Low Prevalence of HLA-G Antibodies in Lung Transplant Patients Detected using MAIPA-Adapted Protocol
Abstract
1. Introduction
2. Results
2.1. Population Characteristics
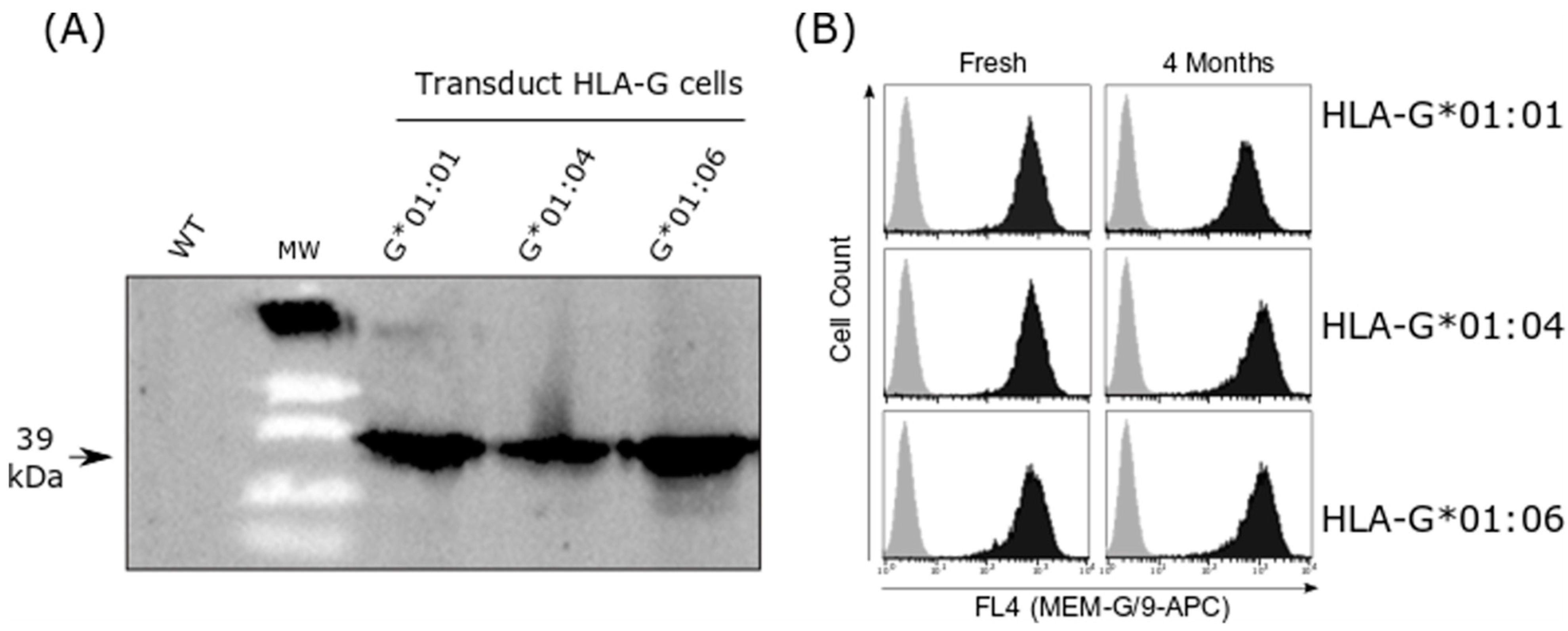
2.2. HLA-G Cell Line Transduction
2.3. HLA-G Antibody Detection in Healthy Donors
2.4. HLA-G Antibody Detection in Lung Transplantation Recipients
3. Discussion
4. Materials and Methods
4.1. Samples
4.2. HLA-G and HLA-G K562 Cell Line Transduction and Expression Assessment
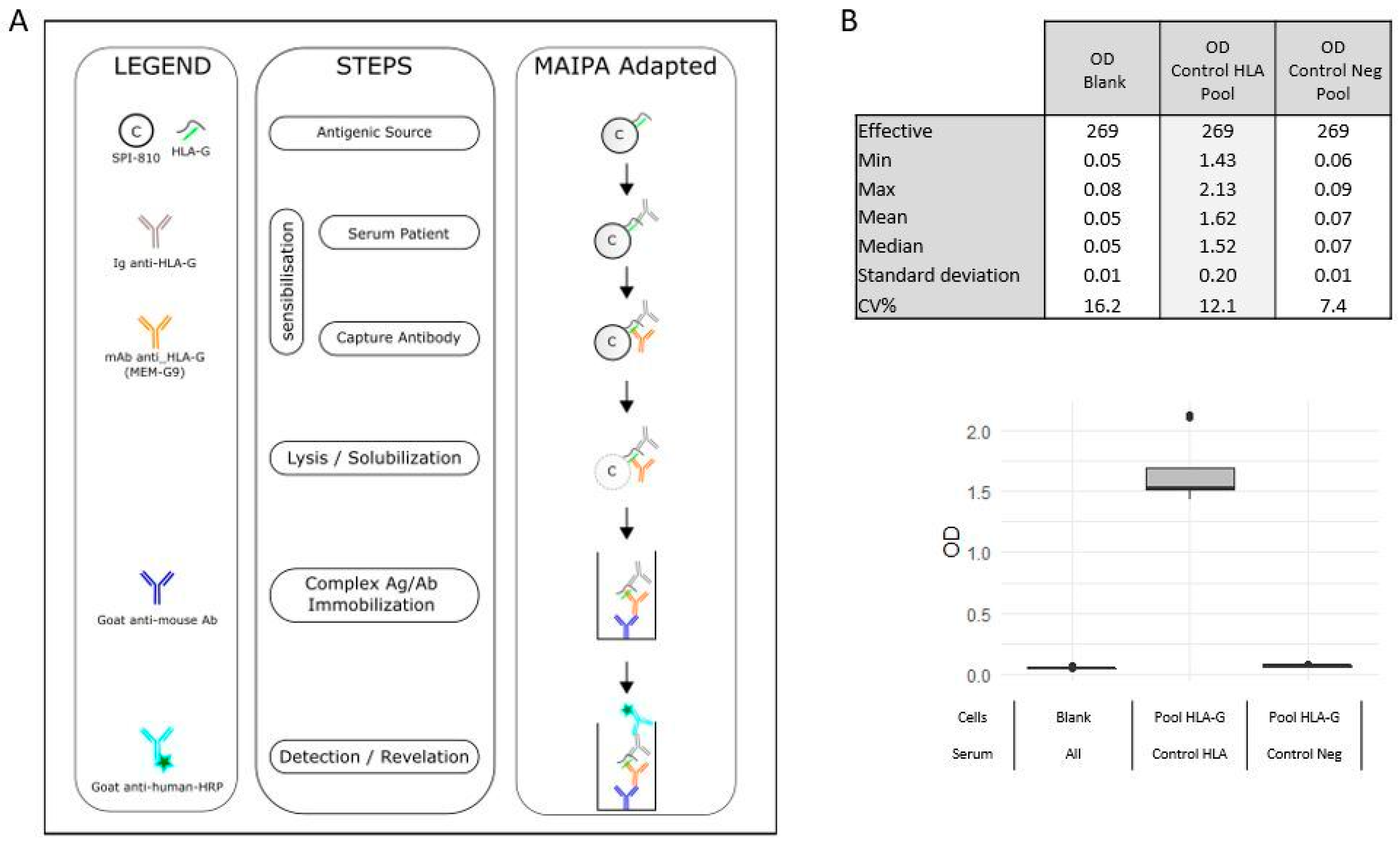
4.3. HLA-G Antibodies Detection
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tabarelli, W.; Bonatti, H.; Tabarelli, D.; Eller, M.; Müller, L.; Ruttmann, E.; Lass-Flörl, C.; Larcher, C.; Geltner, C. Long term complications following 54 consecutive lung transplants. J. Thorac. Dis. 2016, 8, 1234–1244. [Google Scholar] [CrossRef] [PubMed]
- Moreau, A.; Varey, E.; Anegon, I.; Cuturi, M.-C. Effector Mechanisms of Rejection. Cold Spring Harb. Perspect. Med. 2013, 3, a015461. [Google Scholar] [CrossRef] [PubMed]
- Banan, B.; Xu, Z.; Gunasekaran, M.; Mohanakumar, T. Role of alloimmunity and autoimmunity in allograft rejection. Clin. Transpl. 2013, 39, 325–332. [Google Scholar]
- Bharat, A.; Saini, D.; Steward, N.; Hachem, R.; Trulock, E.P.; Patterson, G.A.; Meyers, B.F.; Mohanakumar, T. Antibodies to Self-Antigens Predispose to Primary Lung Allograft Dysfunction and Chronic Rejection. Ann. Thorac. Surg. 2010, 90, 1094–1101. [Google Scholar] [CrossRef] [PubMed]
- Rao, U.; Sharma, M.; Mohanakumar, T.; Ahn, C.; Gao, A.; Kaza, V. Prevalence of antibodies to lung self-antigens (Kα1 tubulin and collagen V) and donor specific antibodies to HLA in lung transplant recipients and implications for lung transplant outcomes: Single center experience. Transpl. Immunol. 2019, 54, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Dragun, D.; Catar, R.; Philippe, A. Non-HLA antibodies in solid organ transplantation: Recent concepts and clinical relevance. Curr. Opin. Organ Transplant. 2013, 18, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, R.; Bansal, S.; Rahman, M.; Sharma, M.; Liu, W.; Bharat, A.; Hachem, R.; Omar, A.; Smith, M.A.; Mohanakumar, T. The role of donor-derived exosomes in lung allograft rejection. Hum. Immunol. 2019, 80, 588–594. [Google Scholar] [CrossRef]
