Transcriptome Shock in Developing Embryos of a Brassica napus and Brassica rapa Hybrid
Abstract
1. Introduction
2. Results
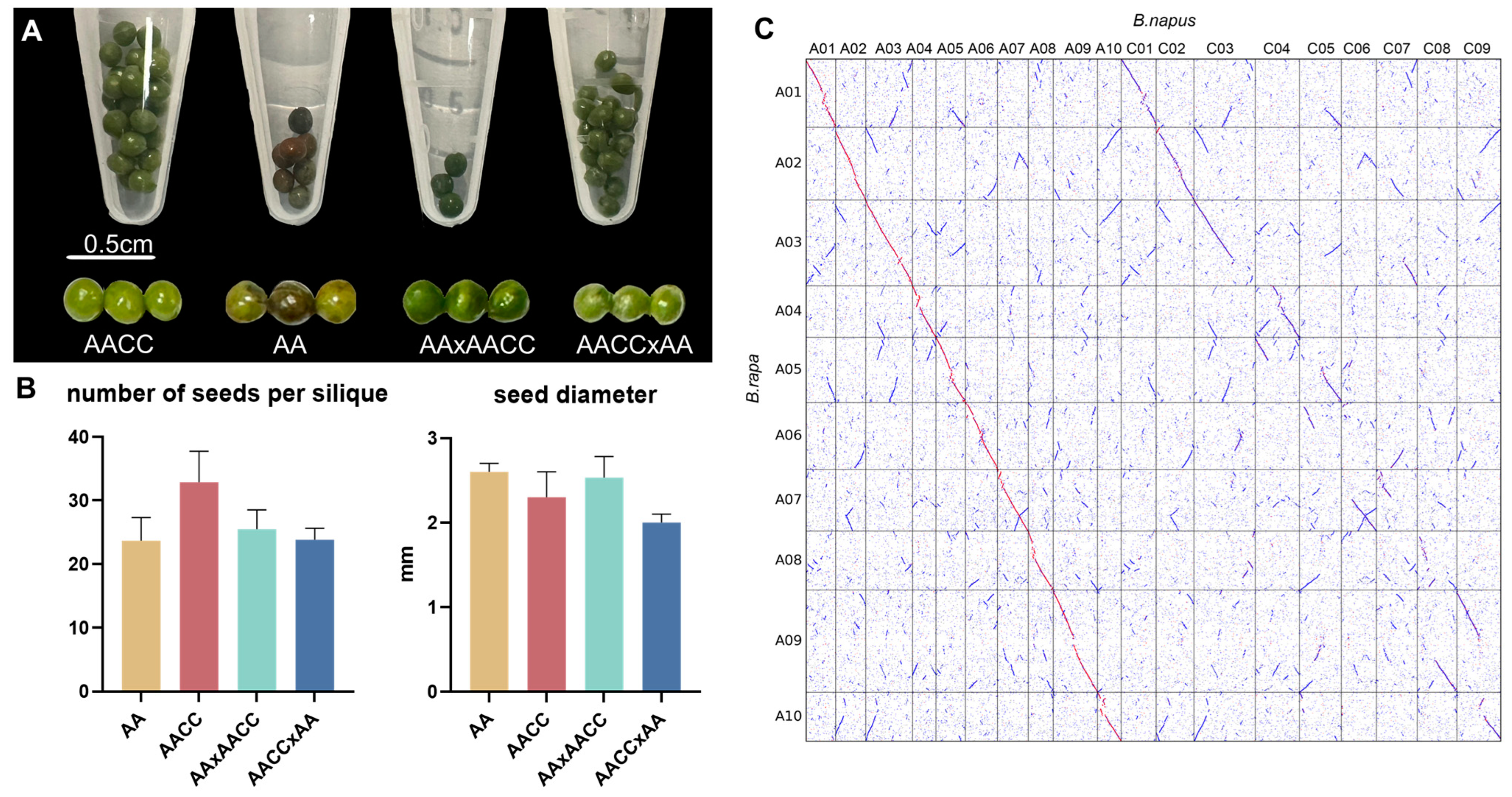
2.1. Phenotypic Observations of the Parents and Their Hybrid
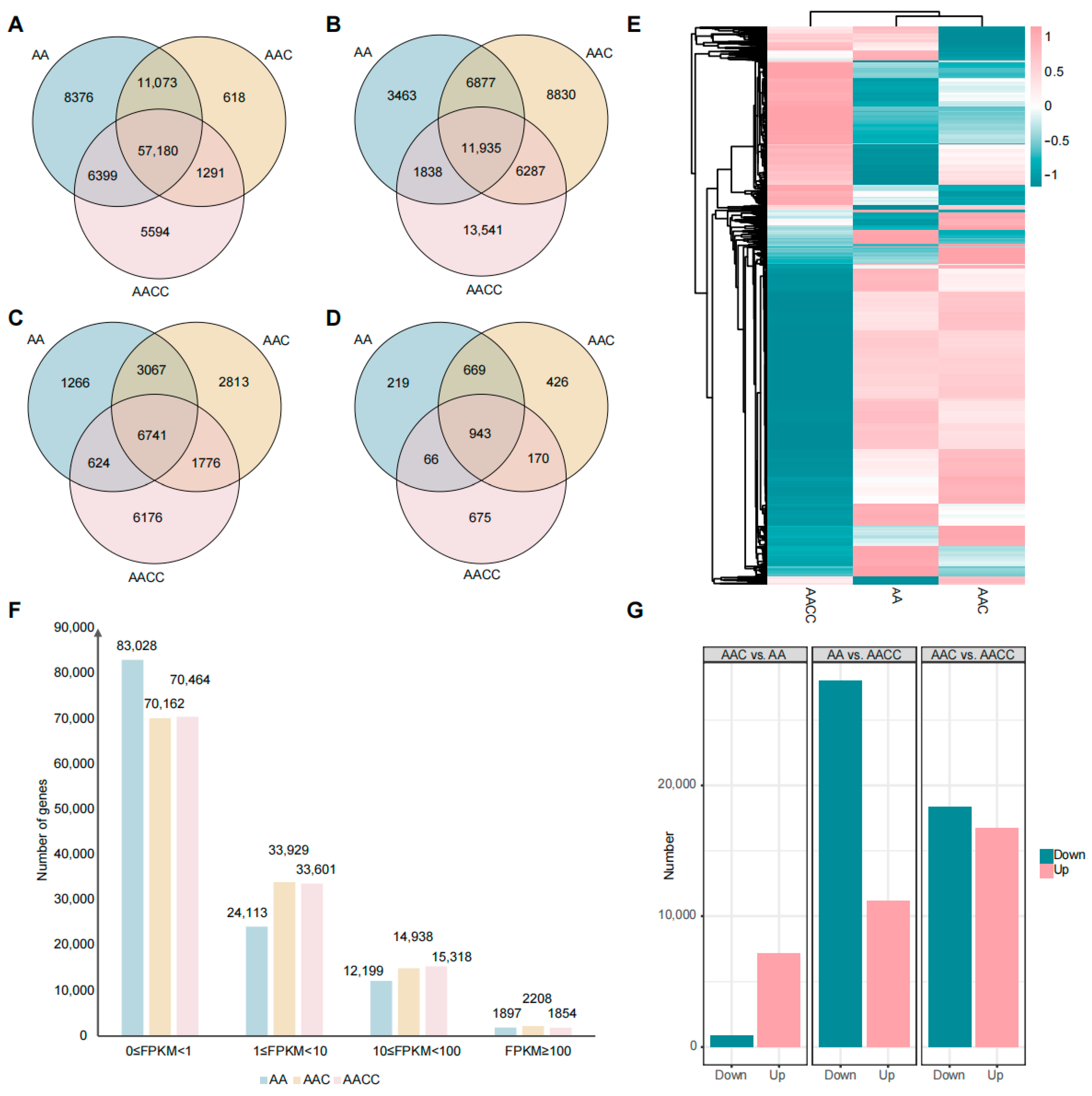
2.2. Global Gene Expression Analysis of the Parents and Interspecific Hybrids
2.3. Differential Expression Profiling of the Parents and F1 Hybrids
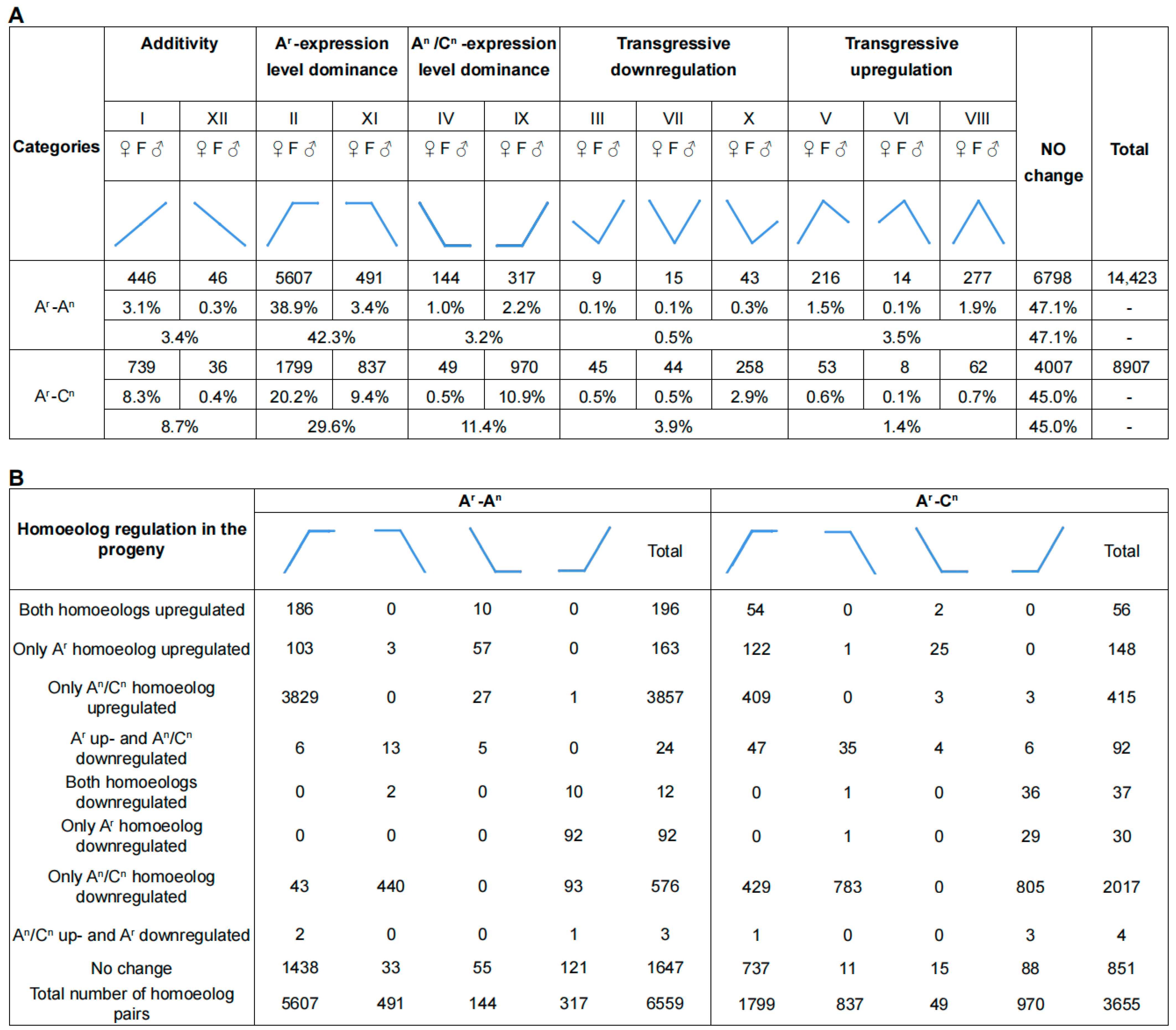
2.4. Genome-Wide Unbalanced Biased Expression toward B. rapa in the Hybrids
2.5. Genome-Wide Expression Level Dominance Biased toward AA in the Embryo of AAC
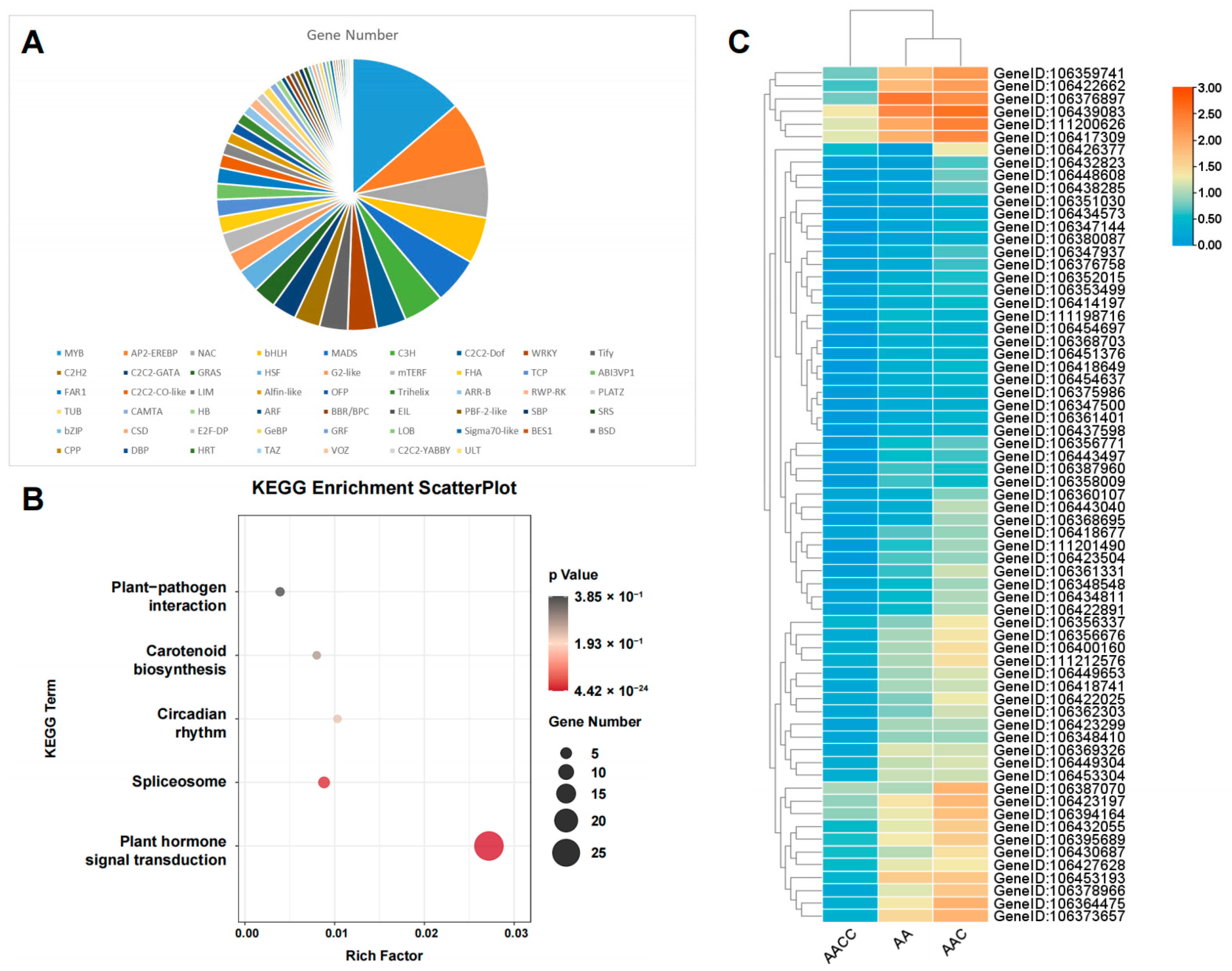
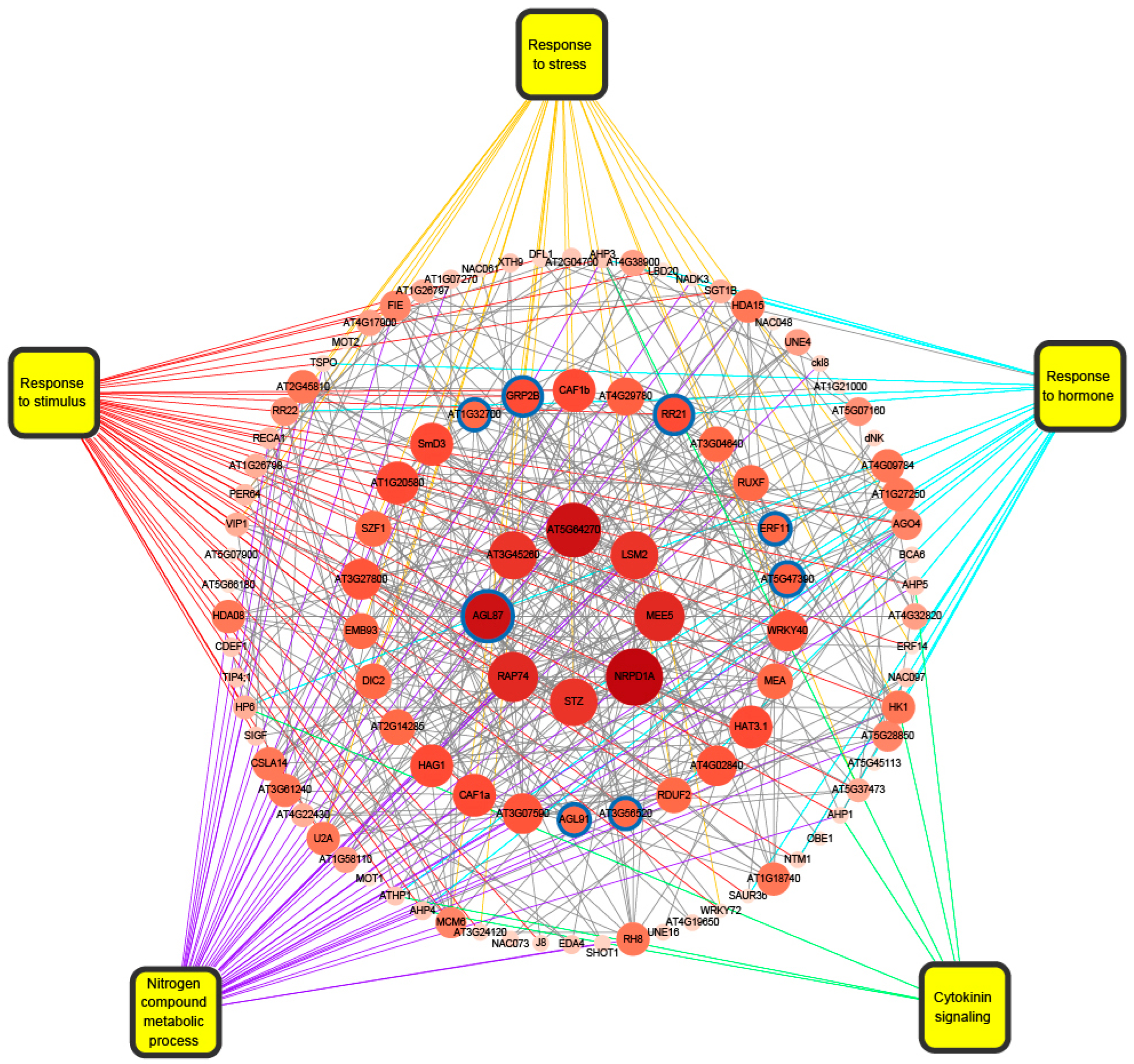
2.6. The Functional Analysis of ELD Genes
2.7. Transcription Factor Analysis of Non-Additive Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Collinearity Analysis
4.3. RNA Isolation, Sequencing, and Data Quality Detection
4.4. RT–qPCR Validations
4.5. Identification of Differentially Expressed Genes and Their GO and KEGG Enrichment Analysis
4.6. Annotation of Homoeologous Gene Pairs
4.7. Analysis of HEB and ELD in the Hybrids
4.8. Prediction and Analysis of Transcription Factors of ELD Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Renny-Byfield, S.; Wendel, J.F. Doubling down on Genomes: Polyploidy and Crop plants. Am. J. Bot. 2014, 101, 1711–1725. [Google Scholar] [CrossRef] [PubMed]
- Rothfels, C.J. Polyploid phylogenetics. New Phytol. 2021, 230, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Kopecky, D.; Martin, A.; Smykal, P. Interspecific hybridization and plant breeding: From historical retrospective through work of Mendel to current crops. Czech. J. Genet. Plant Breed. 2022, 58, 113–126. [Google Scholar] [CrossRef]
