Genetic Concordance in Primary Cutaneous Melanoma and Matched Metastasis: A Systematic Review and Meta-Analysis
Abstract
:1. Introduction
2. Methods
2.1. Search Strategy and Eligibility Criteria
2.2. Data Extraction and Quality Assessment
2.3. Outcome of Interest
2.4. Pathway Analysis
2.5. Statistical Analysis
3. Results
3.1. Search Results and Studies Characteristics
3.2. The Genetic Concordance between Primary Cutaneous Melanoma and Matched Distant Metastasis
3.3. The Genetic Concordance between Primary Cutaneous Melanoma and Matched Distant Metastasis According to Metastatic Site and Gender
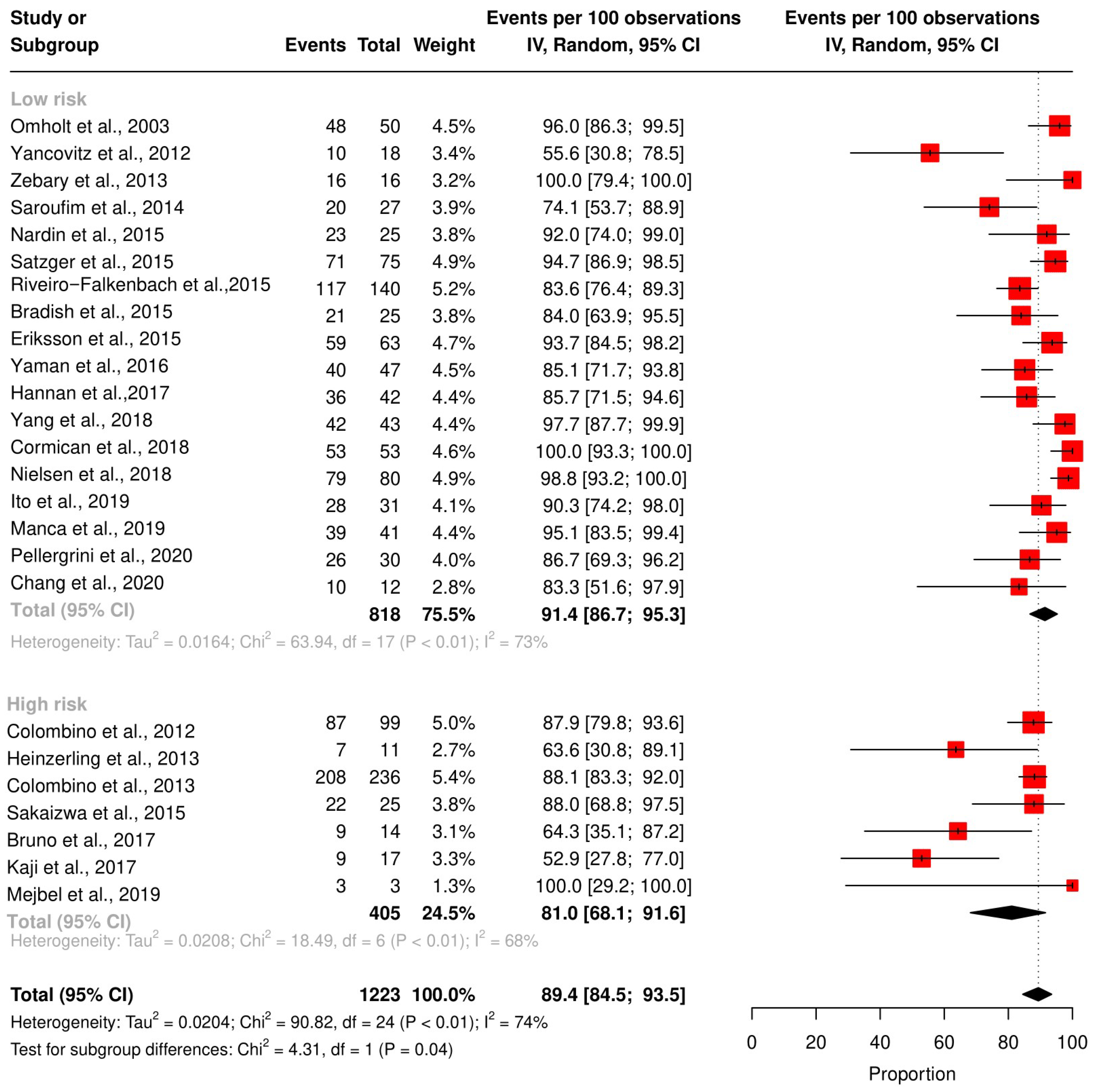
3.4. Meta-Analysis of the Concordance Rate for BRAF and NRAS
3.5. Subgroup Analysis According to the Technique and QUIPS Tool Score
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whiteman, D.C.; Green, A.C.; Olsen, C.M. The growing burden of invasive melanoma: Projections of incidence rates and numbers of new cases in six susceptible populations through 2031. J. Investig. Dermatol. 2016, 136, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Garbe, C.; Keim, U.; Amaral, T.; Berking, C.; Eigentler, T.K.; Flatz, L.; Gesierich, A.; Leiter, U.; Stadler, R.; Sunderkötter, C. Prognosis of patients with primary melanoma stage I and II according to American Joint Committee on Cancer Version 8 Validated in two independent cohorts: Implications for adjuvant treatment. J. Clin. Oncol. 2022, 40, 3741. [Google Scholar] [CrossRef] [PubMed]
- Ertekin, S.S.; Podlipnik, S.; Riquelme-Mc Loughlin, C.; Barreiro-Capurro, A.; Arance, A.; Carrera, C.C.; Malvehy, J.; Puig, S. Initial stage of cutaneous primary melanoma plays a key role in the pattern and timing of disease recurrence. Acta Derm. Venereol. 2021, 101, adv00502. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Louwman, M.; Wakkee, M.; Van der Veldt, A.; Grünhagen, D.; Verhoef, C.; Mooyaart, A.; Nijsten, T.; Hollestein, L. Primary melanoma characteristics of metastatic disease: A nationwide cancer registry study. Cancers 2021, 13, 4431. [Google Scholar] [CrossRef]
- Bedikian, A.Y.; DeConti, R.C.; Conry, R.; Agarwala, S.; Papadopoulos, N.; Kim, K.B.; Ernstoff, M. Phase 3 study of docosahexaenoic acid–paclitaxel versus dacarbazine in patients with metastatic malignant melanoma. Ann. Oncol. 2011, 22, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Garbe, C.; Amaral, T.; Peris, K.; Hauschild, A.; Arenberger, P.; Bastholt, L.; Bataille, V.; Del Marmol, V.; Dréno, B.; Fargnoli, M.C. European consensus-based interdisciplinary guideline for melanoma. Part 2: Treatment–Update 2019. Eur. J. Cancer 2020, 126, 159–177. [Google Scholar] [CrossRef] [PubMed]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef]
- Larkin, J.; Minor, D.; D’Angelo, S.; Neyns, B.; Smylie, M.; Miller Jr, W.H.; Gutzmer, R.; Linette, G.; Chmielowski, B.; Lao, C.D. Overall survival in patients with advanced melanoma who received nivolumab versus investigator’s choice chemotherapy in CheckMate 037: A randomized, controlled, open-label phase III trial. J. Clin. Oncol. 2018, 36, 383. [Google Scholar] [CrossRef]
- Patel, P.M.; Suciu, S.; Mortier, L.; Kruit, W.H.; Robert, C.; Schadendorf, D.; Trefzer, U.; Punt, C.J.A.; Dummer, R.; Davidson, N. Extended schedule, escalated dose temozolomide versus dacarbazine in stage IV melanoma: Final results of a randomised phase III study (EORTC 18032). Eur. J. Cancer 2011, 47, 1476–1483. [Google Scholar] [CrossRef]
