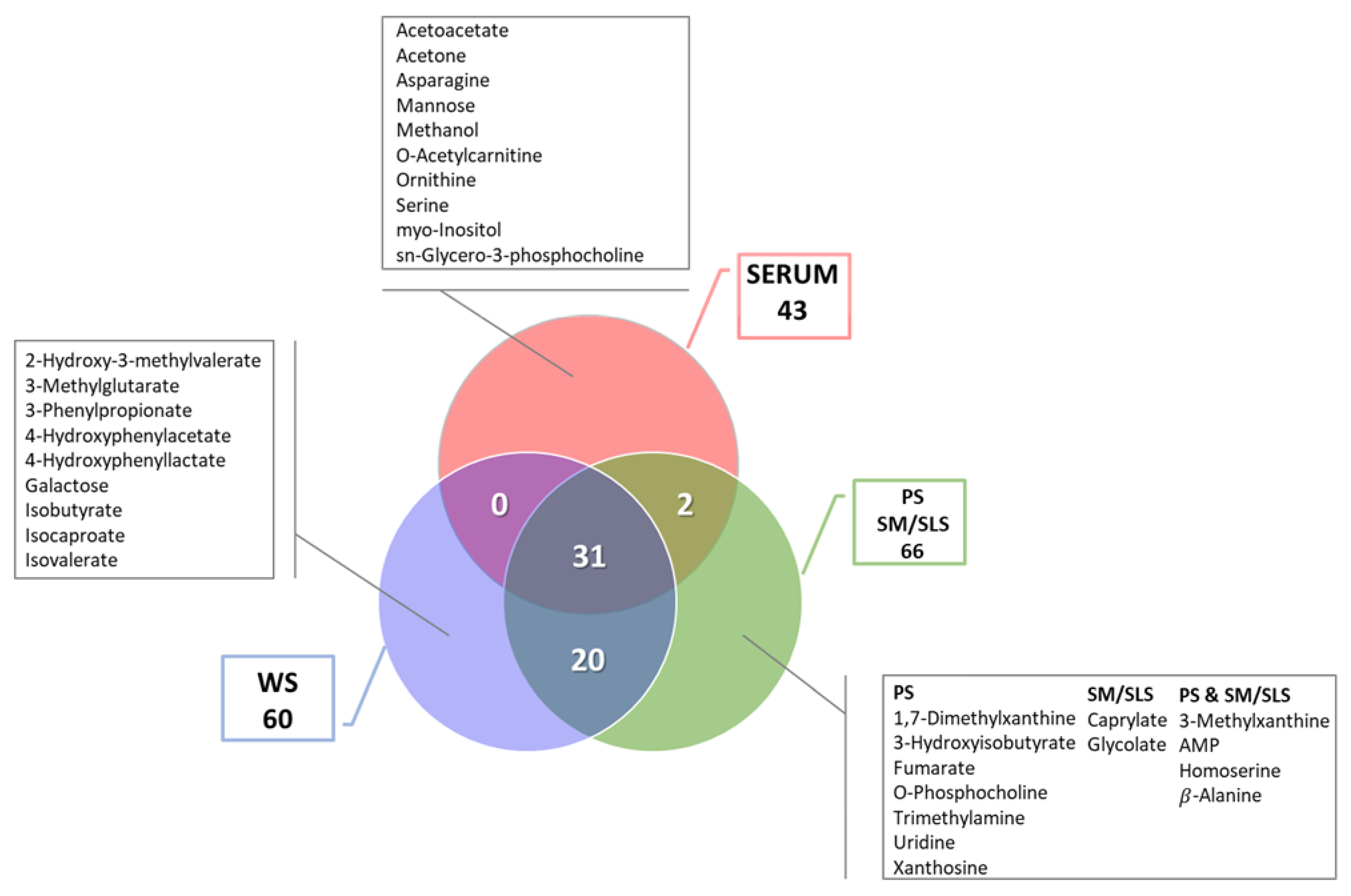
2.1. Sample Collection and Metabolite Profiling
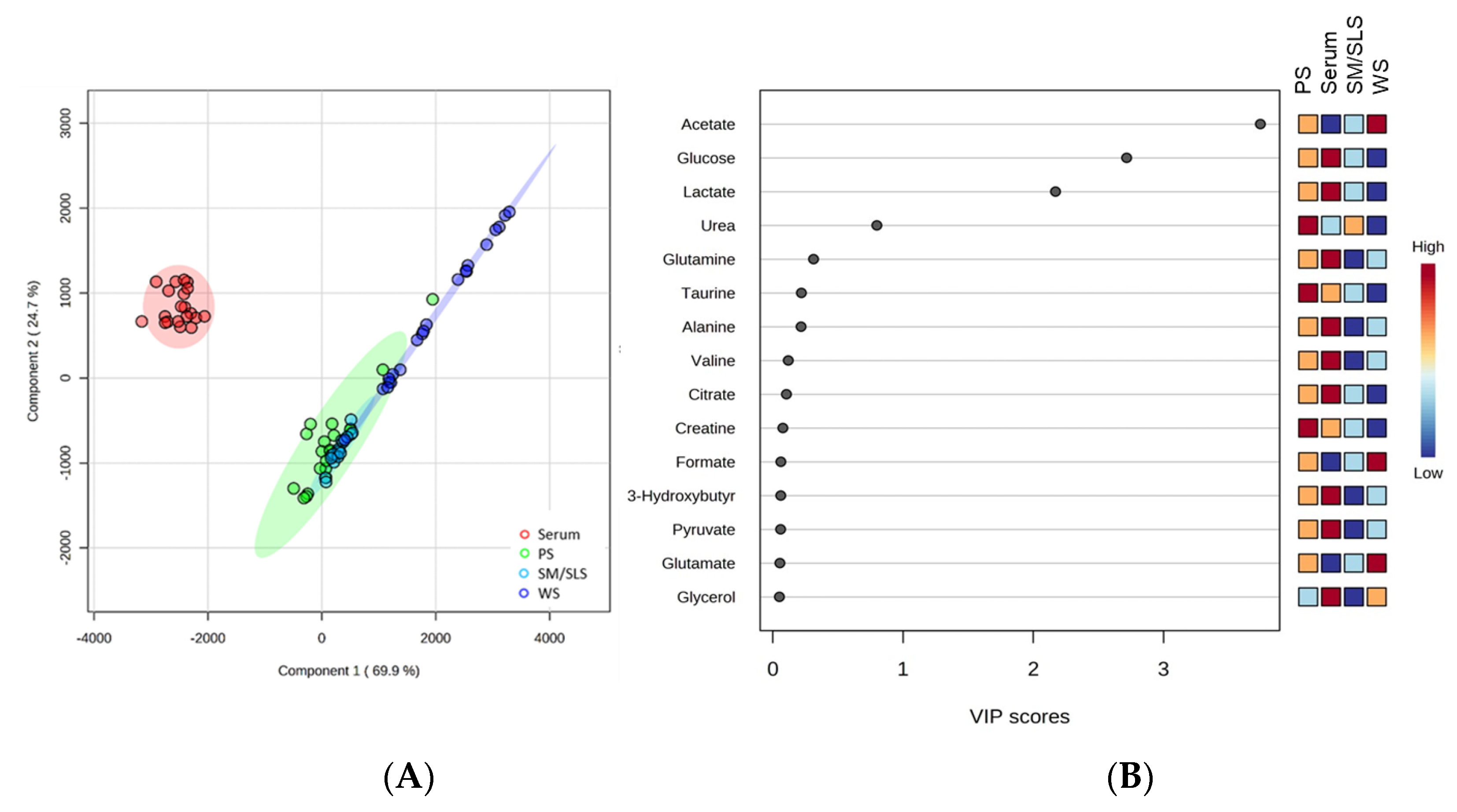
2.2. Comparison of Serum and Salivary Metabolite Profiles
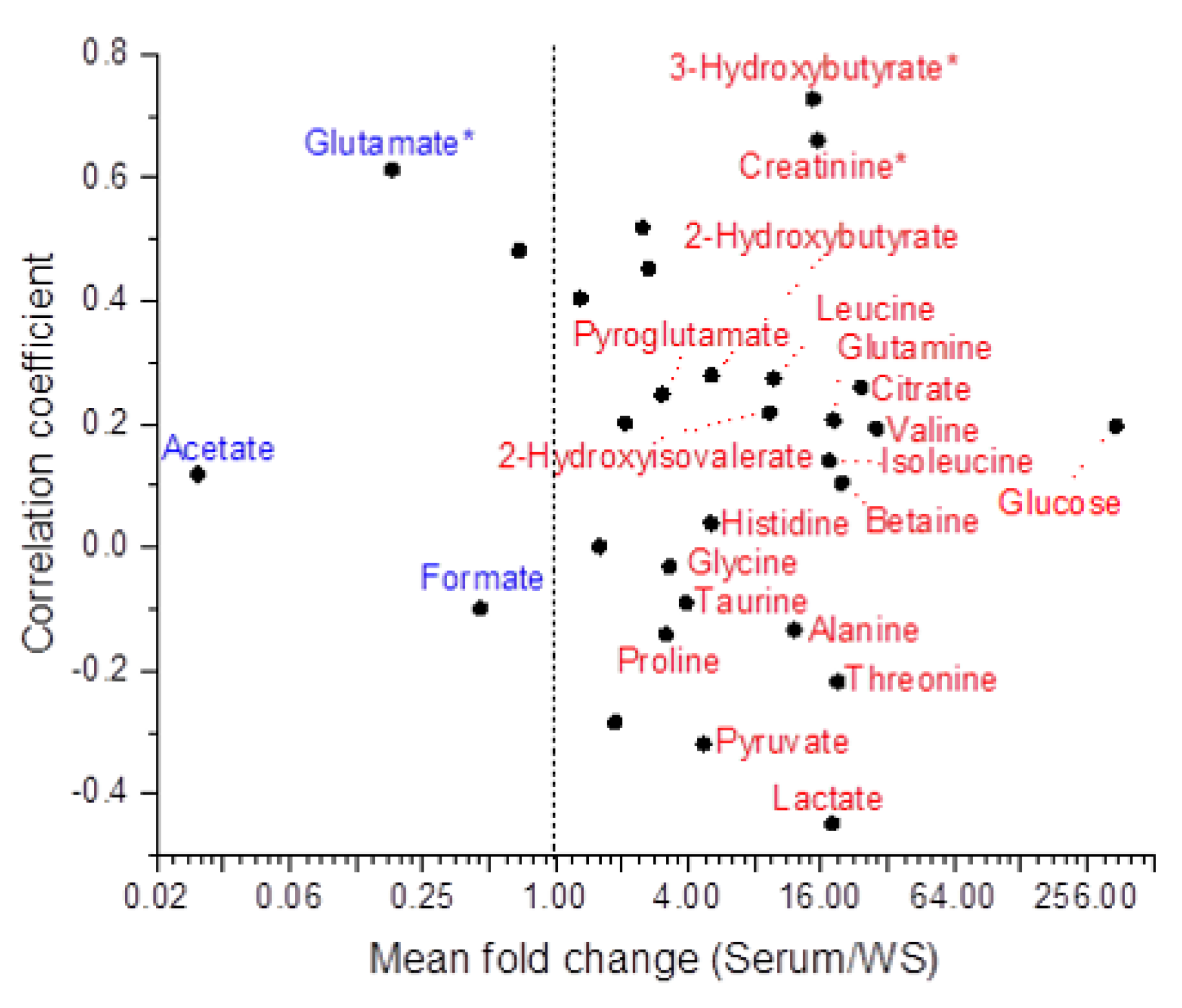
2.3. Comparison of Salivary and Serum Metabolite Concentrations
2.3.1. Metabolites Common to Serum and Saliva Collected from Parotid or Submandibular/Sublingual Glands
2.3.2. Metabolites Common to Serum and WS
3. Discussion
4. Materials and Methods
4.1. Blood Collection, Serum Sample Preparation, and Processing
4.2. Metabolomics Data Analysis
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pappa, E.; Vougas, K.; Zoidakis, J.; Vastardis, H. Proteomic Advances in Salivary Diagnostics. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2020, 1868, 140494. [Google Scholar] [CrossRef]
- Roblegg, E.; Coughran, A.; Sirjani, D. Saliva: An All-Rounder of Our Body. Eur. J. Pharm. Biopharm. 2019, 142, 133–141. [Google Scholar] [CrossRef]
- Gardner, A.; Carpenter, G.; So, P.-W. Salivary Metabolomics: From Diagnostic Biomarker Discovery to Investigating Biological Function. Metabolites 2020, 10, 47. [Google Scholar] [CrossRef]
- Pedersen, A.M.L.; Sørensen, C.E.; Proctor, G.B.; Carpenter, G.H.; Ekström, J. Salivary Secretion in Health and Disease. J. Oral Rehabil. 2018, 45, 730–746. [Google Scholar] [CrossRef]
- Pereira, J.A.M.; Porto-Figueira, P.; Taware, R.; Sukul, P.; Rapole, S.; Câmara, J.S. Unravelling the Potential of Salivary Volatile Metabolites in Oral Diseases. A Review. Molecules 2020, 25, 3098. [Google Scholar] [CrossRef]
- Yoshizawa, J.M.; Schafer, C.A.; Schafer, J.J.; Farrell, J.J.; Paster, B.J.; Wong, D.T.W. Salivary Biomarkers: Toward Future Clinical and Diagnostic Utilities. Clin. Microbiol. Rev. 2013, 26, 781–791. [Google Scholar] [CrossRef]