- Otto, S.P. The evolutionary consequences of polyploidy. Cell 2007, 131, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Botet, R.; Keurentjes, J.J.B. The Role of Transcriptional Regulation in Hybrid Vigor. Front. Plant Sci. 2020, 11, 410. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Dwivedi, K. Induced polyploidization in Brassica campestris L. (Brassicaceae). Cytol. Genet. 2014, 48, 103–110. [Google Scholar] [CrossRef]
- Qin, J.; Mo, R.; Li, H.; Ni, Z.; Sun, Q.; Liu, Z. The Transcriptional and Splicing Changes Caused by Hybridization Can Be Globally Recovered by Genome Doubling during Allopolyploidization. Mol. Biol. Evol. 2021, 38, 2513–2519. [Google Scholar] [CrossRef]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef]
- Hegarty, M.J.; Barker, G.L.; Wilson, I.D.; Abbott, R.J.; Edwards, K.J.; Hiscock, S.J. Transcriptome shock after interspecific hybridization in Senecio is ameliorated by genome duplication. Curr. Biol. 2006, 16, 1652–1659. [Google Scholar] [CrossRef]
- Yoo, M.J.; Liu, X.X.; Pires, J.C.; Soltis, P.S.; Soltis, D.E. Nonadditive Gene Expression in Polyploids. Annu. Rev. Genet. 2014, 48, 485–517. [Google Scholar] [CrossRef] [PubMed]
- Quan, C.; Chen, G.; Li, S.; Jia, Z.; Yu, P.; Tu, J.; Shen, J.; Yi, B.; Fu, T.; Dai, C.; et al. Transcriptome shock in interspecific F1 allotriploid hybrids between Brassica species. J. Exp. Bot. 2022, 73, 2336–2353. [Google Scholar] [CrossRef] [PubMed]
- Grover, C.E.; Gallagher, J.P.; Szadkowski, E.P.; Yoo, M.J.; Flagel, L.E.; Wendel, J.F. Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol. 2012, 196, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, Y.G. Does one subgenome become dominant in the formation and evolution of a polyploid? Ann. Bot. 2023, 131, 11–16. [Google Scholar] [CrossRef]