- Sibaud, V. Dermatologic reactions to immune checkpoint inhibitors: Skin toxicities and immunotherapy. Am. J. Clin. Dermatol. 2018, 19, 345–361. [Google Scholar] [CrossRef]
- Robert, C.; Thomas, L.; Bondarenko, I.; O’Day, S.; Weber, J.; Garbe, C.; Lebbe, C.; Baurain, J.-F.; Testori, A.; Grob, J.-J. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N. Engl. J. Med. 2011, 364, 2517–2526. [Google Scholar] [CrossRef] [PubMed]
- Sosman, J.A.; Kim, K.B.; Schuchter, L.; Gonzalez, R.; Pavlick, A.C.; Weber, J.S.; McArthur, G.A.; Hutson, T.E.; Moschos, S.J.; Flaherty, K.T. Survival in BRAF V600–mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med. 2012, 366, 707–714. [Google Scholar] [CrossRef]
- Ko, J.M.; Fisher, D.E. A new era: Melanoma genetics and therapeutics. J. Pathol. 2011, 223, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Dummer, R.; Schadendorf, D.; Ascierto, P.A.; Arance, A.; Dutriaux, C.; Di Giacomo, A.M.; Rutkowski, P.; Del Vecchio, M.; Gutzmer, R.; Mandala, M. Binimetinib versus dacarbazine in patients with advanced NRAS-mutant melanoma (NEMO): A multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2017, 18, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Landras, A.; Reger de Moura, C.; Villoutreix, B.O.; Battistella, M.; Sadoux, A.; Dumaz, N.; Menashi, S.; Fernández-Recio, J.; Lebbé, C.; Mourah, S. Novel treatment strategy for NRAS-mutated melanoma through a selective inhibitor of CD147/VEGFR-2 interaction. Oncogene 2022, 41, 2254–2264. [Google Scholar] [CrossRef]
- Guo, J.; Si, L.; Kong, Y.; Flaherty, K.T.; Xu, X.; Zhu, Y.; Corless, C.L.; Li, L.; Li, H.; Sheng, X. Phase II, open-label, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J. Clin. Oncol. 2011, 29, 2904–2909. [Google Scholar] [CrossRef]
- Hodi, F.S.; Corless, C.L.; Giobbie-Hurder, A.; Fletcher, J.A.; Zhu, M.; Marino-Enriquez, A.; Friedlander, P.; Gonzalez, R.; Weber, J.S.; Gajewski, T.F. Imatinib for melanomas harboring mutationally activated or amplified KIT arising on mucosal, acral, and chronically sun-damaged skin. J. Clin. Oncol. 2013, 31, 3182. [Google Scholar] [CrossRef]
- Mehnert, J.M.; Kluger, H.M. Driver mutations in melanoma: Lessons learned from bench-to-bedside studies. Curr. Oncol. Rep. 2012, 14, 449–457. [Google Scholar] [CrossRef]
- Pham, D.D.M.; Guhan, S.; Tsao, H. KIT and melanoma: Biological insights and clinical implications. Yonsei Med. J. 2020, 61, 562. [Google Scholar] [CrossRef]
- Valachis, A.; Ullenhag, G.J. Discrepancy in BRAF status among patients with metastatic malignant melanoma: A meta-analysis. Eur. J. Cancer 2017, 81, 106–115. [Google Scholar] [CrossRef]
- en de Nederlandse, N.G.N. Richtlijn Melanoom: Modulaire revisie 2016. Ned. Tijdschr. Oncol. 2016, 13, 218–220. [Google Scholar]
- Hayden, J.A.; van der Windt, D.A.; Cartwright, J.L.; Côté, P.; Bombardier, C. Assessing bias in studies of prognostic factors. Ann. Intern. Med. 2013, 158, 280–286. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g: Profiler—Interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, gkad347. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, D.; Wang, Y.; Han, Y. Using Freeman-Tukey Double Arcsine Transformation in Meta-analysis of Single Proportions. Aesthetic Plast. Surg. 2023, 47, 83–84. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, G. meta: An R package for meta-analysis. R News 2007, 7, 40–45. [Google Scholar]
- Viechtbauer, W. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 2010, 36, 1–48. [Google Scholar] [CrossRef]