- Hyvärinen, E.; Kashyap, B.; Kullaa, A.M. Oral Sources of Salivary Metabolites. Metabolites 2023, 13, 498. [Google Scholar] [CrossRef]
- Ghannam, M.G.; Singh, P. Anatomy, Head and Neck, Salivary Glands; StatPearls Publishing: St. Petersburg, FL, USA, 2023. [Google Scholar]
- D’Agostino, C.; Elkashty, O.A.; Chivasso, C.; Perret, J.; Tran, S.D.; Delporte, C. Insight into Salivary Gland Aquaporins. Cells 2020, 9, 1547. [Google Scholar] [CrossRef]
- Proctor, G.B.; Shaalan, A.M. Disease-Induced Changes in Salivary Gland Function and the Composition of Saliva. J. Dent. Res. 2021, 100, 1201–1209. [Google Scholar] [CrossRef]
- Porcheri, C.; Mitsiadis, T. Physiology, Pathology and Regeneration of Salivary Glands. Cells 2019, 8, 976. [Google Scholar] [CrossRef]
- Pittman, T.W.; Decsi, D.B.; Punyadeera, C.; Henry, C.S. Saliva-Based Microfluidic Point-of-Care Diagnostic. Theranostics 2023, 13, 1091–1108. [Google Scholar] [CrossRef]
- Lima, D.P.; Diniz, D.G.; Moimaz, S.A.S.; Sumida, D.H.; Okamoto, A.C. Saliva: Reflection of the Body. Int. J. Infect. Dis. 2010, 14, e184–e188. [Google Scholar] [CrossRef]
- Shah, S. Salivaomics: The Current Scenario. J. Oral Maxillofac. Pathol. 2018, 22, 375. [Google Scholar] [CrossRef]
- Bel’skaya, L.V.; Sarf, E.A.; Kosenok, V.K. Age and Gender Characteristics of the Biochemical Composition of Saliva: Correlations with the Composition of Blood Plasma. J. Oral Biol. Craniofac. Res. 2020, 10, 59–65. [Google Scholar] [CrossRef]
- Loo, J.A.; Yan, W.; Ramachandran, P.; Wong, D.T. Comparative Human Salivary and Plasma Proteomes. J. Dent. Res. 2010, 89, 1016–1023. [Google Scholar] [CrossRef]