- Brugière, O.; Thabut, G.; Krawice-Radanne, I.; Rizzo, R.; Dauriat, G.; Danel, C.; Suberbielle, C.; Mal, H.; Stern, M.; Schilte, C.; et al. Role of HLA-G as a Predictive Marker of Low Risk of Chronic Rejection in Lung Transplant Recipients: A Clinical Prospective Study. Am. J. Transplant. 2015, 15, 461–471. [Google Scholar] [CrossRef]
- Di Cristofaro, J.; Reynaud-Gaubert, M.; Carlini, F.; Roubertoux, P.; Loundou, A.; Basire, A.; Frassati, C.; Thomas, P.; Gomez, C.; Picard, C. HLA-G*01:04∼UTR3 Recipient Correlates With Lower Survival and Higher Frequency of Chronic Rejection After Lung Transplantation: HLA-G and LTx Outcome. Am. J. Transplant. 2015, 15, 2413–2420. [Google Scholar] [CrossRef]
- Rizzo, R.; Bortolotti, D.; Bolzani, S.; Fainardi, E. HLA-G Molecules in Autoimmune Diseases and Infections. Front. Immunol. 2014, 5, 592. [Google Scholar] [CrossRef] [PubMed]
- Morandi, F.; Pistoia, V. Interactions between HLA-G and HLA-E in Physiological and Pathological Conditions. Front. Immunol. 2014, 5, 394. [Google Scholar] [CrossRef] [PubMed]
- Grant, E.J.; Nguyen, A.T.; Lobos, C.A.; Szeto, C.; Chatzileontiadou, D.S.M.; Gras, S. The unconventional role of HLA-E: The road less traveled. Mol. Immunol. 2020, 120, 101–112. [Google Scholar] [CrossRef]
- Brugière, O.; Thabut, G.; Pretolani, M.; Krawice-Radanne, I.; Dill, C.; Herbreteau, A.; Poras, I.; Moreau, P.; Colombat, M.; Danel, C.; et al. Immunohistochemical Study of HLA-G Expression in Lung Transplant Recipients. Am. J. Transplant. 2009, 9, 1427–1438. [Google Scholar] [CrossRef]
- White, S.R.; Floreth, T.; Liao, C.; Bhorade, S.M. Association of Soluble HLA-G with Acute Rejection Episodes and Early Development of Bronchiolitis Obliterans in Lung Transplantation. PLoS ONE 2014, 9, e103643. [Google Scholar] [CrossRef]
- Carlini, F.; Traore, K.; Cherouat, N.; Roubertoux, P.; Buhler, S.; Cortey, M.; Simon, S.; Doumbo, O.; Chiaroni, J.; Picard, C.; et al. HLA-G UTR Haplotype Conservation in the Malian Population: Association with Soluble HLA-G. PLoS ONE 2013, 8, e82517. [Google Scholar] [CrossRef][Green Version]