- Omholt, K.; Platz, A.; Kanter, L.; Ringborg, U.; Hansson, J. NRAS and BRAF Mutations Arise Early during Melanoma Pathogenesis and Are Preserved throughout Tumor Progression. Clin. Cancer Res. 2003, 9, 6483–6488. [Google Scholar]
- Colombino, M.; Capone, M.; Lissia, A.; Cossu, A.; Rubino, C.; De Giorgi, V.; Massi, D.; Fonsatti, E.; Staibano, S.; Nappi, O.; et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J. Clin. Oncol. 2012, 30, 2522–2529. [Google Scholar] [CrossRef]
- Yancovitz, M.; Litterman, A.; Yoon, J.; Ng, E.; Shapiro, R.L.; Berman, R.S.; Pavlick, A.C.; Darvishian, F.; Christos, P.; Mazumdar, M.; et al. Intra- and inter-tumor heterogeneity of BRAFV600Emutations in primary and metastatic melanoma. PLoS ONE 2012, 7, e29336. [Google Scholar] [CrossRef]
- Colombino, M.; Lissia, A.; Capone, M.; De Giorgi, V.; Massi, D.; Stanganelli, I.; Fonsatti, E.; Maio, M.; Botti, G.; Caracò, C.; et al. Heterogeneous distribution of BRAF/NRAS mutations among Italian patients with advanced melanoma. J. Transl. Med. 2013, 11, 202. [Google Scholar] [CrossRef]
- Heinzerling, L.; Baiter, M.; Kühnapfel, S.; Schuler, G.; Keikavoussi, P.; Agaimy, A.; Kiesewetter, F.; Hartmann, A.; Schneider-Stock, R. Mutation landscape in melanoma patients clinical implications of heterogeneity of BRAF mutations. Br. J. Cancer 2013, 109, 2833–2841. [Google Scholar] [CrossRef] [PubMed]
- Zebary, A.; Omholt, K.; Vassilaki, I.; Höiom, V.; Lindén, D.; Viberg, L.; Kanter-Lewensohn, L.; Johansson, C.H.; Hansson, J. KIT, NRAS, BRAF and PTEN mutations in a sample of Swedish patients with acral lentiginous melanoma. J. Dermatol. Sci. 2013, 72, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Saroufim, M.; Habib, R.H.; Gerges, R.; Saab, J.; Loya, A.; Amr, S.S.; Sheikh, S.; Satti, M.; Oberkanins, C.; Khalifeh, I. Comparing BRAF mutation status in matched primary and metastatic cutaneous melanomas: Implications on optimized targeted therapy. Exp. Mol. Pathol. 2014, 97, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Bradish, J.R.; Richey, J.D.; Post, K.M.; Meehan, K.; Sen, J.D.; Malek, A.J.; Katona, T.M.; Warren, S.; Logan, T.F.; Fecher, L.A.; et al. Discordancy in BRAF mutations among primary and metastatic melanoma lesions: Clinical implications for targeted therapy. Mod. Pathol. 2015, 28, 480–486. [Google Scholar] [CrossRef]
- Eriksson, H.; Zebary, A.; Vassilaki, I.; Omholt, K.; Ghaderi, M.; Hansson, J. BRAFV600E protein expression in primary cutaneous malignant melanomas and paired metastases. JAMA Dermatol. 2015, 151, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Nardin, C.; Puzenat, E.; Prétet, J.L.; Algros, M.P.; Doussot, A.; Puyraveau, M.; Mougin, C.; Aubin, F. BRAF mutation screening in melanoma: Is sentinel lymph node reliable? Melanoma Res. 2015, 25, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Riveiro-Falkenbach, E.; Villanueva, C.A.; Garrido, M.C.; Ruano, Y.; García-Martín, R.M.; Godoy, E.; Ortiz-Romero, P.L.; Ríos-Martín, J.J.; Santos-Briz, A.; Rodríguez-Peralto, J.L. Intra- and inter-tumoral homogeneity of BRAF V600E mutations in melanoma tumors. J. Investig. Dermatol. 2015, 135, 3078–3085. [Google Scholar] [CrossRef]
- Sakaizawa, K.; Ashida, A.; Uchiyama, A.; Ito, T.; Fujisawa, Y.; Ogata, D.; Matsushita, S.; Fujii, K.; Fukushima, S.; Shibayama, Y.; et al. Clinical characteristics associated with BRAF, NRAS and KIT mutations in Japanese melanoma patients. J. Dermatol. Sci. 2015, 80, 33–37. [Google Scholar] [CrossRef]
