Nanoscale Bilirubin Analysis in Translational Research and Precision Medicine by the Recombinant Protein HUG
Abstract
:1. Introduction
2. Results
2.1. Preparation and Quality Controls of Standard Bilirubin Solutions
2.1.1. Principles
2.1.2. Validation of Bilirubin Concentrations in Standard Solutions
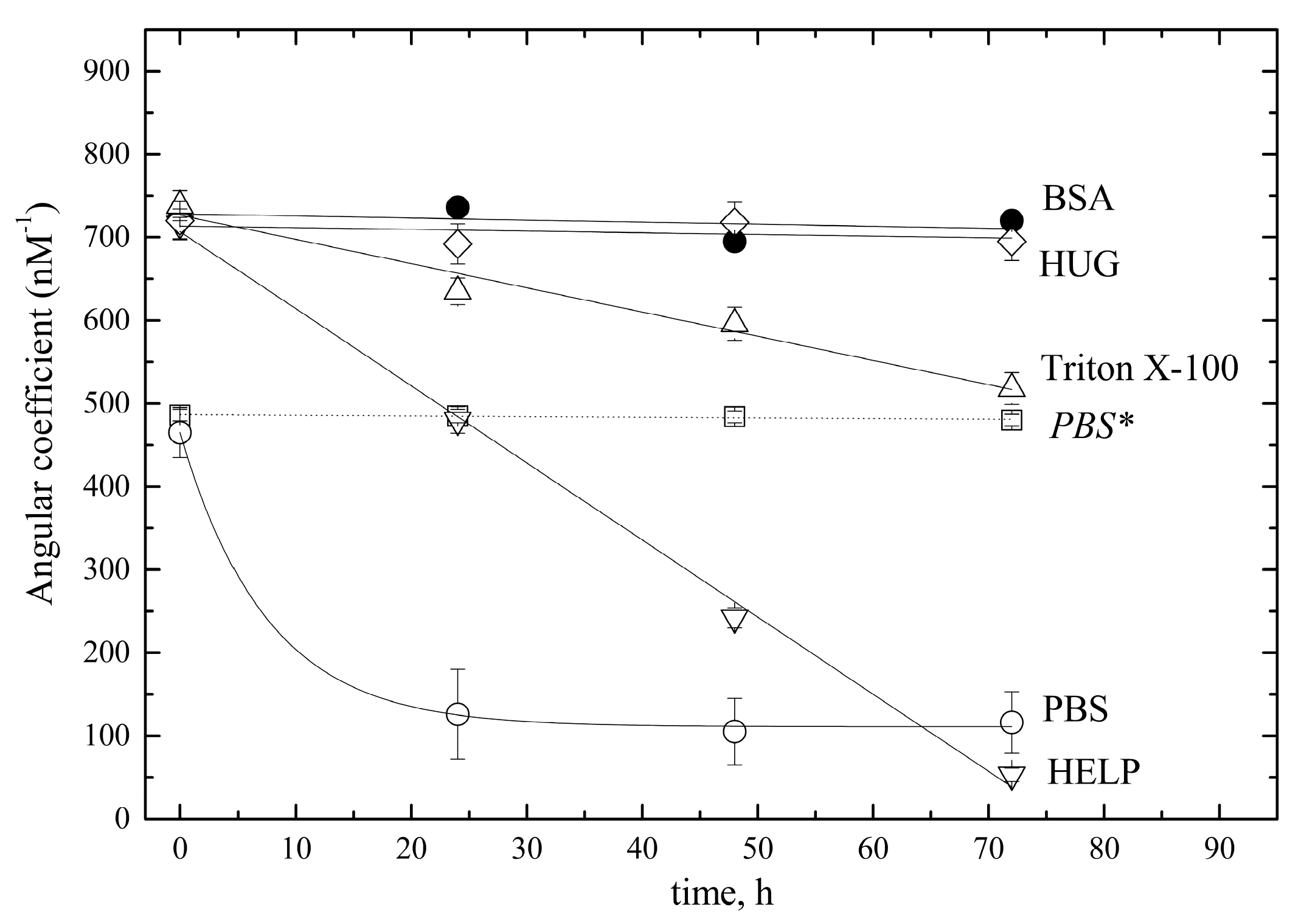
2.1.3. Stability of Bilirubin Standard Solutions
- Stability of the 5 mM stock solution
- Stability of 10 µM pre-calibrator solutions
2.2. Nanoscale HUG-Based Fluorometric Assay of Bilirubin
2.2.1. Basic Requirements
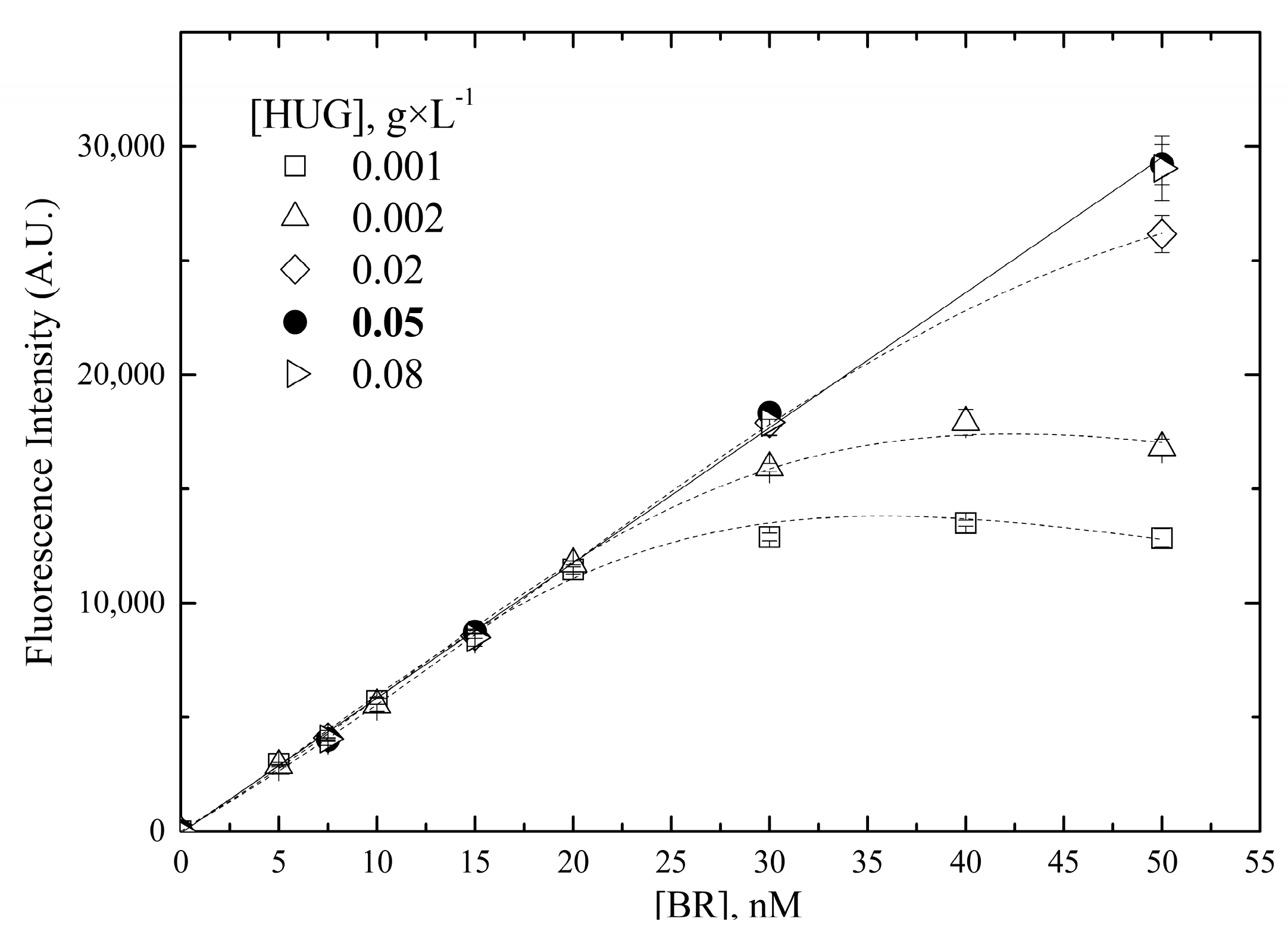
- Concentration of HUG
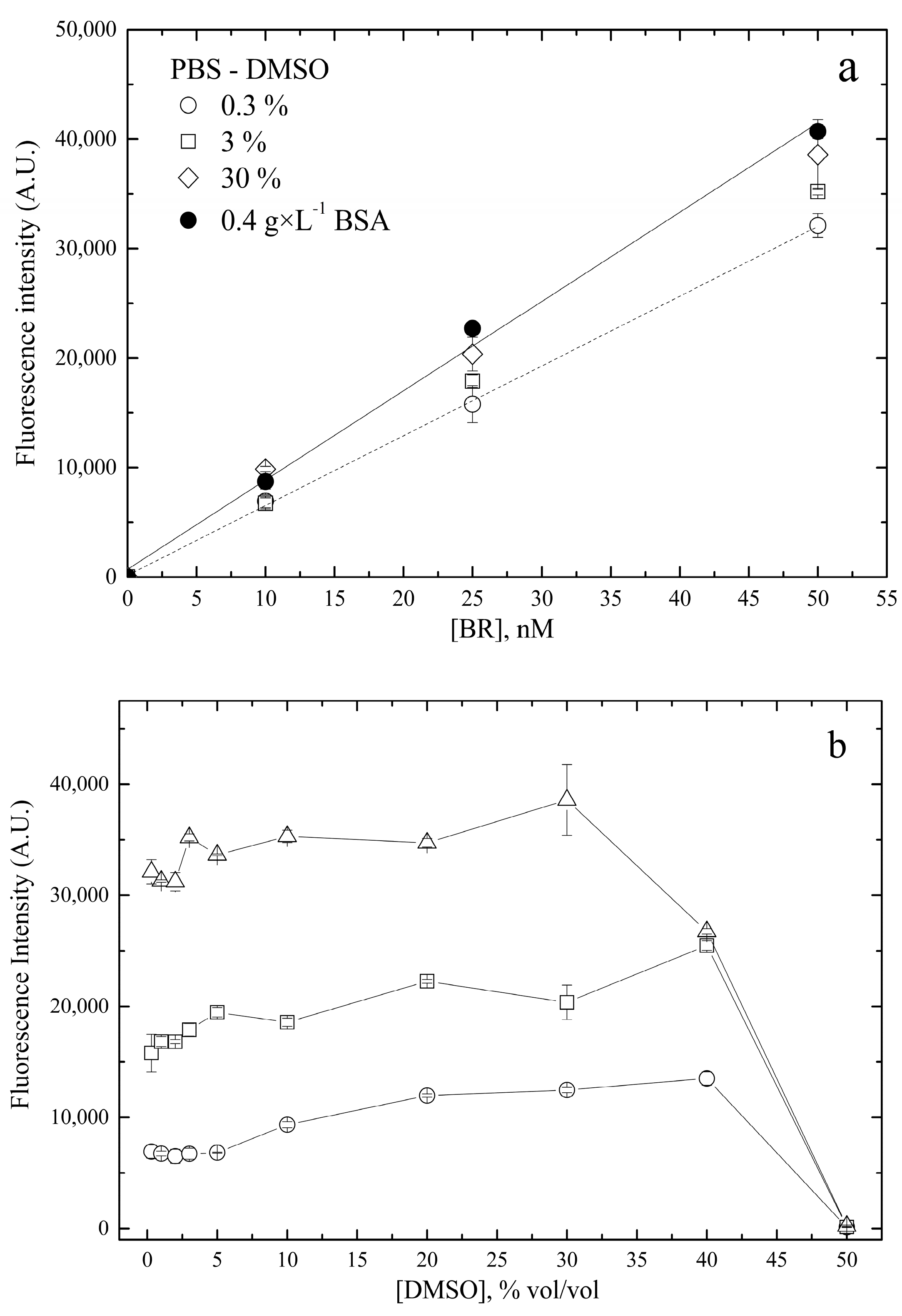
- Concentration of BSA
2.2.2. Linear Range of the Fluorometric Assay
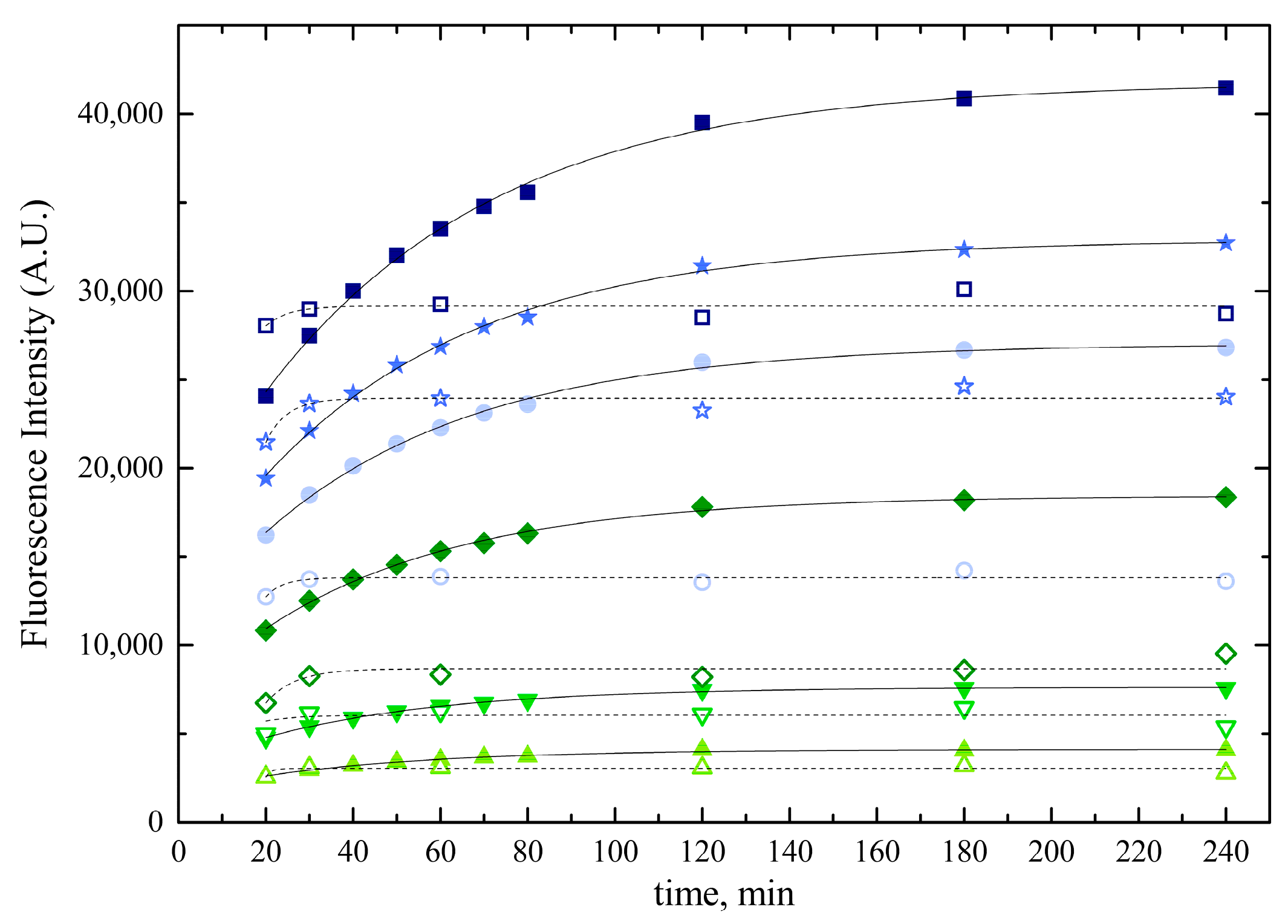
2.2.3. Optimal Reaction Time and Temperature
2.2.4. Limits of Nanoscale Bilirubin Detection and Quantitation
2.2.5. Solubility and Stability of Nanoscale Bilirubin Solutions
2.2.6. Accuracy and Precision
2.2.7. Robustness
2.3. Direct Analysis of Bilirubin in Human Plasma by the HUG Assay
3. Discussion
3.1. Advantages and Limitations of the HUG Assay
3.2. Domains of Application in Precision Medicine
4. Materials and Methods
4.1. Materials
4.2. Standard Bilirubin Solutions
4.2.1. Stock Solution
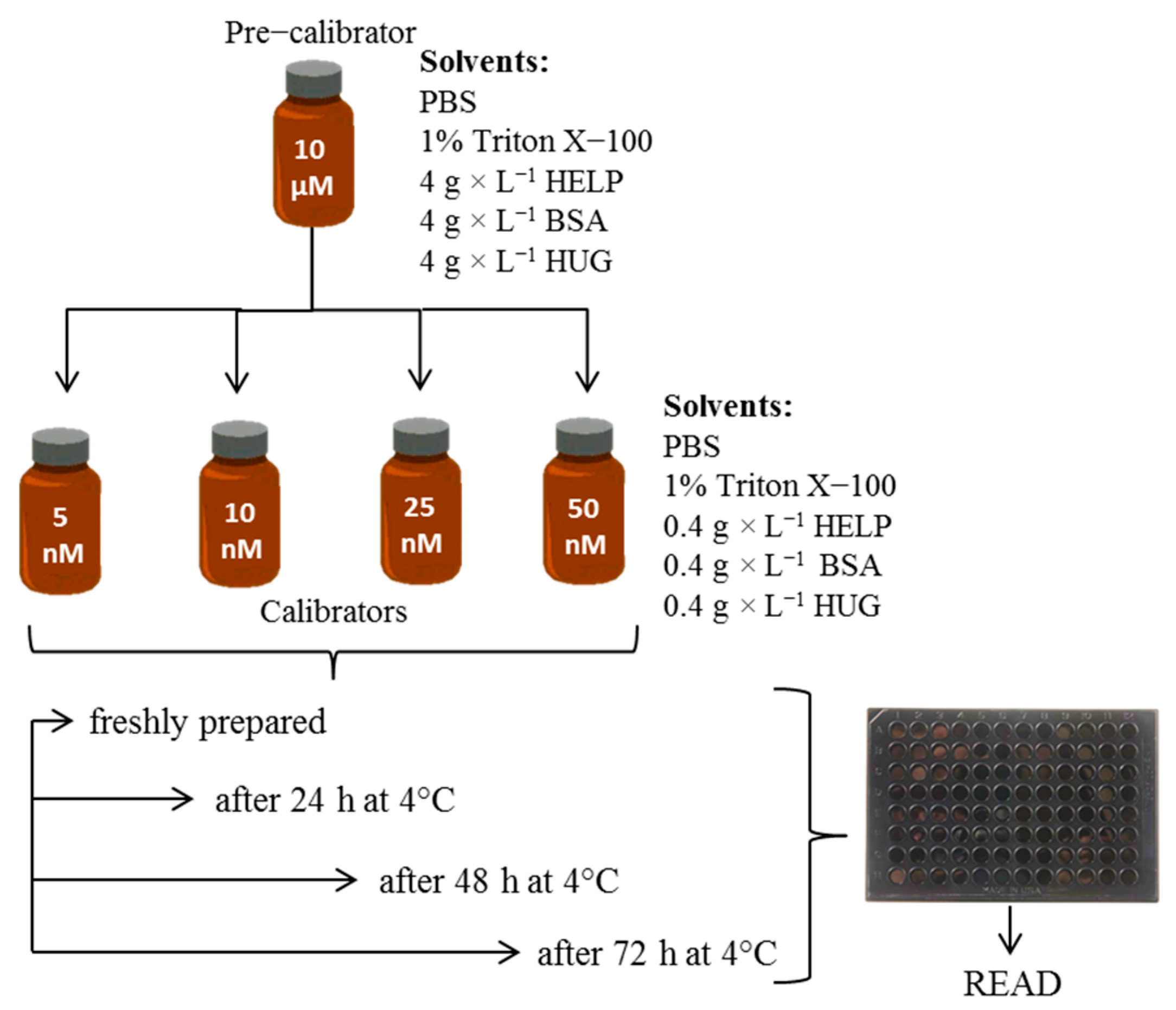
4.2.2. Pre-Calibrator Solutions
4.2.3. Calibrator Solutions
4.3. Spectrophotometric Measurements of Bilirubin Standard Solutions
4.4. Colorimetric Measurements of Bilirubin Standard Solutions
4.5. Fluorometric Measurements of Bilirubin Standard Solutions
4.6. Study Subjects and Ethics Statement
4.7. Bilirubin Fluorometric Measurements in Human Plasma Samples
4.8. DNA Extraction and Genotyping
4.9. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A

| Nominal Concentration (NC, nM) | Determined Concentration (DC, nM) | Relative Error, % | ||
|---|---|---|---|---|
| Day 1 | Day 2 | Day 1 | Day 2 | |
| 5 | 4.97 ± 0.10 | 5.19 ± 0.16 | 1 | −4 |
| 5 | 5.27 ± 0.11 | 5.16 ± 0.29 | −5 | −5 |
| 10 | 10.16 ± 0.34 | 10.83 ± 0.41 | −2 | −8 |
| 10 | 10.27 ± 0.11 | 10.85 ± 0.15 | −3 | −8 |
| 25 | 23.53 ± 0.48 | 27.32 ± 0.25 | 6 | −9 |
| 25 | 25.21 ± 0.20 | 25.45 ± 0.20 | −1 | −2 |
| 50 | 47.60 ± 0.22 | 49.14 ± 0.40 | 5 | 2 |
| 50 | 45.84 ± 0.38 | 50.27 ± 0.35 | 8 | −1 |
| Nominal Concentration (NC, nM) | Determined Concentration (DC ± sd, nM) | Coefficient of Variation (%) | ||
|---|---|---|---|---|
| Day 1 | Day 2 | Day 1 | Day 2 | |
| 5 | 6.32 ± 0.37 | 4.83 ± 0.08 | 5.8 | 1.7 |
| 10 | 11.08 ± 0.46 | 10.07 ± 0.23 | 4.1 | 2.3 |
| 25 | 25.81 ± 0.49 | 24.92 ± 1.08 | 1.9 | 4.3 |
| 50 | 51.26 ± 1.21 | 50.10 ± 1.42 | 2.4 | 2.8 |
| Sample | No Spike (0 nM) | Low Spike (5 nM) | Medium Spike (20 nM) | High Spike (50 nM) | |
|---|---|---|---|---|---|
| BSA 4 g × L−1 | 0 | 5.7 | 19.6 | 45.3 | |
| 1 | 11.8 | 6.3 | 18.3 | 38.2 | |
| 2 | 23.9 | 7.9 | 21.1 | 42.9 | |
| 3 | 10.4 | 4.5 | 19.1 | 36.7 | |
| 4 | 18.0 | 6.2 | 16.0 | 40.3 | |
| 5 | 17.1 | 5.3 | 17.5 | 41.4 | |
| 6 | 19.4 | 5.8 | 16.1 | 41.2 | |