- Castelli, E.C.; Ramalho, J.; Porto, I.O.P.; Lima, T.H.A.; Felà cio, L.P.; Sabbagh, A.; Donadi, E.A.; Mendes-Junior, C.T. Insights into HLA-G Genetics Provided by Worldwide Haplotype Diversity. Front. Immunol. 2014, 5, 476. [Google Scholar] [CrossRef]
- Rousseau, P.; Le Discorde, M.; Mouillot, G.; Marcou, C.; Carosella, E.D.; Moreau, P. The 14 bp Deletion-Insertion polymorphism in the 3′ UT region of the HLA-G gene influences HLA-G mRNA stability. Hum. Immunol. 2003, 64, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Di Cristofaro, J.; El Moujally, D.; Agnel, A.; Mazières, S.; Cortey, M.; Basire, A.; Chiaroni, J.; Picard, C. HLA-G haplotype structure shows good conservation between different populations and good correlation with high, normal and low soluble HLA-G expression. Hum. Immunol. 2013, 74, 203–206. [Google Scholar] [CrossRef]
- Jucaud, V.; Ravindranath, M.H.; Terasaki, P.I.; Morales-Buenrostro, L.E.; Hiepe, F.; Rose, T.; Biesen, R. Serum antibodies to human leucocyte antigen (HLA)-E, HLA-F and HLA-G in patients with systemic lupus erythematosus (SLE) during disease flares: Clinical relevance of HLA-F autoantibodies. Clin. Exp. Immunol. 2016, 183, 326–340. [Google Scholar] [CrossRef] [PubMed]
- Negrini, S.; Contini, P.; Pupo, F.; Greco, M.; Murdaca, G.; Puppo, F. Expression of membrane-bound human leucocyte antigen-G in systemic sclerosis and systemic lupus erythematosus. Hum. Immunol. 2020, 81, 162–167. [Google Scholar] [CrossRef]
- de Freitas Dutra, V.; Costa, T.H.; Santos, L.D.; Sirianni, M.F.M.; Aravechia, M.G.; Kutner, J.M.; Bub, C.B. Platelet antibodies identification: Comparison between two laboratory tests. Hematol. Transfus. Cell Ther. 2022, 44, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Morales-Buenrostro, L.E.; Terasaki, P.I.; Marino-Vázquez, L.A.; Lee, J.-H.; El-Awar, N.; Alberú, J. “Natural” Human Leukocyte Antigen Antibodies Found in Nonalloimmunized Healthy Males. Transplantation 2008, 86, 1111–1115. [Google Scholar] [CrossRef] [PubMed]
- Ravindranath, M.H.; Kaneku, H.; El-Awar, N.; Morales-Buenrostro, L.E.; Terasaki, P.I. Antibodies to HLA-E in Nonalloimmunized Males: Pattern of HLA-Ia Reactivity of Anti–HLA-E–Positive Sera. J. Immunol. 2010, 185, 1935–1948. [Google Scholar] [CrossRef]