- Li, M.; Wang, R.; Wu, X.; Wang, J. Homoeolog expression bias and expression level dominance (ELD) in four tissues of natural allotetraploid Brassica napus. BMC Genom. 2020, 21, 330. [Google Scholar] [CrossRef]
- Zhang, L.; He, J.; He, H.; Wu, J.; Li, M. Genome-wide unbalanced expression bias and expression level dominance toward Brassica oleracea in artificially synthesized intergeneric hybrids of Raphanobrassica. Hortic. Res. 2021, 8, 246. [Google Scholar] [CrossRef]
- Chen, Z.J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 2007, 58, 377–406. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Wendel, J.F. Cis-trans controls and regulatory novelty accompanying allopolyploidization. New Phytol. 2019, 221, 1691–1700. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Moose, S.P.; Hudson, M.E. Transposable elements, mRNA expression level and strand-specificity of small RNAs are associated with non-additive inheritance of gene expression in hybrid plants. BMC Plant Biol. 2015, 15, 168. [Google Scholar] [CrossRef]
- Cox, M.P.; Dong, T.; Shen, G.; Dalvi, Y.; Scott, D.B.; Ganley, A.R.D. An Interspecific Fungal Hybrid Reveals Cross-Kingdom Rules for Allopolyploid Gene Expression Patterns. PLoS Genet. 2014, 10, e1004180. [Google Scholar] [CrossRef]
- Alger, E.I.; Edger, P.P. One subgenome to rule them all: Underlying mechanisms of subgenome dominance. Curr. Opin. Plant Biol. 2020, 54, 108–113. [Google Scholar] [CrossRef]
- Tang, M.; Li, J.; Hu, X.; Sun, L.; Helal, M.M.U.; Chen, J.; Zhang, Y. Asymmetric Divergence in Transmitted SNPs of DNA Replication/Transcription and Their Impact on Gene Expression in Polyploid Brassica napus. Front. Genet. 2021, 12, 756172. [Google Scholar] [CrossRef]