| 7 | 28.0 | 4.9 | 17.3 | 45.1 | |
| Sample (n) | Spike Level | Expected | Observed | Recovery % | p-value |
| Plasma (7) | Low (5 nM) | 5.4 (±0.5) | 5.3 (±0.8) | 99.6 | p > 0.999 |
| Med (20 nM) | 19.7 (±5.2) | 17.9 (±1.8) | 91.1 | p = 0.620 | |
| High (50 nM) | 44.8 (±3.4) | 41.5 (±2.3) | 92.9 | p = 0.170 |
| Key Features | Results | ||||
|---|---|---|---|---|---|
| 1 | A priori, establish the acceptable limit of agreement | The limit of agreement should be <LoQ of the diazo method, which is the benchmark method. | |||
| 2 | The data structure | Paired measurements, each of them in four replicates; each measurement was in different subject. Data were obtained from a random sample of 36 human subjects. There was no prior knowledge of their total bilirubinemia, which was measured for the first time in this study by two methods. | |||
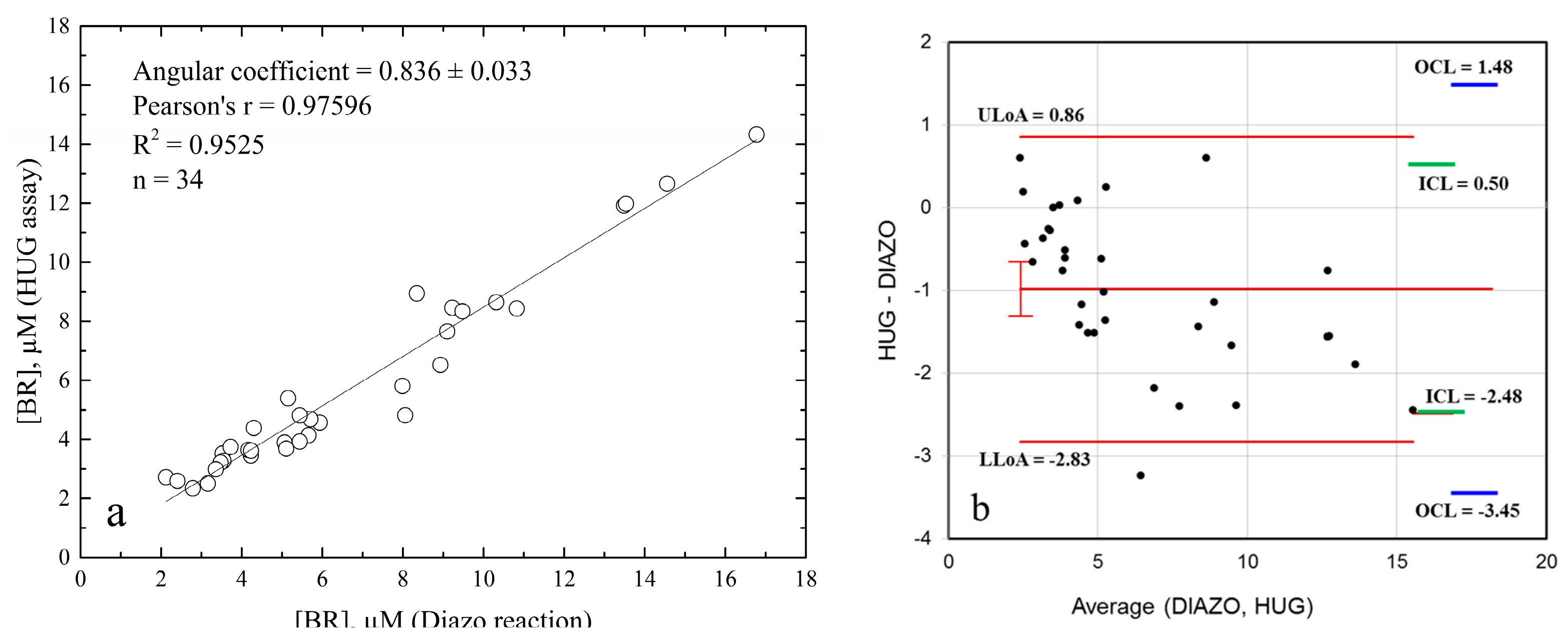
| 3 | Estimate the repeatability of the measures | HUG assay: RSD = 1%, n = 34; Diazo reaction: RSD = 5.3%, n = 34; Relative standard deviations of technical replicates. | |||
| 4 | Plot the data and inspect for absence of trend and constant variance | 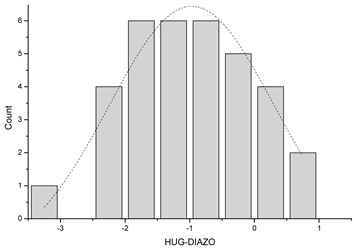 The dotted line represents Gaussian distribution. Shapiro–Wilk test for normal distribution accepted normality (p = 0.878). | |||
| 5 | If necessary, transform the data | Not needed. | |||
| 6 | Plot and report numerically the mean of the differences (bias) | −0.99 units (µM) | |||
| 7 | Give an estimate of the precision | The mean of the differences is −0.986; The standard deviation is 0.940. | |||
| 8 | Calculate and indicate the limits of agreement (LoAs) of the differences | Upper LoA = 0.86; Lower LoA = −2.83. | |||
| 9 | Provide an estimate of precision | 95% CI for mean of differences from −1.314 to −0.657. | |||
| 10 | Ensure that the range of the mean values is sufficient | The range of the mean total bilirubinemia values in our study group was 2.35–14.33 µM (HUG assay) or 2.12–16.78 µM (diazo assay), comprising the reference intervals of bilirubin concentrations in human sera [43], as well as the min–max range (1.5–22.6 µM) in the 226 subjects that were analyzed for group difference. | |||
| 11 | Variance between and within subjects | Inner confidence limits: there is a 2.5% probability that 95% of the differences lie between 0.518 and −2489 units (µM); Outer confidence limits: there is a 97.5% probability that 95% of the differences lie between 1.48 and −3.45 units (µM); Variance within subjects was RSD = 1% (HUG assay) and RSD = 5.3% (diazo reaction). | |||
| 12 | Software or computing processes used | Worksheet calculators attached to Carkeet’s ‘A Review of the Use of Confidence Intervals for Bland–Altman Limits of Agreement’, Optometry and Vision Science, January 2020. | |||
| 13 | Statistical assumptions made, such as normality of the data | No. | Statistic (W) | p-value | Decision at level (5%) |
| 34 | 0.976 | 0.651 | Cannot reject normality | ||