- Satzger, I.; Marks, L.; Kerick, M.; Klages, S.; Berking, C.; Herbst, R.; Völker, B.; Schacht, V.; Timmermann, B.; Gutzmer, R. Allele frequencies of BRAFV600 mutations in primary melanomas and matched metastases and their relevance for BRAF inhibitor therapy in metastatic melanoma. Oncotarget 2015, 6, 37895–37905. [Google Scholar] [CrossRef]
- Yaman, B.; Kandiloglu, G.; Akalin, T. BRAF-V600 Mutation Heterogeneity in Primary and Metastatic Melanoma: A Study with Pyrosequencing and Immunohistochemistry. Am. J. Dermatopathol. 2016, 38, 113–120. [Google Scholar] [CrossRef]
- Bruno, W.; Martinuzzi, C.; Andreotti, V.; Pastorino, L.; Spagnolo, F.; Dalmasso, B.; Cabiddu, F.; Gualco, M.; Ballestrero, A.; Bianchi-Scarrà, G.; et al. Heterogeneity and frequency of BRAF mutations in primary melanoma: Comparison between molecular methods and immunohistochemistry. Oncotarget 2017, 8, 8069–8082. [Google Scholar] [CrossRef] [PubMed]
- Hannan, E.J.; O’Leary, D.P.; Macnally, S.P.; Kay, E.W.; Farrell, M.A.; Morris, P.G.; Power, C.P.; Hill, A.D.K. The significance of BRAF V600E mutation status discordance between primary cutaneous melanoma and brain metastases. Medicine 2017, 96, e8404. [Google Scholar] [CrossRef] [PubMed]
- Kaji, T.; Yamasaki, O.; Takata, M.; Otsuka, M.; Hamada, T.; Morizane, S.; Asagoe, K.; Yanai, H.; Hirai, Y.; Umemura, H.; et al. Comparative study on driver mutations in primary and metastatic melanomas at a single Japanese institute: A clue for intra- and inter-tumor heterogeneity. J. Dermatol. Sci. 2017, 85, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, L.B.; Dabrosin, N.; Sloth, K.; Bønnelykke–Behrndtz, M.L.; Steiniche, T.; Lade-Keller, J. Concordance in BRAF V600E status over time in malignant melanoma and corresponding metastases. Histopathology 2018, 72, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Leone, D.A.; Biswas, A.; Deng, A.; Jukic, D.; Singh, R.; Sundram, U.; Mahalingam, M. Concordance of somatic mutation profiles (BRAF, NRAS, and TERT) and tumoral PD-L1 in matched primary cutaneous and metastatic melanoma samples. Hum. Pathol. 2018, 82, 206–214. [Google Scholar] [CrossRef]
- Cormican, D.; Kennedy, C.; Murphy, S.; Werner, R.; Power, D.G.; Heffron, C.C.B.B. High concordance of BRAF mutational status in matched primary and metastatic melanoma. J. Cutan. Pathol. 2019, 46, 117–122. [Google Scholar] [CrossRef]
- Ito, T.; Kaku-Ito, Y.; Murata, M.; Ichiki, T.; Kuma, Y.; Tanaka, Y.; Ide, T.; Ohno, F.; Wada-Ohno, M.; Yamada, Y.; et al. Intra-and inter-tumor braf heterogeneity in acral melanoma: An immunohistochemical analysis. Int. J. Mol. Sci. 2019, 20, 6191. [Google Scholar] [CrossRef]
- Manca, A.; Paliogiannis, P.; Colombino, M.; Casula, M.; Lissia, A.; Botti, G.; Caracò, C.; Ascierto, P.A.; Sini, M.C.; Palomba, G.; et al. Mutational concordance between primary and metastatic melanoma: A next-generation sequencing approach. J. Transl. Med. 2019, 17, 289. [Google Scholar] [CrossRef]
- Mejbel, H.A.; Arudra, S.K.C.; Pradhan, D.; Torres-Cabala, C.A.; Nagarajan, P.; Tetzlaff, M.T.; Curry, J.L.; Ivan, D.; Duose, D.Y.; Luthra, R.; et al. Immunohistochemical and molecular features of melanomas exhibiting intratumor and intertumor histomorphologic heterogeneity. Cancers 2019, 11, 1714. [Google Scholar] [CrossRef]