- Grenzi, P.C.; de Marco, R.; Silva, R.Z.R.; Campos, É.F.; Gerbase-DeLima, M. Antibodies against denatured HLA class II molecules detected in luminex-single antigen assay. Hum. Immunol. 2013, 74, 1300–1303. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Pham, T.; Ozawa, M.; Terasaki, P.I. Antibodies to HLA-E may account for the non-donor-specific anti-HLA class-Ia antibodies in renal and liver transplant recipients. Int. Immunol. 2012, 24, 43–57. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fournel, S.; Huc, X.; Aguerre-Girr, M.; Solier, C.; Legros, M.; Praud-Brethenou, C.; Moussa, M.; Chaouat, G.; Berrebi, A.; Bensussan, A.; et al. Comparative reactivity of different HLA-G monoclonal antibodies to soluble HLA-G molecules. Tissue Antigens 2000, 55, 510–518. [Google Scholar] [CrossRef]
- Zhao, L.; Teklemariam, T.; Hantash, B.M. Reassessment of HLA-G isoform specificity of MEM-G/9 and 4H84 monoclonal antibodies. Tissue Antigens 2012, 80, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, A.; Meguro, M.; Yamazaki, R.; Watanabe, H.; Takahashi, A.; Kuroki, K.; Maenaka, K. Evaluation of the Reactivity and Receptor Competition of HLA-G Isoforms toward Available Antibodies: Implications of Structural Characteristics of HLA-G Isoforms. Int. J. Mol. Sci. 2019, 20, 5947. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Ravindranath, N.M.; Selvan, S.R.; Hilali, F.E.; Amato-Menker, C.J.; Filippone, E.J. Cell Surface B2m-Free Human Leukocyte Antigen (HLA) Monomers and Dimers: Are They Neo-HLA Class and Proto-HLA? Biomolecules 2023, 13, 1178. [Google Scholar] [CrossRef]
- Naldini, L.; Blömer, U.; Gage, F.H.; Trono, D.; Verma, I.M. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. USA 1996, 93, 11382–11388. [Google Scholar] [CrossRef]
- Hayashi, T.; Amakishi, E. Detection of anti-human platelet antibodies against integrin a2ß1 using cell lines. Blood Transfus. 2014, 12, 273–280. [Google Scholar] [CrossRef]
- Celik, A.A.; Simper, G.S.; Huyton, T.; Blasczyk, R.; Bade-Döding, C. HLA-G mediated immune regulation is impaired by a single amino acid exchange in the alpha 2 domain. Hum. Immunol. 2018, 79, 453–462. [Google Scholar] [CrossRef] [PubMed]
| Cohort | LTRs | Healthy | |
|---|---|---|---|
| LTRs | With Positive Serum | Blood | |
| Patient/donor | |||