- Heller, D.; Helmerhorst, E.J.; Oppenheim, F.G. Saliva and Serum Protein Exchange at the Tooth Enamel Surface. J. Dent. Res. 2017, 96, 437–443. [Google Scholar] [CrossRef]
- Lau, W.W.; Hardt, M.; Zhang, Y.H.; Freire, M.; Ruhl, S. The Human Salivary Proteome Wiki: A Community-Driven Research Platform. J. Dent. Res. 2021, 100, 1510–1519. [Google Scholar] [CrossRef]
- Kim, S.; Song, Y.; Kim, S.; Kim, S.; Na, H.; Lee, S.; Chung, J.; Kim, S. Identification of a Biomarker Panel for Diagnosis of Early Childhood Caries Using Salivary Metabolic Profile. Metabolites 2023, 13, 356. [Google Scholar] [CrossRef]
- Li, Z.; Mu, Y.; Guo, C.; You, X.; Liu, X.; Li, Q.; Sun, W. Analysis of the Saliva Metabolic Signature in Patients with Primary Sjögren’s Syndrome. PLoS ONE 2022, 17, e0269275. [Google Scholar] [CrossRef]
- Nijakowski, K.; Zdrojewski, J.; Nowak, M.; Gruszczyński, D.; Knoll, F.; Surdacka, A. Salivary Metabolomics for Systemic Cancer Diagnosis: A Systematic Review. Metabolites 2022, 13, 28. [Google Scholar] [CrossRef]
- Meleti, M.; Quartieri, E.; Antonelli, R.; Pezzi, M.E.; Ghezzi, B.; Viani, M.V.; Setti, G.; Casali, E.; Ferrari, E.; Ciociola, T.; et al. Metabolic Profiles of Whole, Parotid and Submandibular/Sublingual Saliva. Metabolites 2020, 10, 318. [Google Scholar] [CrossRef]
- Quartieri, E.; Casali, E.; Ferrari, E.; Ghezzi, B.; Gallo, M.; Spisni, A.; Meleti, M.; Pertinhez, T.A. Sample Optimization for Saliva 1H-NMR Metabolic Profiling. Anal. Biochem. 2022, 640, 114412. [Google Scholar] [CrossRef]
- Proctor, G.B. The Physiology of Salivary Secretion. Periodontol 2000 2016, 70, 11–25. [Google Scholar] [CrossRef]