- Vuylsteke, M.; van Eeuwijk, F.; Van Hummelen, P.; Kuiper, M.; Zabeau, M. Genetic analysis of variation in gene expression in Arabidopsis thaliana. Genetics 2005, 171, 1267–1275. [Google Scholar] [CrossRef]
- Yin, L.; Xu, G.; Yang, J.; Zhao, M. The Heterogeneity in the Landscape of Gene Dominance in Maize is Accompanied by Unique Chromatin Environments. Mol. Biol. Evol. 2022, 39, msac198. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Shi, Y.; Qi, Y.; Yu, H.; Zou, C.; Cao, X.; Sun, C.; Chen, B.; Wang, H. Epigenetic and Genetic Contribution for Expression Bias of Homologous Alleles in Polyploid Sugarcane. Agronomy 2022, 12, 2852. [Google Scholar] [CrossRef]
- Shao, L.; Xing, F.; Xu, C.; Zhang, Q.; Che, J.; Wang, X.; Song, J.; Li, X.; Xiao, J.; Chen, L.-L.; et al. Patterns of genome-wide allele-specific expression in hybrid rice and the implications on the genetic basis of heterosis. Proc. Natl. Acad. Sci. USA 2019, 116, 5653–5658. [Google Scholar] [CrossRef]
- Hu, G.; Koh, J.; Yoo, M.-J.; Chen, S.; Wendel, J.F. Gene-Expression Novelty in Allopolyploid Cotton: A Proteomic Perspective. Genetics 2015, 200, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, N.A. Genome Dominance and Interaction at the Gene Expression Level in Allohexaploid Wheat. Plant Cell 2014, 26, 1834. [Google Scholar] [CrossRef]
- Yoo, M.J.; Szadkowski, E.; Wendel, J.F. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 2013, 110, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Pea, G.; Ferron, S.; Gianfranceschi, L.; Krajewski, P.; Pe, M.E. Gene expression non-additivity in immature ears of a heterotic F-1 maize hybrid. Plant Sci. 2008, 174, 17–24. [Google Scholar] [CrossRef]