| Number | 35 | 12 | 90 |
| Age | |||
| mean | 40 | 36.5 | 39.5 |
| min-max | 19–66 | 21–66 | 18–69 |
| <40 years | 18 | 7 | 53 |
| ≥40 years | 17 | 5 | 37 |
| Sexe | |||
| M | 17 | 5 | 41 |
| F | 18 | 7 | 49 |
| Pregnancy | ND | ND | 30 |
| First pregancy | ND | ND | |
| >4 pregnancies | ND | ND | |
| Blood Group | |||
| A | 12 | 4 | 35 |
| B | 7 | 3 | 7 |
| AB | 2 | 2 | 8 |
| O | 14 | 3 | 40 |
| Pathology | |||
| Emphysema | 7 | 3 | |
| Cystic Fibrosis | 17 | 5 | |
| Fibrosis | 9 | 3 | |
| Bronchial dysplasia | 1 | 1 | |
| Histocytosis | 1 | 0 | |
| Type of LTx | |||
| Single LTx | 9 | 3 | |
| Bilateral LTx | 26 | 9 | |
| Mismatch HLA | |||
| 0 | 5 | 2 | |
| 1–4 | 2 | 1 | |
| 5–8 | 28 | 9 | |
| CMV Status | |||
| Absence mismatch | 25 | 8 | |
| D+ and R− without seroconversion | 1 | 0 | |
| D+ and R− with seroconversion | 4 | 1 | |
| D− and R+ | 5 | 3 | |
| Type of rejection | |||
| Acute (DSA) | 7 | 2 | |
| Chronic (DSA) | 7 | 1 | |
| HLA antibodies | |||
| DSA after LTx | 26 | 8 | |
| Ab No DSA after LTx | 11 | 5 | |
| DO | 2 | 1 | |
| M1 | 5 | 2 | |
| M3 | 6 | 2 | |
| M12 | 2 | 0 | |
| HLA-G genotyping | |||
| G*01:01,*-- | 21 | 6 | |
| G*01:01,G*01:06 | 4 | 3 | |
| G*01:01,G*01:04 | 6 | 2 | |
| G*01:01,G*01:05N | 2 | 0 | |
| G*01:01,G*01:03 | 1 | 0 | |
| G*01:03,G*01:06 | 1 | 1 | |
| Women | Men | Post-Partum Women | Total | |
|---|---|---|---|---|
| Effective | 49 | 41 | 16 | 105 |
| Serum reactivity vs. Pool HLA-G cells | ||||
| Negative | 49 | 40 | 16 | 103 |
| Positive | 1 | 1 | 0 | 2 |
| Serum reactivity (screnning OD > 0.150 and S/N ratio > 1.5) vs. identification | ||||
| Negative | 0 | 1 | 0 | 1 |
| Positive | 1 | 0 | 0 | 1 |
| Collection Dates (D: Day; M: Month) | Characteristic of Pathology | Typage HLA-G | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OD | D15 | M1 | M3 | M12 | |||||||||||||||
| Patients | OD | OD Pool | OD | OD Pool | OD | OD Pool | OD | OD Pool | OD | OD Pool | Disease | Acute Rejection | Chronic Rejection | DSA | CMV Status | Type of LTx | HLA | HLA-G | Haplotypes UTR |
| WT Cells | HLA-G Cells | WT Cells | HLA-G Cells | WT Cells | HLA-G Cells | WT Cells | HLA-G Cells | WT Cells | HLA-G Cells | Mismatch | Genotyping | ||||||||