- Sakanaka, A.; Kuboniwa, M.; Katakami, N.; Furuno, M.; Nishizawa, H.; Omori, K.; Taya, N.; Ishikawa, A.; Mayumi, S.; Tanaka Isomura, E.; et al. Saliva and Plasma Reflect Metabolism Altered by Diabetes and Periodontitis. Front. Mol. Biosci. 2021, 8, 742002. [Google Scholar] [CrossRef]
- Spick, M.; Lewis, H.-M.; Frampas, C.F.; Longman, K.; Costa, C.; Stewart, A.; Dunn-Walters, D.; Greener, D.; Evetts, G.; Wilde, M.J.; et al. An Integrated Analysis and Comparison of Serum, Saliva and Sebum for COVID-19 Metabolomics. Sci. Rep. 2022, 12, 11867. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, H.; Zhu, J.; Liao, Z.; Wang, S.; Liu, W. Correlations of Salivary and Blood Glucose Levels among Six Saliva Collection Methods. Int. J. Environ. Res. Public Health 2022, 19, 4122. [Google Scholar] [CrossRef]
- Goodson, J.M. Disease Reciprocity between Gingivitis and Obesity. J. Periodontol. 2020, 91, S26–S34. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef]
- Tzimas, K.; Pappa, E. Saliva Metabolomic Profile in Dental Medicine Research: A Narrative Review. Metabolites 2023, 13, 379. [Google Scholar] [CrossRef]
- Miyazaki, T.; Honda, A.; Ikegami, T.; Iwamoto, J.; Monma, T.; Hirayama, T.; Saito, Y.; Yamashita, K.; Matsuzaki, Y. Simultaneous Quantification of Salivary 3-Hydroxybutyrate, 3-Hydroxyisobutyrate, 3-Hydroxy-3-Methylbutyrate, and 2-Hydroxybutyrate as Possible Markers of Amino Acid and Fatty Acid Catabolic Pathways by LC–ESI–MS/MS. Springerplus 2015, 4, 494. [Google Scholar] [CrossRef]
- Jasim, H.; Carlsson, A.; Hedenberg-Magnusson, B.; Ghafouri, B.; Ernberg, M. Saliva as a Medium to Detect and Measure Biomarkers Related to Pain. Sci. Rep. 2018, 8, 3220. [Google Scholar] [CrossRef]