- Wu, J.; Lin, L.; Xu, M.; Chen, P.; Liu, D.; Sun, Q.; Ran, L.; Wang, Y. Homoeolog expression bias and expression level dominance in resynthesized allopolyploid Brassica napus. BMC Genom. 2018, 19, 586. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.H.; Meng, J.L. RFLP and AFLP analysis of inter- and intraspecific variation of Brassica rapa and B. napus shows that B. rapa is an important genetic resource for B. napus improvement. Acta Genet. Sin. 2006, 33, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Bocianowski, J.; Niemann, J.; Nowosad, K. Genotype-by-environment interaction for seed quality traits in interspecific cross-derived Brassica lines using additive main effects and multiplicative interaction model. Euphytica 2019, 215, 7. [Google Scholar] [CrossRef]
- Sohn, S.-I.; Thamilarasan, S.K.; Pandian, S.; Oh, Y.-J.; Kang, H.-J.; Shin, E.-K. Characteristics and Fitness Analysis through Interspecific Hybrid Progenies of Transgenic Brassica napus and B. rapa L. ssp. Int. J. Mol. Sci. 2022, 23, 10512. [Google Scholar] [CrossRef]
- Zhang, W. Piecing together the puzzles of allopolyploid evolution in six Brassica crops. Plant Physiol. 2021, 186, 198–200. [Google Scholar] [CrossRef] [PubMed]
- Sohn, S.-I.; Thamilarasan, S.K.; Pandian, S.; Oh, Y.-J.; Ryu, T.-H.; Lee, G.-S.; Shin, E.-K. Interspecific Hybridization of Transgenic Brassica napus and Brassica rapa—An Overview. Genes 2022, 13, 1442. [Google Scholar] [CrossRef] [PubMed]
- Hauser, T.P.; Ostergard, H. Precocious germination of Brassica rapa x B-napus seeds within pods. Hereditas 1999, 130, 89–93. [Google Scholar] [CrossRef]