| 1 | NR | NR | 0.055 | 0.066 | 0.054 | 0.058 | 0.052 | 0.073 | 0.055 | 0.056 | Histiocytosis | Yes | D15 | 0 | bilateral | 8 | G*01:01,*-- | UTR2,UTR4 | |
| 2 | 0.099 | 0.082 | 0.059 | 0.071 | 0.071 | 0.061 | 0.085 | 0.058 | 0.07 | 0.06 | Cystic Fibrosis | 0 | bilateral | 7 | G*01:01,*-- | UTR1,UTR1 | |||
| 3 | 0.062 | 0.058 | 0.051 | 0.063 | NR | NR | 0.074 | 0.06 | 0.059 | 0.058 | Fibrosis | M1 | 0 | bilateral | 8 | G*01:01,*-- | UTR1,UTR1 | ||
| 4 | 0.115 | 0.136 | 0.154 | 0.127 | 0.115 | 0.106 | 0.06 | 0.063 | 0.084 | 0.092 | Fibrosis | M12 | D15 | 3 | bilateral | 0 | G*01:01,*-- | UTR1,UTR1 | |
| 5 | 0.149 | 0.182 | 0.064 | 0.08 | 0.09 | 0.108 | 0.076 | 0.082 | 0.071 | 0.107 | Emphysema | 0 | single | 3 | G*01:01,*-- | UTR1,UTR4 | |||
| 6 | 0.078 | 0.077 | NR | NR | 0.058 | 0.061 | 0.052 | 0.059 | 0.051 | 0.056 | Cystic fibrosis | 3 | bilateral | 8 | G*01:01,*-- | UTR1,UTR2 | |||
| 7 | 0.06 | 0.065 | 0.056 | 0.058 | 0.051 | 0.051 | 0.061 | 0.067 | 0.061 | 0.067 | Emphysema | 2 | bilateral | 6 | G*01:01,G*01:06 | UTR2,UTR6 | |||
| 8 | NR | NR | NR | NR | NR | NR | 0.068 | 0.072 | NR | NR | Cystic fibrosis | M1 | 0 | bilateral | 6 | G*01:01,G*01:04 | UTR1,UTR3 | ||
| 9 | 0.062 | 0.105 | 0.096 | 0.09 | 0.262 | 0.325 | 0.058 | 0.076 | 0.057 | 0.088 | Emphysema | D0 | 0 | bilateral | 6 | G*01:01,G*01:06 | UTR2,UTR2 | ||
| 10 | 0.645 | 0.349 | 0.064 | 0.057 | 0.167 | 0.081 | 0.074 | 0.077 | 0.072 | 0.068 | Cystic fibrosis | M3 | 0 | bilateral | 6 | G*01:01,G*01:04 | UTR3,UTR6 | ||
| 11 | 0.385 | 0.394 | 0.065 | 0.074 | NR | NR | NR | NR | NR | NR | Emphysema | D15 | 0 | bilateral | 0 | G*01:01,G*01:06 | UTR2,UTR2 | ||
| 12 | 0.05 | 0.063 | 0.061 | 0.066 | 0.059 | 0.07 | 0.061 | 0.06 | 0.052 | 0.059 | Cystic fibrosis | M1 | D15 | 0 | single | 8 | G*01:01,G*01:05N | UTR4,UTR3 | |
| 13 | 0.102 | 0.103 | 0.064 | 0.062 | 0.088 | 0.084 | 0.07 | 0.057 | 0.066 | 0.072 | Cystic fibrosis | M1 | 0 | single | 6 | G*01:01,*-- | UTR1,UTR2 | ||
| 14 | 0.35 | 0.357 | NR | NR | 0.057 | 0.06 | 0.061 | 0.059 | 0.09 | 0.068 | Fibrosis | M1 | 0 | bilateral | 8 | G*01:01,G*01:04 | UTR3,UTR7 | ||
| 15 | 0.086 | 0.078 | 0.054 | 0.057 | 0.064 | 0.06 | 0.068 | 0.064 | 0.057 | 0.078 | Cystic fibrosis | M1 | 0 | bilateral | 7 | G*01:01,*-- | UTR1,UTR2 | ||