- Teruya, T.; Goga, H.; Yanagida, M. Human Age-Declined Saliva Metabolic Markers Determined by LC–MS. Sci. Rep. 2021, 11, 18135. [Google Scholar] [CrossRef]
- Pandya, D. Assessment and Correlation of Urea and Creatinine Levels in Saliva and Serum of Patients with Chronic Kidney Disease, Diabetes and Hypertension—A Research Study. J. Clin. Diagn. Res. 2016, 10, ZC58–ZC62. [Google Scholar] [CrossRef]
- Lasisi, T.J.; Raji, Y.R.; Salako, B.L. Salivary Creatinine and Urea Analysis in Patients with Chronic Kidney Disease: A Case Control Study. BMC Nephrol. 2016, 17, 10. [Google Scholar] [CrossRef]
- Temilola, D.O.; Bezuidenhout, K.; Erasmus, R.T.; Stephen, L.; Davids, M.R.; Holmes, H. Salivary Creatinine as a Diagnostic Tool for Evaluating Patients with Chronic Kidney Disease. BMC Nephrol. 2019, 20, 387. [Google Scholar] [CrossRef]
- Nam, J.H.; Lee, H.S.; Kim, J.; Kim, J.; Chu, M.K. Salivary Glutamate Is Elevated in Individuals with Chronic Migraine. Cephalalgia 2018, 38, 1485–1492. [Google Scholar] [CrossRef]
- Gallo, M.; Matteucci, S.; Alaimo, N.; Pitti, E.; Orsale, M.V.; Summa, V.; Cicero, D.O.; Monteagudo, E. A Novel Method Using Nuclear Magnetic Resonance for Plasma Protein Binding Assessment in Drug Discovery Programs. J. Pharm. Biomed. Anal. 2019, 167, 21–29. [Google Scholar] [CrossRef]
- Gallo, M.; Giovati, L.; Magliani, W.; Pertinhez, T.A.; Conti, S.; Ferrari, E.; Spisni, A.; Ciociola, T. Metabolic Plasticity of Candida Albicans in Response to Different Environmental Conditions. J. Fungi 2022, 8, 723. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Cho, H.W.; Kim, S.B.; Jeong, M.K.; Park, Y.; Miller, N.G.; Ziegler, T.R.; Jones, D.P. Discovery of Metabolite Features for the Modelling and Analysis of High-Resolution NMR Spectra. Int. J. Data Min. Bioinform. 2008, 2, 176. [Google Scholar] [CrossRef]