| At the 0.05 level, the data were significantly drawn from a normally distributed population. | |||||
| SNP | Genotype | HWE | N (%) | Median (25–75%) | p-Value |
|---|---|---|---|---|---|
| BLVRA rs699512 | AA | 0.050 | 147 (65.6) | 5.53 (4.02–7.83) | Ref. |
| GA | 63 (28.1) | 4.94 (3.54–6.84) | 0.191 | ||
| GG | 14 (6.3) | 5.65 (5.18–6.56) | 0.815 | ||
| GA + GG | 77 (34.4) | 5.42 (4.04–6.70) | 0.288 | ||
| UGT1A1 rs8175347 * | –– | 0.823 | 85 (38.3) | 4.57 (3.77–5.98) | Ref. |
| TA– | 106 (47.7) | 5.59 (4.02–7.36) | 0.008 | ||
| TATA | 31 (14.0) | 9.38 (5.87–13.71) | <0.001 | ||
| TA– + TATA | 137 (61.7) | 6.02 (4.31–8.07) | <0.001 | ||
| HMOX1 rs2071746 | AA | 0.056 | 73 (32.6) | 5.23 (3.77–7.66) | Ref. |
| TA | 97 (43.3) | 5.61 (4.28–7.77) | 0.195 | ||
| TT | 54 (24.1) | 5.31 (4.03–6.98) | 0.755 | ||
| TA + TT | 151 (67.4) | 5.57 (4.14–7.41) | 0.295 | ||
| HMOX1 rs2071747 | GG | 0.122 | 210 (93.8) | 5.43 (3.97–7.47) 6.02 (4.42–7.23) | Ref. |
| GC | 13 (5.8) | 6.02 (4.42–7.23) | 0.518 | ||
| CC | 1 (0.4) | 2.37 (2.37–2.37) | 0.133 | ||
| GC + CC | 14 (6,3) | 5.44 (4.34–7.13) | 0.816 | ||
| HMOX2 rs2270363 ** | GG | 0.161 | 129 (57.8) | 5.37 (3.95–7.33) | Ref. |
| GA | 76 (34.1) | 5.40 (4.02–7.53) | 0.956 | ||
| AA | 18 (8.1) | 5.83 (4.35–7.83) | 0.520 | ||
| GA + AA | 94 (42,2) | 5.57 (4.05–7.63) | 0.782 | ||
| HMOX2 rs1051308 | AA | 0.346 | 113 (50.4) | 5.53 (3.95–7.75) | Ref. |
| AG | 88 (39.3) | 5.26 (4.02–7.23) | 0.793 | ||
| GG | 23 (10.3) | 5.53 (4.24–7.41) | 0.940 | ||
| AG + GG | 111 (49,6) | 5.26 (4.02–7.28) | 0.845 |
References
- Li, C.; Stocker, R. Heme oxygenase and iron: From bacteria to humans. Redox Rep. 2009, 14, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Tift, M.S.; Alves de Souza, R.W.; Weber, J.; Heinrich, E.C.; Villafuerte, F.C.; Malhotra, A.; Otterbein, L.E.; Simonson, T.S. Adaptive Potential of the Heme Oxygenase/Carbon Monoxide Pathway During Hypoxia. Front. Physiol. 2020, 11, 886. [Google Scholar] [CrossRef] [PubMed]
- Asad, S.F.; Singh, S.; Ahmad, A.; Khan, N.U.; Hadi, S.M. Prooxidant and antioxidant activities of bilirubin and its metabolic precursor biliverdin: A structure–activity study. Chem. Biol. Interact. 2001, 137, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Ryter, S.W. Heme oxygenase-1: An anti-inflammatory effector in cardiovascular, lung, and related metabolic disorders. Antioxidants 2022, 11, 555. [Google Scholar] [CrossRef] [PubMed]
- Campbell, N.K.; Fitzgerald, H.K.; Dunne, A. Regulation of inflammation by the antioxidant haem oxygenase 1. Nat. Rev. Immunol. 2021, 21, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Lightner, D.A.; Holmes, D.L.; McDonagh, A.F. On the acid dissociation constants of bilirubin and biliverdin. pKa values from 13C NMR spectroscopy. J. Biol. Chem. 1996, 271, 2397–2405. [Google Scholar] [CrossRef]
- Zucker, S.D.; Goessling, W.; Hoppin, A.G. Unconjugated bilirubin exhibits spontaneous diffusion through model lipid bilayers and native hepatocyte membranes. J. Biol. Chem. 1999, 274, 10852–10862. [Google Scholar] [CrossRef]