- Daurova, A.; Daurov, D.; Volkov, D.; Zhapar, K.; Raimbek, D.; Shamekova, M.; Zhambakin, K. Doubled haploids of interspecific hybrids between Brassica napus and Brassica rapa for canola production with valuable breeding traits. Ocl Oilseeds Fats Crops Lipids 2020, 27, 45. [Google Scholar] [CrossRef]
- Hauser, T.P.; Shaw, R.G.; Ostergard, H. Fitness of F-1 hybrids between weedy Brassica rapa and oilseed rape (B-napus). Heredity 1998, 81, 429–435. [Google Scholar] [CrossRef][Green Version]
- Lin, F.; Cao, J.; Yuan, J.; Liang, Y.; Li, J. Integration of Light and Brassinosteroid Signaling during Seedling Establishment. Int. J. Mol. Sci. 2021, 22, 12971. [Google Scholar] [CrossRef]
- Jurasek, M.; Drasar, P. About the Miracle of Nature from Rapeseed Pollen. Chem. Listy 2022, 116, 223–227. [Google Scholar] [CrossRef]
- Kutschera, U.; Wang, Z.-Y. Brassinosteroid action in flowering plants: A Darwinian perspective. J. Exp. Bot. 2012, 63, 3511–3522. [Google Scholar] [CrossRef] [PubMed]
- Day, R.C.; Herridge, R.P.; Ambrose, B.A.; Macknight, R.C. Transcriptome Analysis of Proliferating Arabidopsis Endosperm Reveals Biological Implications for the Control of Syncytial Division, Cytokinin Signaling, and Gene Expression Regulation. Plant Physiol. 2008, 148, 1964–1984. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Chen, J.-H.; Zhang, W.; Yang, G.; Yu, L.-J.; Li, D.-M.; Li, B.; Sheng, H.-M.; Zhang, H.; An, L.-Z. BYPASS1-LIKE, A DUF793 Family Protein, Participates in Freezing Tolerance via the CBF Pathway in Arabidopsis. Front. Plant Sci. 2019, 10, 807. [Google Scholar] [CrossRef] [PubMed]
- Meng, W.J.; Cheng, Z.J.; Sang, Y.L.; Zhang, M.M.; Rong, X.F.; Wang, Z.W.; Tang, Y.Y.; Zhang, X.S. Type-B ARABIDOPSIS RESPONSE REGULATORs Specify the Shoot Stem Cell Niche by Dual Regulation of WUSCHEL. Plant Cell 2017, 29, 1357–1372. [Google Scholar] [CrossRef]