| Metabolite | SERUM vs. PS Mean Fold Change | Metabolite | SERUM vs. SM/SLS Mean Fold Change | Metabolite | SERUM vs. WS Mean Fold Change |
|---|---|---|---|---|---|
| Aspartate | 0.7 | Formate | 0.7 | Aspartate | 0.7 |
| Formate | 0.8 | Glutamate * | 0.9 | Choline * | 1.3 |
| Taurine | 0.9 | Creatine * | 1.5 | Glycerol | 1.6 |
| Choline * | 1.0 | Urea * | 2.1 | Tyrosine * | 1.9 |
| Urea | 1.4 | Aspartate | 2.7 | Creatine | 2.1 |
| Glycerol | 1.7 | Lysine a | 2.5 | ||
| Tyrosine | 1.9 | Phenylalanine | 2.6 | ||
| Arginine | 2.1 | ||||
| Lactate | 2.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrari, E.; Gallo, M.; Spisni, A.; Antonelli, R.; Meleti, M.; Pertinhez, T.A. Human Serum and Salivary Metabolomes: Diversity and Closeness. Int. J. Mol. Sci. 2023, 24, 16603. https://doi.org/10.3390/ijms242316603
Ferrari E, Gallo M, Spisni A, Antonelli R, Meleti M, Pertinhez TA. Human Serum and Salivary Metabolomes: Diversity and Closeness. International Journal of Molecular Sciences. 2023; 24(23):16603. https://doi.org/10.3390/ijms242316603
Chicago/Turabian StyleFerrari, Elena, Mariana Gallo, Alberto Spisni, Rita Antonelli, Marco Meleti, and Thelma A. Pertinhez. 2023. "Human Serum and Salivary Metabolomes: Diversity and Closeness" International Journal of Molecular Sciences 24, no. 23: 16603. https://doi.org/10.3390/ijms242316603
APA StyleFerrari, E., Gallo, M., Spisni, A., Antonelli, R., Meleti, M., & Pertinhez, T. A. (2023). Human Serum and Salivary Metabolomes: Diversity and Closeness. International Journal of Molecular Sciences, 24(23), 16603. https://doi.org/10.3390/ijms242316603