- Chang, G.A.; Wiggins, J.M.; Corless, B.C.; Syeda, M.M.; Tadepalli, J.S.; Blake, S.; Fleming, N.; Darvishian, F.; Pavlick, A.; Berman, R.; et al. TERT, BRAF, and NRAS Mutational Heterogeneity between Paired Primary and Metastatic Melanoma Tumors. J. Investig. Dermatol. 2020, 140, 1609–1618. [Google Scholar] [CrossRef]
- Pellegrini, C.; Cardelli, L.; De Padova, M.; Di Nardo, L.; Ciciarell, V.; Rocco, T.; Cipolloni, G.; Clementi, M.; Cortellini, A.; Ventura, A.; et al. Intra-patient heterogeneity of BRAF and NRAS molecular alterations in primary melanoma and metastases. Acta Derm. Venereol. 2020, 100, adv00040. [Google Scholar] [CrossRef] [PubMed]
- Demunter, A.; Stas, M.; Degreef, H.; De Wolf-Peeters, C.; Van den Oord, J.J. Analysis of N- and K-ras mutations in the distinctive tumor progression phases of melanoma. J. Investig. Dermatol. 2001, 117, 1483–1489. [Google Scholar] [CrossRef] [PubMed]
- Omholt, K.; Karsberg, S.; Platz, A.; Kanter, L.; Ringborg, U.; Hansson, J. Screening of N-ras codon 61 mutations in paired primary and metastatic cutaneous melanomas: Mutations occur early and persist throughout tumor progression. Clin. Cancer Res. 2002, 8, 3468–3474. [Google Scholar] [PubMed]
- Akslen, L.A.; Angelini, S.; Straume, O.; Bachmann, I.M.; Molven, A.; Hemminki, K.; Kumar, R. BRAF and NRAS mutations are frequent in nodular melanoma but are not associated with tumor cell proliferation or patient survival. J. Investig. Dermatol. 2005, 125, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Uhara, H.; Ashida, A.; Koga, H.; Ogawa, E.; Uchiyama, A.; Uchiyama, R.; Hayashi, K.; Kiniwa, Y.; Okuyama, R. NRAS mutations in primary and metastatic melanomas of Japanese patients. Int. J. Clin. Oncol. 2014, 19, 544–548. [Google Scholar] [CrossRef]
- Davis, E.J.; Johnson, D.B.; Sosman, J.A.; Chandra, S. Melanoma: What do all the mutations mean? Cancer 2018, 124, 3490–3499. [Google Scholar] [CrossRef]
- Zhang, T.; Dutton-Regester, K.; Brown, K.M.; Hayward, N.K. The genomic landscape of cutaneous melanoma. Pigment. Cell Melanoma Res. 2016, 29, 266–283. [Google Scholar] [CrossRef]
- Vergara, I.A.; Mintoff, C.P.; Sandhu, S.; McIntosh, L.; Young, R.J.; Wong, S.Q.; Colebatch, A.; Cameron, D.L.; Kwon, J.L.; Wolfe, R. Evolution of late-stage metastatic melanoma is dominated by aneuploidy and whole genome doubling. Nat. Commun. 2021, 12, 1434. [Google Scholar] [CrossRef]
- Cherepakhin, O.S.; Argenyi, Z.B.; Moshiri, A.S. Genomic and transcriptomic underpinnings of melanoma genesis, progression, and metastasis. Cancers 2021, 14, 123. [Google Scholar] [CrossRef]
- Castle, J.C.; Uduman, M.; Pabla, S.; Stein, R.B.; Buell, J.S. Mutation-derived neoantigens for cancer immunotherapy. Front. Immunol. 2019, 10, 1856. [Google Scholar] [CrossRef]
- Tímár, J.; Vizkeleti, L.; Doma, V.; Barbai, T.; Rásó, E. Genetic progression of malignant melanoma. Cancer Metastasis Rev. 2016, 35, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, Z. Meta-analysis of epidermal growth factor receptor and KRAS gene status between primary and corresponding metastatic tumours of non-small cell lung cancer. Clin. Oncol. 2015, 27, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Pinzani, P.; Santucci, C.; Mancini, I.; Simi, L.; Salvianti, F.; Pratesi, N.; Massi, D.; De Giorgi, V.; Pazzagli, M.; Orlando, C. BRAFV600E detection in melanoma is highly improved by COLD-PCR. Clin. Chim. Acta 2011, 412, 901–905. [Google Scholar] [CrossRef]
- Barbano, R.; Pasculli, B.; Coco, M.; Fontana, A.; Copetti, M.; Rendina, M.; Valori, V.M.; Graziano, P.; Maiello, E.; Fazio, V.M. Competitive allele-specific TaqMan PCR (Cast-PCR) is a sensitive, specific and fast method for BRAF V600 mutation detection in Melanoma patients. Sci. Rep. 2015, 5, 18592. [Google Scholar] [CrossRef] [PubMed]