- Brodersen, R. Bilirubin. Solubility and interaction with albumin and phospholipid. J. Biol. Chem. 1979, 254, 2364–2369. [Google Scholar] [CrossRef]
- Hahm, J.S.; Ostrow, J.D.; Mukerjee, P.; Celic, L. Ionization and self-association of unconjugated bilirubin, determined by rapid solvent partition from chloroform, with further studies of bilirubin solubility. J. Lipid Res. 1992, 33, 1123–1137. [Google Scholar] [CrossRef]
- Overbeek, J.T.G.; Vink, C.L.J.; Deenstra, H. The solubility of bilirubin. Recl. Trav. Chim. Pays-Bas 1955, 74, 81–84. [Google Scholar] [CrossRef]
- Fog, J.; Bugge-Asperheim, B. Stability of bilirubin. Nature 1964, 203, 756–757. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, A.F. Photolysis and photoisomerization of bilirubin in serum specimens exposed to room lighting. Clin. Chim. Acta 2008, 393, 130. [Google Scholar] [CrossRef] [PubMed]
- Peters, T., Jr. Serum albumin. Adv. Protein Chem. 1985, 37, 161–245. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, A.; Pedersen, A.O.; Brodersen, R. Spectroscopic properties of bilirubin-human serum albumin complexes: A stoichiometric analysis. Arch. Biochem. Biophys. 1986, 244, 273–284. [Google Scholar] [CrossRef]
- Goncharova, I.; Orlov, S.; Urbanová, M. The location of the high-and low-affinity bilirubin-binding sites on serum albumin: Ligand-competition analysis investigated by circular dichroism. Biophys. Chem. 2013, 180–181, 55–65. [Google Scholar] [CrossRef]
- Sofronescu, A.G.; Loebs, T.; Zhu, Y. Effects of temperature and light on the stability of bilirubin in plasma samples. Clin. Chim. Acta 2012, 413, 463–466. [Google Scholar] [CrossRef]
- Hulzebos, C.V.; Vitek, L.; Coda Zabetta, C.D.; Dvořák, A.; Schenk, P.; van der Hagen, E.A.E.; Cobbaert, C.; Tiribelli, C. Diagnostic methods for neonatal hyperbilirubinemia: Benefits, limitations, requirements, and novel developments. Pediatr. Res. 2021, 90, 277–283. [Google Scholar] [CrossRef]
- Ngashangva, L.; Bachu, V.; Goswami, P. Development of new methods for determination of bilirubin. J. Pharm. Biomed. Anal. 2019, 162, 272–285. [Google Scholar] [CrossRef]
- Rawal, R.; Kharangarh, P.R.; Dawra, S.; Tomar, M.; Gupta, V.; Pundir, C.S. A comprehensive review of bilirubin determination methods with special emphasis on biosensors. Process Biochem. 2020, 89, 165–174. [Google Scholar] [CrossRef]
- Narwal, V.; Batra, B.; Kalra, V.; Jalandra, R.; Ahlawat, J.; Hooda, R.; Sharma, M.; Rana, J.S. Bilirubin detection by different methods with special emphasis on biosensing: A review. Sens. Bio-Sens. Res. 2021, 33, 100436. [Google Scholar] [CrossRef]
- Guo, Y.; Wei, C.; Wang, R.; Xiang, Y. Nanomaterials for fluorescent assay of bilirubin. Anal. Biochem. 2023, 666, 115078. [Google Scholar] [CrossRef] [PubMed]
- Iwatani, S.; Nakamura, H.; Kurokawa, D.; Yamana, K.; Nishida, K.; Fukushima, S.; Koda, T.; Nishimura, N.; Nishio, H.; Iijima, K.; et al. Fluorescent protein-based detection of unconjugated bilirubin in newborn serum. Sci. Rep. 2016, 6, 28489. [Google Scholar] [CrossRef] [PubMed]