- Kwon, Y.; Kim, J.H.; Nguyen, H.N.; Jikumaru, Y.; Kamiya, Y.; Hong, S.-W.; Lee, H. A novel Arabidopsis MYB-like transcription factor, MYBH, regulates hypocotyl elongation by enhancing auxin accumulation. J. Exp. Bot. 2013, 64, 3911–3922. [Google Scholar] [CrossRef]
- Robles, P.; Micol, J.L.; Quesada, V. Arabidopsis MDA1, a Nuclear-Encoded Protein, Functions in Chloroplast Development and Abiotic Stress Responses. PLoS ONE 2012, 7, e42924. [Google Scholar] [CrossRef]
- Song, W.; Koh, S.; Czako, M.; Marton, L.; Drenkard, E.; Becker, J.M.; Stacey, G. Antisense Expression of the Peptide Transport Gene AtPTR2-B Delays Flowering and Arrests Seed Development in Transgenic Arabidopsis Plants. Plant Physiol. 1997, 114, 927–935. [Google Scholar] [CrossRef][Green Version]
- Meinke, D.W. Genome-wide identification of EMBRYO-DEFECTIVE (EMB) genes required for growth and development in Arabidopsis. New Phytol. 2020, 226, 306–325. [Google Scholar] [CrossRef]
- Liu, M.; Yuan, L.; Liu, N.Y.; Shi, D.Q.; Liu, J.; Yang, W.C. GAMETOPHYTIC FACTOR 1, Involved in Pre-mRNA Splicing, Is Essential for Megagametogenesis and Embryogenesis in Arabidopsis. J. Integr. Plant Biol. 2009, 51, 261–271. [Google Scholar] [CrossRef]
- Zhu, D.Z.; Zhao, X.F.; Liu, C.Z.; Ma, F.F.; Wang, F.; Gao, X.-Q.; Zhang, X.S. Interaction between RNA helicase ROOT INITIATION DEFECTIVE 1 and GAMETOPHYTIC FACTOR 1 is involved in female gametophyte development in Arabidopsis. J. Exp. Bot. 2016, 67, 5757–5768. [Google Scholar] [CrossRef]
- Perea-Resa, C.; Hernández-Verdeja, T.; López-Cobollo, R.; Castellano, M.d.M.; Salinas, J. LSM Proteins Provide Accurate Splicing and Decay of Selected Transcripts to Ensure Normal Arabidopsis Development. Plant Cell 2012, 24, 4930–4947. [Google Scholar] [CrossRef]