- Colomba, E.; Hélias-Rodzewicz, Z.; Von Deimling, A.; Marin, C.; Terrones, N.; Pechaud, D.; Surel, S.; Côté, J.-F.; Peschaud, F.; Capper, D. Detection of BRAF p. V600E mutations in melanomas: Comparison of four methods argues for sequential use of immunohistochemistry and pyrosequencing. J. Mol. Diagn. 2013, 15, 94–100. [Google Scholar] [CrossRef]
- Yakout, N.M.; Abdallah, D.M.; Abdelmonsif, D.A.; Kholosy, H.M.; Talaat, I.M.; Elsakka, O. BRAFV600E mutational status assessment in cutaneous melanocytic neoplasms in a group of the Egyptian population. Cancer Cell Int. 2023, 23, 17. [Google Scholar] [CrossRef] [PubMed]
- Thiel, A.; Moza, M.; Kytölä, S.; Orpana, A.; Jahkola, T.; Hernberg, M.; Virolainen, S.; Ristimäki, A. Prospective immunohistochemical analysis of BRAF V600E mutation in melanoma. Hum. Pathol. 2015, 46, 169–175. [Google Scholar] [CrossRef]
- Ardito, F.; Razionale, F.; Salvatore, L.; Cenci, T.; Vellone, M.; Basso, M.; Panettieri, E.; Calegari, M.A.; Tortora, G.; Martini, M. Discordance of KRAS mutational status between primary tumors and liver metastases in colorectal cancer: Impact on long-term survival following radical resection. Cancers 2021, 13, 2148. [Google Scholar] [CrossRef]
- Bhullar, D.S.; Barriuso, J.; Mullamitha, S.; Saunders, M.P.; O’Dwyer, S.T.; Aziz, O. Biomarker concordance between primary colorectal cancer and its metastases. eBioMedicine 2019, 40, 363–374. [Google Scholar] [CrossRef]
- Abate-Daga, D.; Ramello, M.C.; Smalley, I.; Forsyth, P.A.; Smalley, K.S.M. The biology and therapeutic management of melanoma brain metastases. Biochem. Pharmacol. 2018, 153, 35–45. [Google Scholar] [CrossRef]
- Röckmann, H.; Schadendorf, D. Drug resistance in human melanoma: Mechanisms and therapeutic opportunities. Oncol. Res. Treat. 2003, 26, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Wu, X.-Y.; Yang, Z.-Y.; Threapleton, D.E.; Yuan, J.-Q.; Yu, Y.-Y.; Tang, J.-L. Concordant analysis of KRAS, BRAF, PIK3CA mutations and PTEN expression between primary colorectal cancer and matched metastases. Sci. Rep. 2015, 5, 8065. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Takata, M.; Murata, H.; Goto, Y.; Kido, K.; Ferrone, S.; Saida, T. Polyclonality of BRAF mutations in acquired melanocytic nevi. JNCI J. Natl. Cancer Inst. 2009, 101, 1423–1427. [Google Scholar] [CrossRef] [PubMed]
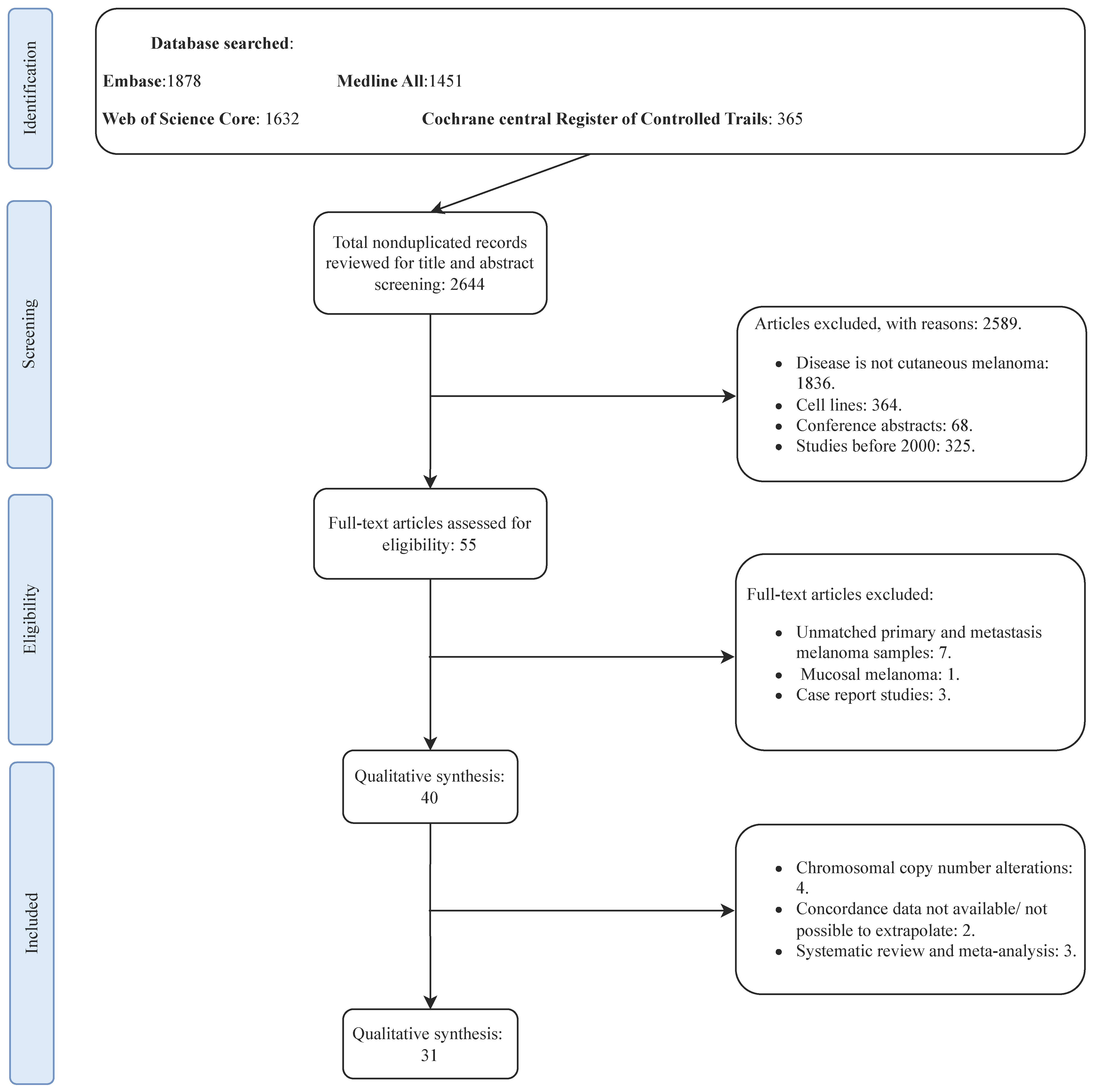
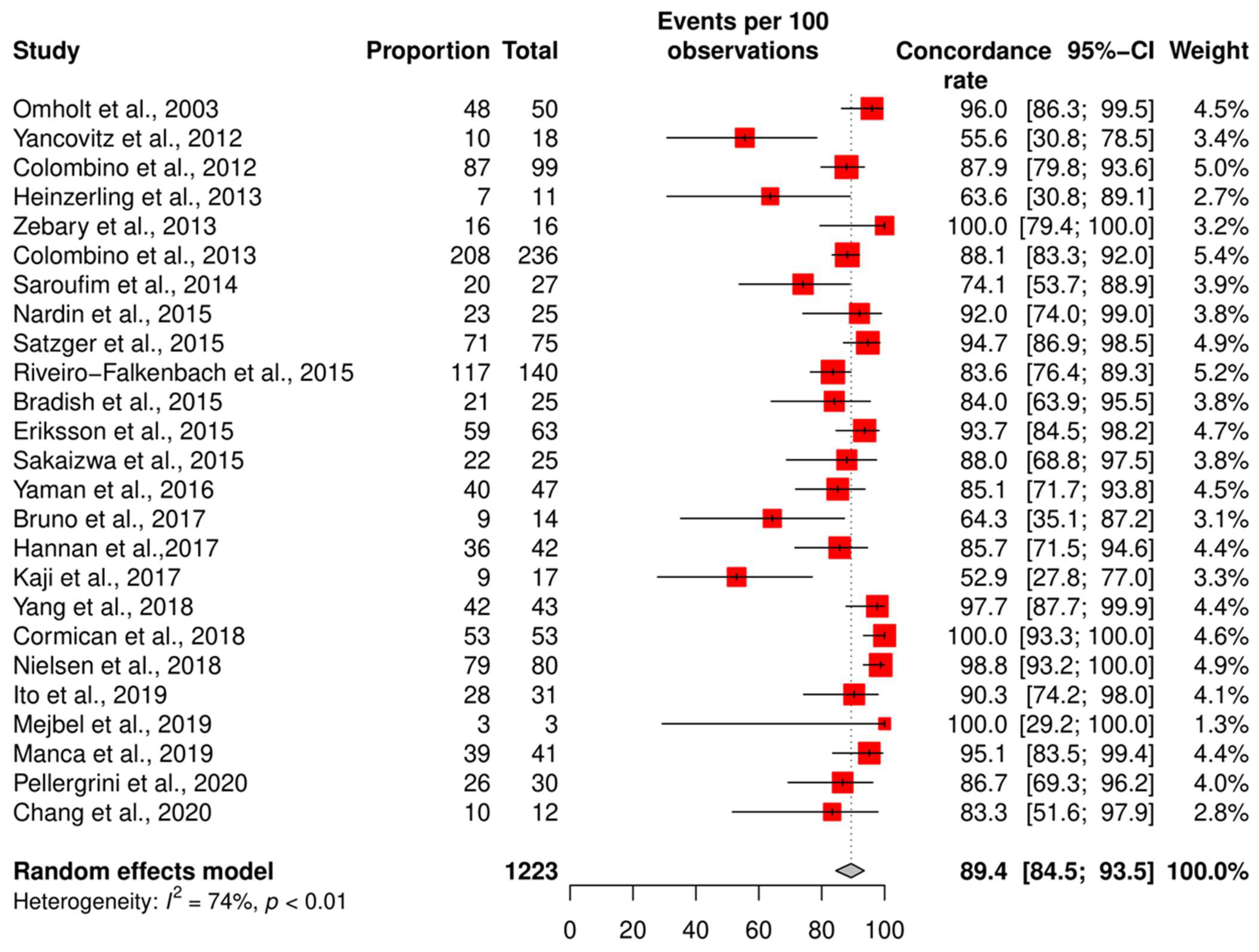
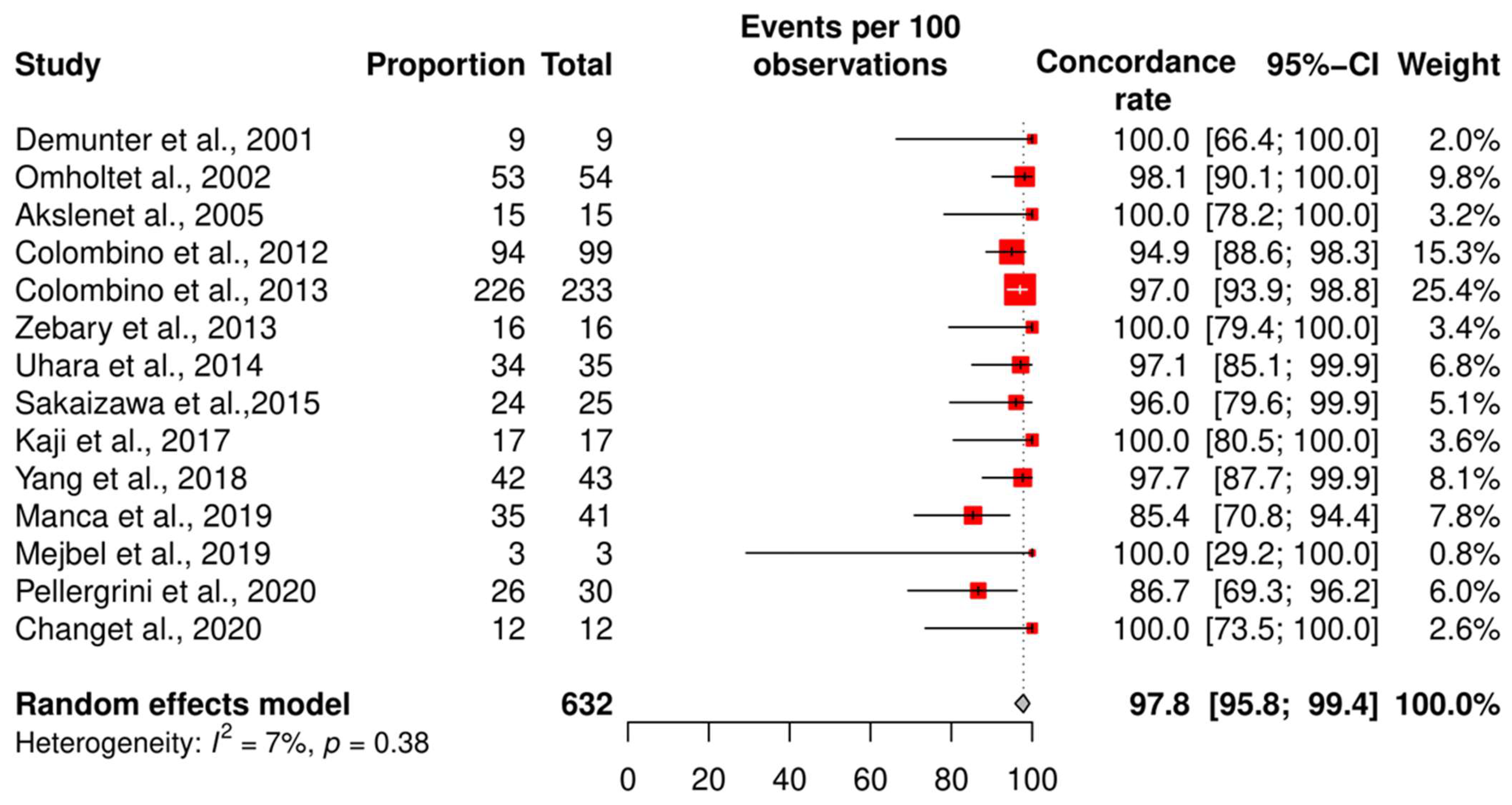
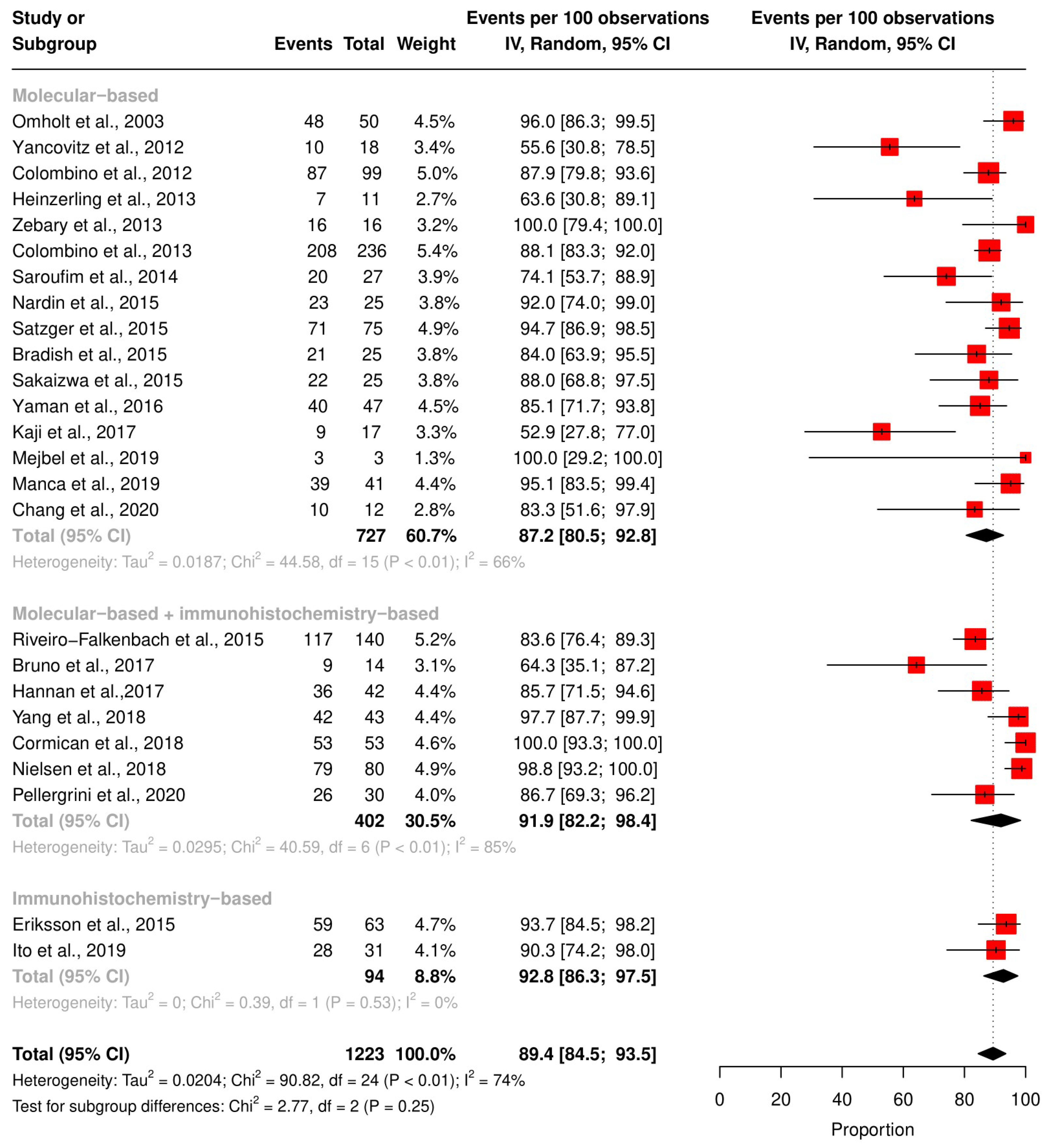

| Study | Country | Technique | Total Cohort (n) | Patients with Concordant Status (n) | Risk of Bias Score (Study Quality) |
|---|---|---|---|---|---|
| Omholt et al., 2003 [27] | Sweden | PCR | 50 | 48 | 5 (high) |
| Yancovitz et al., 2012 [29] | USA | MS-PCR | 18 | 10 | 4 (high) |
| Colombino et al., 2012 [28] | Italy | ADS | 99 | 87 | 2.5 (low) |
| Heinzerling et al., 2013 [31] | Germany | PCR | 11 | 7 | 1 (low) |
| Zebary et al., 2013 [32] | Sweden | PCR | 16 | 16 | 4 (high) |
| Colombino et al., 2013 [30] | Italy | ADS | 236 | 208 | 3 (low) |
| Saroufim et al., 2014 [33] | Lebanon | PCR | 27 | 20 | 4.5 (high) |
| Nardin et al., 2015 [36] | France | Pyrosequencing | 25 | 23 | 6 (high) |
| Satzger et al., 2015 [39] | Germany | Ultra-deep NGS | 75 | 71 | 6 (high) |