- Bandiera, A.; Corich, L.; Tommasi, S.; De Bortoli, M.; Pelizzo, P.; Stebel, M.; Paladin, D.; Passamonti, S. Human elastin-like polypeptides as a versatile platform for exploitation of ultrasensitive bilirubin detection by UnaG. Biotechnol. Bioeng. 2020, 117, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Sist, P.; Bandiera, A.; Urbani, R.; Passamonti, S. Macromolecular and Solution Properties of the Recombinant Fusion Protein HUG. Biomacromolecules 2022, 23, 3336–3348. [Google Scholar] [CrossRef] [PubMed]
- Klauke, R.; Kytzia, H.-J.; Weber, F.; Grote-Koska, D.; Brand, K.; Schumann, G. Reference measurement procedure for total bilirubin in serum re-evaluated and measurement uncertainty determined. Clin. Chim. Acta 2018, 481, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Faerch, T.; Jacobsen, J. Determination of association and dissociation rate constants for bilirubin-bovine serum albumin. Arch. Biochem. Biophys. 1975, 168, 351–357. [Google Scholar] [CrossRef]
- Williams, K.R.; Adhyaru, B.; Pierce, R.E.; Schulman, S.G. The Binding Constant for Complexation of Bilirubin to Bovine Serum Albumin. An Experiment for the Biophysical Chemistry Laboratory. J. Chem. Educ. 2002, 79, 115. [Google Scholar] [CrossRef]
- Lee, K.S.; Gartner, L.M. Spectrophotometric characteristics of bilirubin. Pediatr. Res. 1976, 10, 782–788. [Google Scholar] [CrossRef]
- Levitt, D.G.; Levitt, M.D. Quantitative assessment of the multiple processes responsible for bilirubin homeostasis in health and disease. Clin. Exp. Gastroenterol. 2014, 7, 307–328. [Google Scholar] [CrossRef]
- Dekker, D.; Dorresteijn, M.J.; Welzen, M.E.B.; Timman, S.; Pickkers, P.; Burger, D.M.; Smits, P.; Wagener, F.A.D.T.G.; Russel, F.G.M. Parenteral bilirubin in healthy volunteers: A reintroduction in translational research. Br. J. Clin. Pharmacol. 2018, 84, 268–279. [Google Scholar] [CrossRef]
- Ederveen, J. A practical approach to biological assay validation. Hoofddorp Prog. 2010, 106, 08090. [Google Scholar]
- ICH Harmonised Guideline. Bioanalytical Method Validation and Study Sample Analysis M10; ICH Harmonised Guideline: Geneva, Switzerland, 2022. [Google Scholar]
- ICH Harmonised Tripartite Guideline. Note for Guidance on Validation of Analytical Procedures: Text and Methodology (CPMP/ICH/381/95); European Medicines Agency: Amsterdam, The Netherlands, 1995. [Google Scholar]
- Kumagai, A.; Ando, R.; Miyatake, H.; Greimel, P.; Kobayashi, T.; Hirabayashi, Y.; Shimogori, T.; Miyawaki, A. A bilirubin-inducible fluorescent protein from eel muscle. Cell 2013, 153, 1602–1611. [Google Scholar] [CrossRef] [PubMed]
- Karnes, H.T.; March, C. Precision, accuracy, and data acceptance criteria in biopharmaceutical analysis. Pharm. Res. 1993, 10, 1420–1426. [Google Scholar] [CrossRef] [PubMed]
- U.S. Department of Health and Human Services; Food and Drug Administration. Bioanalytical Method Validation: Guidance for Industry; Food and Drug Administration: Rockville, MD, USA, 2018.