| 16 | 0.06 | 0.06 | NR | NR | 0.29 | 0.23 | 0.06 | 0.06 | 0.06 | 0.07 | Fibrosis | 2 | single | 7 | G*01:01,*-- | UTR2,UTR6 | |||
| 17 | 0.101 | 0.088 | 0.053 | 0.102 | 0.064 | 0.076 | 0.078 | 0.085 | 0.07 | 0.07 | Fibrosis | D15 | 0 | bilateral | 4 | G*01:01,G*01:03 | UTR1, UTR2 | ||
| 18 | NR | NR | NR | NR | NR | NR | 0.057 | 0.055 | NR | NR | Cystic fibrosis | M1 | 0 | bilateral | 5 | G*01:01,*-- | UTR4,UTR2 | ||
| 19 | 0.728 | 0.564 | 0.134 | 0.104 | 0.112 | 0.094 | 0.081 | 0.073 | 0.08 | 0.074 | Cystic fibrosis | D15 | 0 | bilateral | 8 | G*01:03,G*01:06 | UTR2,UTR5 | ||
| 20 | 0.122 | 0.081 | 0.314 | 0.247 | 0.054 | 0.059 | 0.057 | 0.07 | 0.06 | 0.077 | Cystic fibrosis | 0 | single | 6 | G*01:01,*-- | UTR1,UTR6 | |||
| 21 | 0.078 | 0.068 | 0.071 | 0.058 | NR | NR | 0.051 | 0.059 | 0.07 | 0.093 | Fibrosis | M3 | Yes | M1 | 0 | bilateral | 0 | G*01:01,G*01:04 | UTR4,UTR3 |
| 22 | 0.138 | 0.143 | 0.059 | 0.054 | 0.059 | 0.063 | 0.058 | 0.07 | 0.055 | 0.075 | Emphysema | Yes | D15 | 2 | bilateral | 5 | G*01:01,*-- | UTR1,UTR2 | |
| 23 | 0.194 | 0.209 | 0.116 | 0.178 | 0.118 | 0.301 | 0.117 | 0.192 | NR | NR | Bronchial Dysplasia | Yes | D0 | 3 | bilateral | 7 | G*01:01,*-- | UTR4,UTR6 | |
| 24 | 0.076 | 0.07 | 0.058 | 0.059 | 0.134 | 0.055 | 0.058 | 0.055 | 0.065 | 0.069 | Cystic fibrosis | 0 | bilateral | 0 | G*01:01,G*01:05N | UTR1,UTR2 | |||
| 25 | 0.11 | 0.117 | 0.1 | 0.072 | 0.194 | 0.194 | 0.418 | 0.466 | 0.082 | 0.068 | Fibrosis | 0 | single | 6 | G*01:01,*-- | UTR4,UTR7 | |||
| 26 | NR | NR | 0.077 | 0.081 | NR | NR | NR | NR | 0.056 | 0.068 | cystic Fibrosis | D15 | 0 | bilateral | 6 | G*01:01,*-- | UTR1,UTR2 | ||
| 27 | 0.059 | 0.065 | 0.061 | 0.058 | 0.055 | 0.069 | 0.059 | 0.058 | 0.07 | 0.063 | Emphysema | M12 | Yes | M1 | 0 | single | 7 | G*01:01,*-- | UTR1,UTR2 |
| 28 | 0.169 | 0.208 | 0.057 | 0.059 | 0.125 | 0.074 | 0.061 | 0.074 | 0.06 | 0.063 | Cystic fibrosis | M3 | M1 | 3 | bilateral | 7 | G*01:01,*-- | UTR2,UTR4 | |
| 29 | NR | NR | NR | NR | NR | NR | 0.07 | 0.058 | NR | NR | Cystic fibrosis | 1 | bilateral | 7 | G*01:01,G*01:04 | UTR1,UTR3 | |||
| 30 | 0.073 | 0.081 | NR | NR | 0.078 | 0.096 | 0.058 | 0.067 | NR | NR | Cystic fibrosis | M3 | M1 | 2 | bilateral | 7 | G*01:01,*-- | UTR1,UTR2 | |