| Riveiro-Falkenbach et al., 2015 [37] | Spain | Cobas + IHC | 140 | 117 | 5 (high) |
| Bradish et al., 2015 [34] | USA | PCR | 25 | 21 | 5 (high) |
| Eriksson et al., 2015 [35] | Sweden | IHC | 63 | 59 | 4 (high) |
| Sakaizwa et al., 2015 [38] | Japan | DS | 25 | 22 | 3 (low) |
| Yaman et al., 2016 [40] | Turkey | Pyrosequencing + IHC | 47 | 40 | 5 (high) |
| Bruno et al., 2017 [41] | Italy | PNA, IHC, capillary seq | 14 | 9 | 2 (low) |
| Hannan et al.,2017 [42] | Ireland | PCR and IHC | 42 | 36 | 5.5 (high) |
| Kaji et al., 2017 [43] | Japan | MassARRAY | 17 | 9 | 3.5 (low) |
| Yang et al., 2018 [45] | USA | IHC and direct + Sanger sequencing | 43 | 42 | 6 (high) |
| Cormican et al., 2018 [46] | Ireland | PCR and IHC | 53 | 53 | 5.5 (high) |
| Nielsen et al., 2018 [44] | Denmark | Cobas test and IHC | 80 | 79 | 5.5 (high) |
| Manca et al., 2019 [48] | Italy | Targeted NGS | 41 | 39 | 5.5 (high) |
| Ito et al., 2019 [47] | Japan | IHC | 31 | 28 | 4 (high) |
| Mejbel et al., 2019 [49] | USA | NGS | 3 | 3 | 2.5 (high) |
| Pellergrini et al., 2020 [51] | Italy | PCR and IHC | 30 | 26 | 5.5 (high) |
| Chang et al., 2020 [50] | USA | SNaPshot assays, Sanger sequencing, MS PCR | 12 | 10 | 4 (high) |
| Study | Country | Technique | Total Cohort (n) | Patients with Concordant Status (n) | Risk of Bias Score |
|---|---|---|---|---|---|
| Demunter et al., 2001 [52] | Belgium | DOP-PCR | 9 | 9 | 1 |
| Omholtet al., 2002 [53] | Sweden | PCR and SSCP | 54 | 53 | 5 |
| Akslenet al., 2005 [54] | Germany | SSCP | 15 | 15 | 3 |
| Colombino et al., 2012 [28] | Italy | ADS | 99 | 94 | 2.5 |
| Colombino et al., 2013 [30] | Italy | ADS | 233 | 226 | 3 |
| Zebary et al., 2013 [32] | Sweden | PCR | 16 | 16 | 4 |
| Uhara et al., 2014 [55] | Japan | PCR | 35 | 34 | 2.5 |
| Sakaizawa et al., 2015 [38] | Japan | DS | 25 | 24 | 3 |
| Kaji et al., 2017 [43] | Japan | Sequenom MelaCarta MassARRAY | 17 | 17 | 3.5 |
| Yang et al., 2018 [45] | USA | Direct and Sanger sequencing + IHC | 43 | 42 | 6 |
| Manca et al., 2019 [48] | Italy | Targeted NGS | 41 | 35 | 5.5 |
| Mejbel et al., 2019 [49] | USA | NGS | 3 | 3 | 2.5 |
| Pellergrini et al., 2020 [51] | Italy | PCR and IHC | 30 | 26 | 5.5 |
| Chang et al., 2020 [50] | USA | SNaPshot assays, Sanger sequencing, MS PCR | 12 | 12 | 4 |
| Study Name | Country | Technique | Population Cohort (n) | Patients with Concordant Status (n) | Risk of Bias Score |
|---|---|---|---|---|---|
| Zebary et al., 2013 [32] | Sweden | Sequencing | 16 | 16 | 4 |
| Sakaizawa et al., 2015 [38] | Japan | DS | 25 | 22 | 3 |
| Mejbel et al., 2019 [49] | USA | PCR | 3 | 3 | 2.5 |
| Study | Total Patient | Gene | Mutation | N Mutated Primary | N Mutated Metastasis | N Mutated Samples | N Concordant Patients |
|---|---|---|---|---|---|---|---|
| Chang et al., 2020 [50] | 11 | TERT | promoter (146 C > T) | 4 | 11 * | 15 | 6 |
| Yang et al., 2018 [45] | 41 | TERT | promoter | N/A | N/A | N/A | 28 |
| Kaji et al., 2017 [43] | 17 | CDK4 | R24C | 1 | 2 | 3 | 0 |
| 17 | KRAS | G12A | 1 | 0 | 1 | 0 | |
| 17 | NEK10 | E379K | 1 | 0 | 1 | 0 | |
| 17 | EPHB6 | G404S | 2 | 1 | 3 | 1 | |
| Manca et al., 2019 [48] | 41 | TP53 | V216M | 0 | 2 | 2 | 0 |
| 41 | TP53 | R158C | 0 | 1 | 1 | 0 | |
| 41 | MAP2K1 | Q46Tter | 0 | 1 | 1 | 0 | |
| 41 | MAP2K1 | Q110Ter | 0 | 1 | 1 | 0 | |
| 41 | PTEN | G127 | 0 | 1 | 1 | 0 | |
| 41 | PTEN | Q110ter | 0 | 1 | 1 | 0 | |
| 41 | CCND1 | G103R | 0 | 1 | 1 | 0 | |
| 41 | CDKN2A | G23S | 1 | 0 | 1 | 0 | |