- Marconi, V.C.; Duncan, M.S.; So-Armah, K.; Lo Re, V., 3rd; Lim, J.K.; Butt, A.A.; Goetz, M.B.; Rodriguez-Barradas, M.C.; Alcorn, C.W.; Lennox, J.; et al. Bilirubin is inversely associated with cardiovascular disease among HIV-positive and HIV-negative individuals in VACS (Veterans Aging Cohort Study). J. Am. Heart Assoc. 2018, 7, e007792. [Google Scholar] [CrossRef]
- Vitek, L.; Bellarosa, C.; Tiribelli, C. Induction of Mild Hyperbilirubinemia: Hype or Real Therapeutic Opportunity? Clin. Pharmacol. Ther. 2018, 106, 568–575. [Google Scholar] [CrossRef]
- Lampe, J.W.; Bigler, J.; Horner, N.K.; Potter, J.D. UDP-glucuronosyltransferase (UGT1A1*28 and UGT1A6*2) polymorphisms in Caucasians and Asians: Relationships to serum bilirubin concentrations. Pharmacogenetics 1999, 9, 341–349. [Google Scholar] [CrossRef]
- Bandiera, A.; Sist, P.; Urbani, R. Comparison of thermal behavior of two recombinantly expressed human elastin-like polypeptides for cell culture applications. Biomacromolecules 2010, 11, 3256–3265. [Google Scholar] [CrossRef]
- Shitashima, Y.; Shimozawa, T.; Kumagai, A.; Miyawaki, A.; Asahi, T. Two distinct fluorescence states of the ligand-induced green fluorescent protein UnaG. Biophys. J. 2017, 113, 2805–2814. [Google Scholar] [CrossRef]
- Tonelotto, V.; Smith, K.F.; Slater, K.; O’Reilly, E.; Dillon, E.T.; Tramer, F.; Passamonti, S.; Vanella, L.; Costa-García, M.; Bosch, R.; et al. 1,4-dihydroxy Quininib Activates Ferroptosis Pathways in OMM2.5 cells: Implications for Treatment of Metastatic Uveal Melanoma. Investig. Ophthalmol. Vis. Sci. 2023, 64, 1254. [Google Scholar]
- Pelizzo, P.; Stebel, M.; Medic, N.; Sist, P.; Vanzo, A.; Anesi, A.; Vrhovsek, U.; Tramer, F.; Passamonti, S. Cyanidin 3-glucoside targets a hepatic bilirubin transporter in rats. Biomed. Pharmacother. 2023, 157, 114044. [Google Scholar] [CrossRef]
- Vítek, L. Bilirubin as a predictor of diseases of civilization. Is it time to establish decision limits for serum bilirubin concentrations? Arch. Biochem. Biophys. 2019, 672, 108062. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.H.; Scholtz, H.E.; Schall, R.; Muller, F.O.; Hundt, H.K.; Maree, J.S. The effect of fasting on total serum bilirubin concentrations. Br. J. Clin. Pharmacol. 1995, 39, 169–171. [Google Scholar] [CrossRef] [PubMed]