| 31 | 0.083 | 0.077 | NR | NR | 0.055 | 0.062 | 0.056 | 0.054 | 0.064 | 0.069 | Cystic fibrosis | Yes | M1 | 0 | bilateral | 8 | G*01:01,*-- | UTR7,UTR1 | |
| 32 | 0.07 | 0.101 | 0.067 | 0.068 | 0.094 | 0.079 | 0.139 | 0.154 | 0.071 | 0.061 | Fibrosis | D0 | 3 | single | 6 | G*01:01,G*01:04 | UTR1,UTR3 | ||
| 33 | 0.61 | 0.397 | 0.057 | 0.054 | 0.11 | 0.1 | 0.066 | 0.058 | 0.088 | 0.065 | Cystic fibrosis | M3 | M1 | 0 | bilateral | 0 | G*01:01,G*01:06 | UTR2,UTR2 | |
| 34 | NR | NR | NR | NR | 0.06 | 0.066 | NR | NR | 0.053 | 0.058 | Emphysema | M1 | 0 | single | 5 | G*01:01,*-- | UTR1,UTR7 | ||
| 35 | 0.077 | 0.1 | NR | NR | 0.076 | 0.084 | 0.063 | 0.076 | 0.08 | 0.131 | Fibrosis | Yes | M1 | 0 | bilateral | 7 | G*01:01,*-- | UTR1, UTR1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedini, P.; Hubert, L.; Carlini, F.; Baudey, J.B.; Tous, A.; Jordier, F.; Basire, A.; Bagnis, C.; Reynaud-Gaubert, M.; Coiffard, B.; et al. Low Prevalence of HLA-G Antibodies in Lung Transplant Patients Detected using MAIPA-Adapted Protocol. Int. J. Mol. Sci. 2023, 24, 16479. https://doi.org/10.3390/ijms242216479
Pedini P, Hubert L, Carlini F, Baudey JB, Tous A, Jordier F, Basire A, Bagnis C, Reynaud-Gaubert M, Coiffard B, et al. Low Prevalence of HLA-G Antibodies in Lung Transplant Patients Detected using MAIPA-Adapted Protocol. International Journal of Molecular Sciences. 2023; 24(22):16479. https://doi.org/10.3390/ijms242216479
Chicago/Turabian StylePedini, Pascal, Lucas Hubert, Federico Carlini, Jean Baptiste Baudey, Audrey Tous, Francois Jordier, Agnès Basire, Claude Bagnis, Martine Reynaud-Gaubert, Benjamin Coiffard, and et al. 2023. "Low Prevalence of HLA-G Antibodies in Lung Transplant Patients Detected using MAIPA-Adapted Protocol" International Journal of Molecular Sciences 24, no. 22: 16479. https://doi.org/10.3390/ijms242216479
APA StylePedini, P., Hubert, L., Carlini, F., Baudey, J. B., Tous, A., Jordier, F., Basire, A., Bagnis, C., Reynaud-Gaubert, M., Coiffard, B., Chiaroni, J., Silvy, M., & Picard, C. (2023). Low Prevalence of HLA-G Antibodies in Lung Transplant Patients Detected using MAIPA-Adapted Protocol. International Journal of Molecular Sciences, 24(22), 16479. https://doi.org/10.3390/ijms242216479