| 41 | CDKN2A | R131H | 1 | 0 | 1 | 0 | |
| 41 | PIK3CA | T1031I | 1 | 0 | 1 | 0 | |
| 41 | PIK3CA | G1049S | 1 | 0 | 1 | 0 | |
| 41 | TP53 | E286K | 1 | 2 | 3 | 0 | |
| 41 | TP53 | R196L | 1 | 0 | 1 | 0 | |
| 41 | MAP2K1 | Q383ter | 1 | 0 | 1 | 0 | |
| 41 | MAP2K1 | Q243Ter | 1 | 0 | 1 | 0 | |
| 41 | MAP2K1 | Q354ter | 1 | 1 | 2 | 0 | |
| 41 | RB1 | Q354ter | 1 | 1 | 2 | 0 | |
| 41 | PTEN | G165R | 1 | 0 | 1 | 0 | |
| 41 | CDKN2A | A40V | 1 | 1 | 2 | 1 | |
| 41 | PIK3CA | V344M | 1 | 1 | 2 | 1 | |
| 41 | TP53 | R196Ter | 1 | 1 | 2 | 1 | |
| 41 | TP53 | P278L | 1 | 1 | 2 | 1 | |
| 41 | TP53 | P278S | 1 | 1 | 2 | 1 | |
| 41 | PIK3CA | V344A | 2 | 0 | 2 | 0 | |
| Mejbel et al., 2019 [49] | 3 | RAC1 | P29S | 0 | 1 | 1 | 0 |
| 3 | CTNNB1 | S37F | 1 | 0 | 1 | 0 | |
| 3 | HNFA1 | A269T | 1 | 0 | 1 | 0 | |
| 3 | TP53 | H179Y | 1 | 1 * | 2 | 1 |
| Study | Lymph Node Metastasis | Brain Metastasis | Visceral Metastasis | Subcutaneous Metastasis | Skin Metastasis/ Other Type |
|---|---|---|---|---|---|
| Concordance Rate % (Concordant Cases/Total Cases) | |||||
| Heinzerling et al., 2013 [31] | - | - | - | - | 100 (7/7) |
| Zebary et al., 2013 [32] | 100 (15/15) | - | - | - | 100 (1/1) |
| Colombino et al., 2013 [30] | 90 (109/120) | 92 (22/24) | 93 (37/40) | - | 77 (40/52) |
| Saroufim et al., 2014 [33] | 89 (16/18) | - | - | 67 (4/6) | 50 (2/4) |
| Nardin et al., 2015 [36] | 100 (14/14) | - | 80 (4/5) | 92 (11/12) | |
| Bradish et al., 2015 [34] | - | 50 (2/4) | - | - | 92 (11/12) |
| Yaman et al., 2016 [40] | 83 (34/41) | - | - | - | - |
| Kaji et al., 2017 [43] | 53 (9/17) | - | - | - | - |
| Manca et al., 2019 [48] | 94 (17/18) | - | 100 (3/3) | - | - |
| Pellergrini et al., 2020 [51] | 89 (17/19) | 100 (1/1) | 50 (1/2) | - | 88 (7/8) |
| Total samples in all studies | 262 | 29 | 50 | 18 | 84 |
| Study | Risk of Bias Score | Total Cohort (n) | Total Females (n) | Total Males (n) | Concordant Female Patients (n) | Concordant Male Patients (n) |
|---|---|---|---|---|---|---|
| Heinzerling et al., 2013 [31] | 1 | 11 | 7 | 5 | 4 | 4 |
| Saroufim et al., 2014 [33] | 4.5 | 27 | 7 | 19 | 5 | 15 |
| Bradish et al., 2015 [34] | 5 | 25 | 13 | 11 | 11 | 9 |
| Yaman et al., 2016 [40] | 5 | 47 | 18 | 29 | 14 | 26 |
| Kaji et al., 2017 [43] | 3.5 | 17 | 9 | 8 | 3 | 5 |
| Total | 127 | 54 | 72 | 37 | 59 | |
| Concordance rate % (p-value = 0.19) | 68.5 | 81.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kerkour, T.; Zhou, C.; Hollestein, L.; Mooyaart, A. Genetic Concordance in Primary Cutaneous Melanoma and Matched Metastasis: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2023, 24, 16281. https://doi.org/10.3390/ijms242216281
Kerkour T, Zhou C, Hollestein L, Mooyaart A. Genetic Concordance in Primary Cutaneous Melanoma and Matched Metastasis: A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences. 2023; 24(22):16281. https://doi.org/10.3390/ijms242216281
Chicago/Turabian StyleKerkour, Thamila, Catherine Zhou, Loes Hollestein, and Antien Mooyaart. 2023. "Genetic Concordance in Primary Cutaneous Melanoma and Matched Metastasis: A Systematic Review and Meta-Analysis" International Journal of Molecular Sciences 24, no. 22: 16281. https://doi.org/10.3390/ijms242216281
APA StyleKerkour, T., Zhou, C., Hollestein, L., & Mooyaart, A. (2023). Genetic Concordance in Primary Cutaneous Melanoma and Matched Metastasis: A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences, 24(22), 16281. https://doi.org/10.3390/ijms242216281