- Andersson, C.; Weeke, P.; Fosbøl, E.L.; Brendorp, B.; Køber, L.; Coutinho, W.; Sharma, A.M.; Van Gaal, L.; Finer, N.; James, W.P.T.; et al. Acute effect of weight loss on levels of total bilirubin in obese, cardiovascular high-risk patients: An analysis from the lead-in period of the Sibutramine Cardiovascular Outcome trial. Metabolism 2009, 58, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- McCarty, M.F. “Iatrogenic Gilbert syndrome”—A strategy for reducing vascular and cancer risk by increasing plasma unconjugated bilirubin. Med. Hypotheses 2007, 69, 974–994. [Google Scholar] [CrossRef]
- Vitek, L.; Hinds, T.D.; Stec, D.E.; Tiribelli, C. The physiology of bilirubin: Health and disease equilibrium. Trends Mol. Med. 2023, 29, 315–328. [Google Scholar] [CrossRef]
- Sist, P.; Tramer, F.; Urbani, R.; Bandiera, A.; Passamonti, S. Preparation of Bilirubin Standard Solutions for Assay Calibration. Res. Sq. 2022; preprint. [Google Scholar] [CrossRef]
- Redenšek, S.; Flisar, D.; Kojović, M.; Gregorič Kramberger, M.; Georgiev, D.; Pirtošek, Z.; Trošt, M.; Dolžan, V. Dopaminergic pathway genes influence adverse events related to dopaminergic treatment in Parkinson’s disease. Front. Pharmacol. 2019, 10, 8. [Google Scholar] [CrossRef]
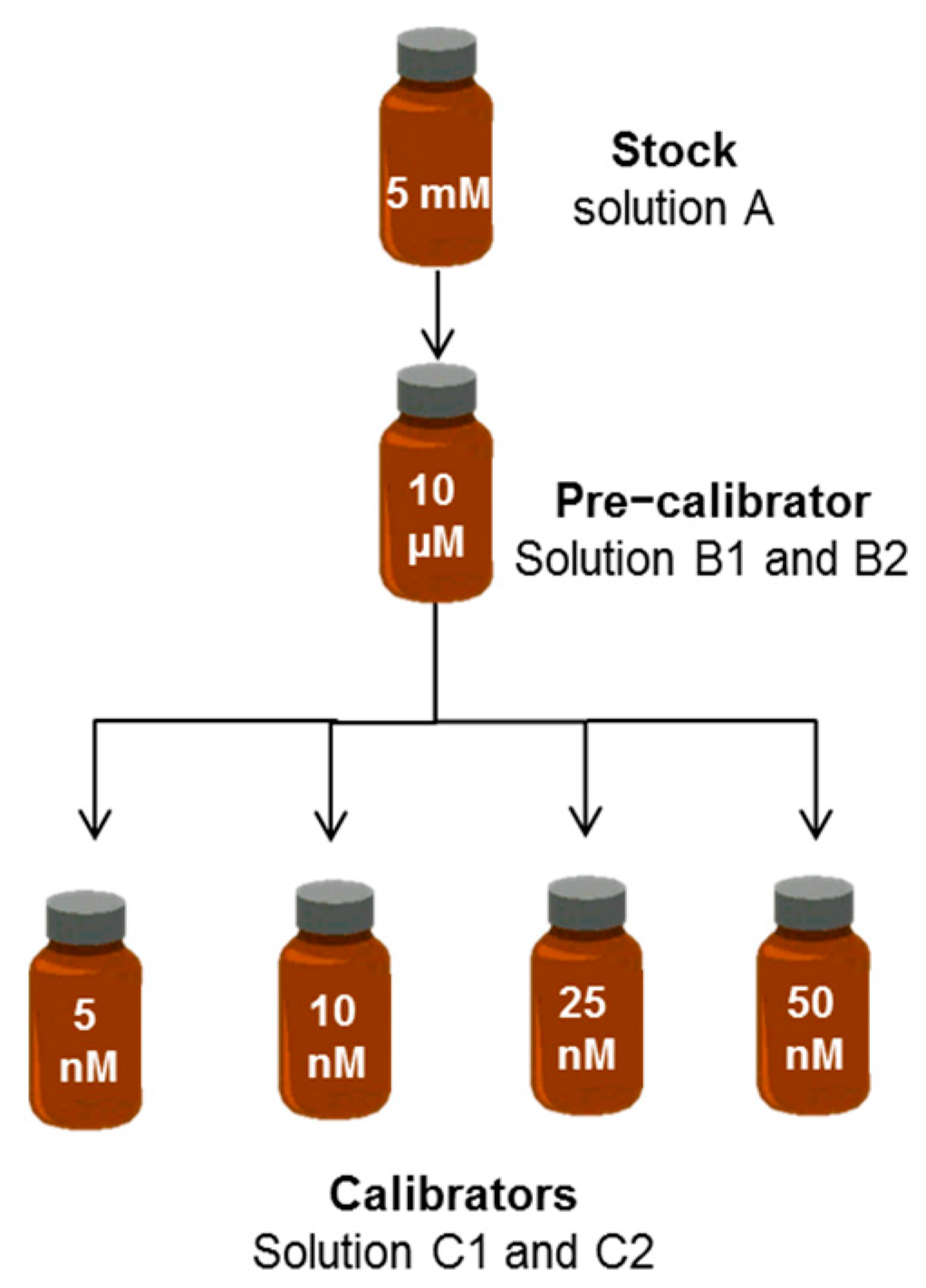


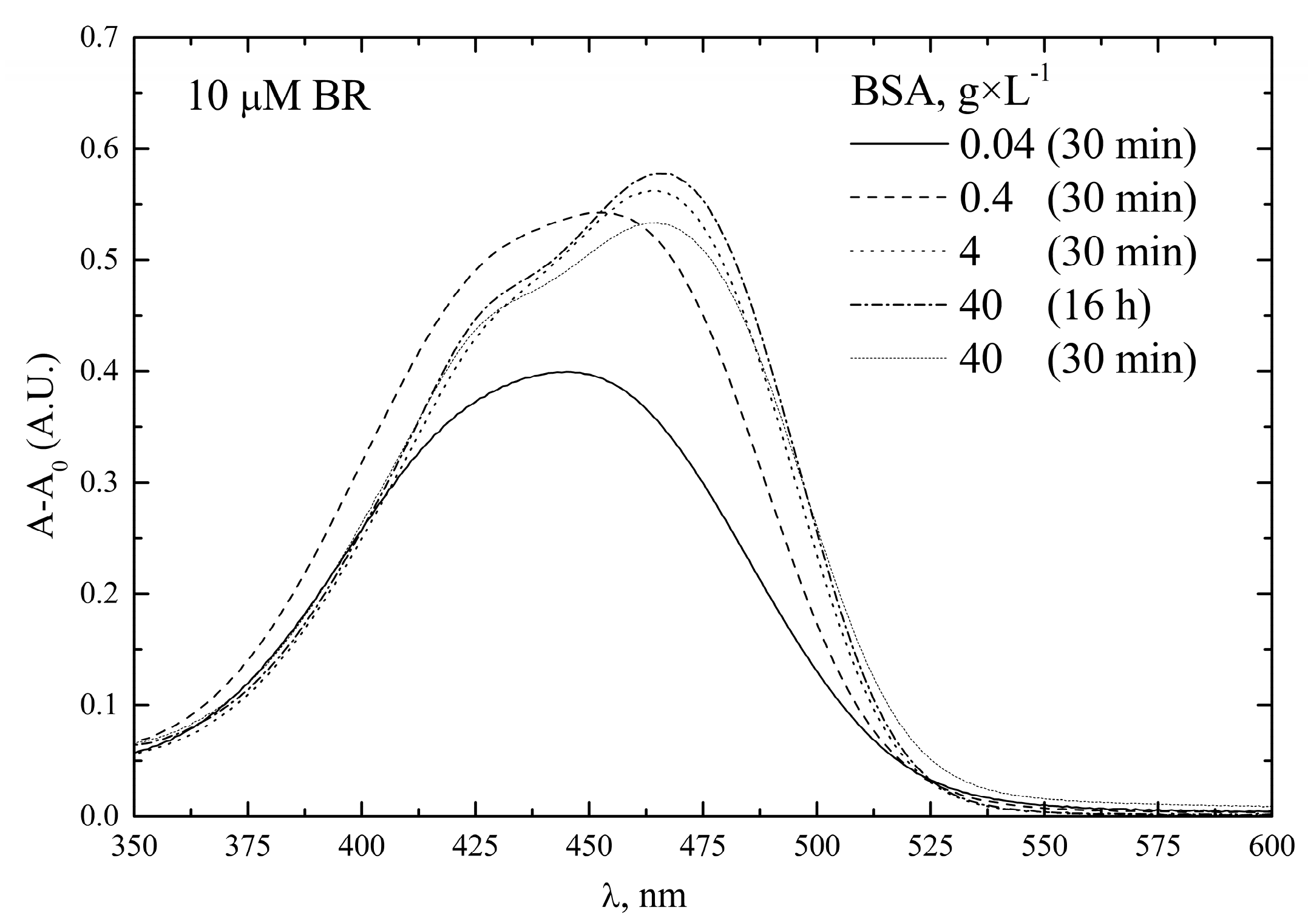
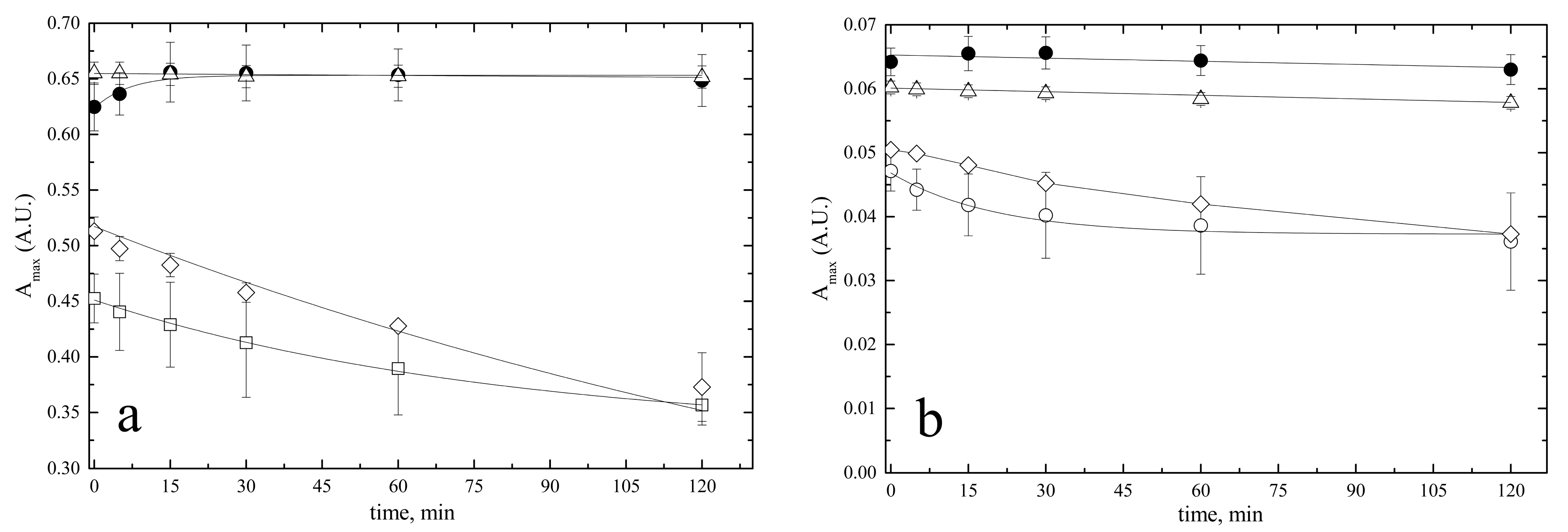
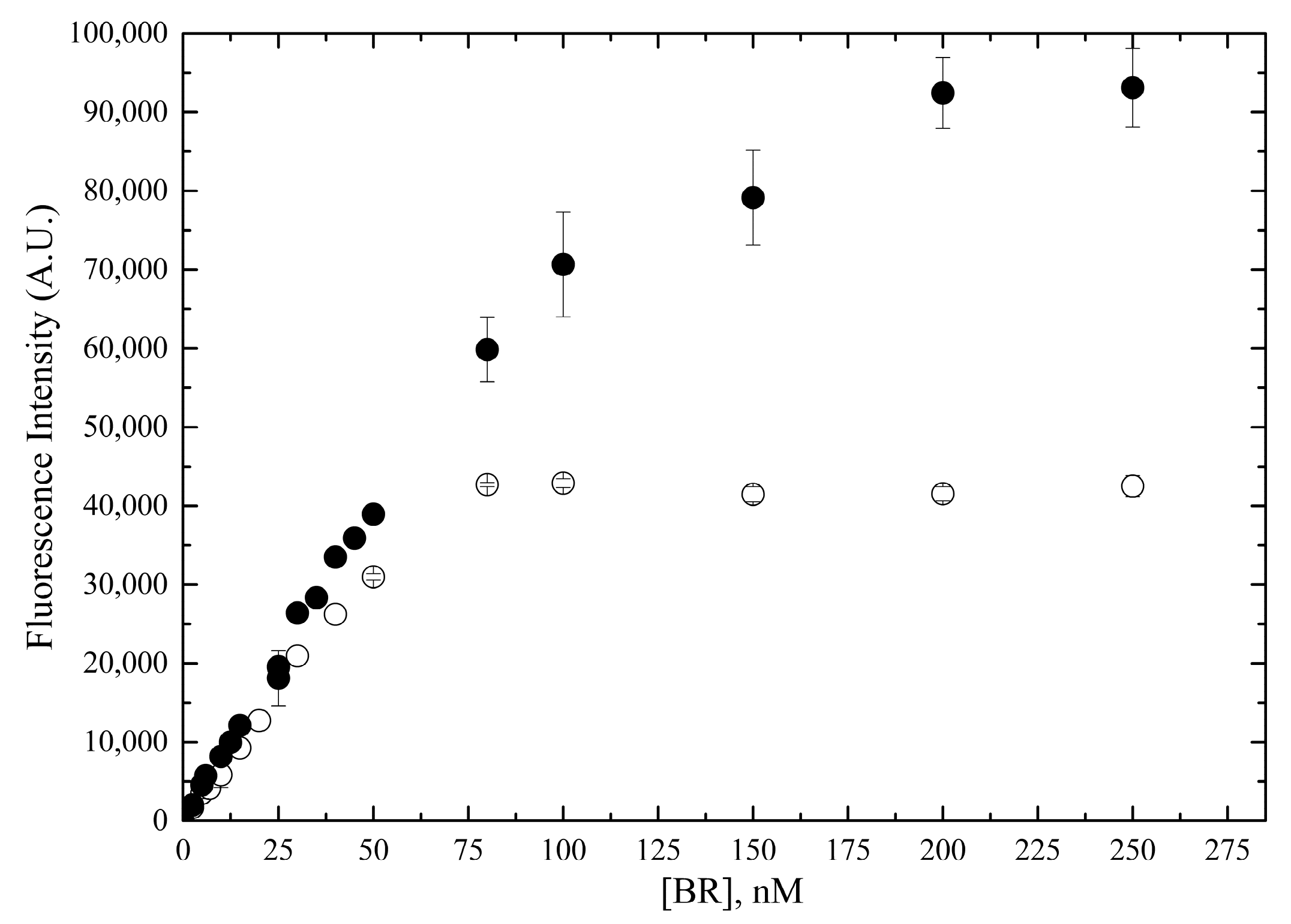

| ID Term | [BR] | Solvent | Quality Control |
|---|---|---|---|
| A. Stock | 5 mM | DMSO | UV–VIS |
| B. Pre-calibrator | 10 µM | PBS–BSA | UV–VIS; diazo reaction |
| C. Calibrator | 50 nM | PBS–BSA | HUG fluorescence |
| DMSO | PBS−BSA | PBS−HSA | PBS | |
|---|---|---|---|---|
| Amax (A.U.) | 0.631 (±0.02) | 0.636 (±0.05) | 0.601 (±0.02) | 0.446 (±0.05) |
| λmax (nm) | 456 | 465 | 465 | 440 |
| ε (cm−1 M−1) | 63,175 (±2859) | 61,079 (±1940) | 60,053 (±1425) | 41,885 (±2558) |
| R2 | 0.9997 | 0.9992 | 0.9989 | 0.9946 |
| LoD (nM) | 240 | 350 | 426 | 840 |
| LoQ (nM) | 720 | 1100 | 1290 | 2600 |
| [BSA] g × L−1 Pre-Calibrator | [BSA] g × L−1 Calibrators | Angular Coefficient (nM−1) |
|---|---|---|
| 40 | 40 | 744 ± 74 |
| 4 | 4 | 815 ± 30 |
| 4 | 0.4 | 785 ± 36 |
| BR (nM) | LoD (nM) | LoQ (nM) |
|---|---|---|
| 0.05–10 | 0.36 | 1.10 |
| 0.5–50 | 1.56 | 4.75 |
| Solvent | Angular Coefficient (nM−1) | R2 | Fluorescence Intensity of 50 nM BR Solution (A.U. ± SD) | Number of Replicates (n) |
|---|---|---|---|---|
| Tris 10 mM pH 8 | 663 | 0.9959 | 34,095 (±1189) | 5 |
| HEPES 10 mM, pH 7.4 | 620 | 0.9911 | 31,313 (±2089) | 3 |
| HBSS, pH 7.4 | 33 | 0.9841 | 1695 (±333) | 2 |
| PBS pH 7.4 | 511 | 0.9994 | 25,390 (± 2868) | 40 |
| PBS pH 8.5 | 523 | 0.9988 | 26,416 (± 1243) | 3 |
| PBS pH 9.5 | 560 | 0.9985 | 28,172 (± 2484) | 3 |
| PBS–Triton X–100 1%, pH 7.4 | 780 | 0.9965 | 40,045 (±2205) | 6 |
| PBS–Triton X–100 1%, pH 8.5 | 750 | 0.9904 | 37,278 (±4863) | 6 |
| PBS–Tween 20 1% | 7.5 | 0.9994 | 379 (±43) | 3 |
| PBS–Na–Taurocholate 0.2 mM | 534 | 0.9999 | 26,662 (±1274) | 3 |
| PBS–DMSO 30% | 757 | 0.9947 | 38,586 (±3190) | 3 |
| PBS–0.4 g × L−1 BSA, pH 7.4 | 785 | 0.9990 | 39,950 (±1702) | 40 |
| PBS–0.4 g × L−1 HSA, pH 7.4 | 718 | 0.9973 | 35,823 (±1862) | 3 |
| Categorical Parameter | Median BR (µM) (25–75%) | p-Value | |
|---|---|---|---|
| Sex | Male | 5.88 (4.44–7.86) | <0.001 |
| Female | 4.92 (3.31–6.36) | ||
| UGT1A1 rs8175347 | –– | 4.57 (3.77–5.98) | Ref. |
| TA– | 5.59 (4.02–7.36) | 0.008 | |
| TATA | 9.38 (5.87–13.71) | <0.001 | |
| TA– + TATA | 6.02 (4.31–8.07) | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sist, P.; Tramer, F.; Bandiera, A.; Urbani, R.; Redenšek Trampuž, S.; Dolžan, V.; Passamonti, S. Nanoscale Bilirubin Analysis in Translational Research and Precision Medicine by the Recombinant Protein HUG. Int. J. Mol. Sci. 2023, 24, 16289. https://doi.org/10.3390/ijms242216289
Sist P, Tramer F, Bandiera A, Urbani R, Redenšek Trampuž S, Dolžan V, Passamonti S. Nanoscale Bilirubin Analysis in Translational Research and Precision Medicine by the Recombinant Protein HUG. International Journal of Molecular Sciences. 2023; 24(22):16289. https://doi.org/10.3390/ijms242216289
Chicago/Turabian StyleSist, Paola, Federica Tramer, Antonella Bandiera, Ranieri Urbani, Sara Redenšek Trampuž, Vita Dolžan, and Sabina Passamonti. 2023. "Nanoscale Bilirubin Analysis in Translational Research and Precision Medicine by the Recombinant Protein HUG" International Journal of Molecular Sciences 24, no. 22: 16289. https://doi.org/10.3390/ijms242216289
APA StyleSist, P., Tramer, F., Bandiera, A., Urbani, R., Redenšek Trampuž, S., Dolžan, V., & Passamonti, S. (2023). Nanoscale Bilirubin Analysis in Translational Research and Precision Medicine by the Recombinant Protein HUG. International Journal of Molecular Sciences, 24(22), 16289. https://doi.org/10.3390/ijms242216289