- Babiychuk, E.; Hoang, K.T.; Vandepoele, K.; Van De Slijke, E.; Geelen, D.; De Jaeger, G.; Obokata, J.; Kushnir, S. The mutation nrpb1-A325V in the largest subunit of RNA polymerase II suppresses compromised growth of Arabidopsis plants deficient in a function of the general transcription factor IIF. Plant J. 2017, 89, 730–745. [Google Scholar] [CrossRef]
- Arjmand, M.P.; Lahiji, H.S.; Golfazani, M.M.; Biglouei, M.H. New insights on the regulatory network of drought-responsive key genes in Arabidopsis thaliana. Genetica 2023, 151, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Nie, S.; Zhang, N.; Tian, M.; Zhang, L. Transcriptome Analysis Reveals a Major Gene Expression Pattern and Important Metabolic Pathways in the Control of Heterosis in Chinese Cabbage. Plants 2023, 12, 1195. [Google Scholar] [CrossRef] [PubMed]
- Niemann, J.; Bocianowski, J.; Stuper-Szablewska, K.; Wojciechowski, T. New Interspecific Brassica Hybrids with High Levels of Heterosis for Fatty Acids Composition. Agriculture 2020, 10, 221. [Google Scholar] [CrossRef]
- Wolko, J.; Dobrzycka, A.; Bocianowski, J.; Bartkowiak-Broda, I. Estimation of heterosis for yield-related traits for single cross and three-way cross hybrids of oilseed rape (Brassica napus L.). Euphytica 2019, 215, 156. [Google Scholar] [CrossRef]
- Bansal, P.; Banga, S.; Banga, S.S. Heterosis as Investigated in Terms of Polyploidy and Genetic Diversity Using Designed Brassica juncea Amphiploid and Its Progenitor Diploid Species. PLoS ONE 2012, 7, e29607. [Google Scholar] [CrossRef]
- Zhang, J.; Li, G.; Li, H.; Pu, X.; Jiang, J.; Chai, L.; Zheng, B.; Cui, C.; Yang, Z.; Zhu, Y.; et al. Transcriptome Analysis of Interspecific Hybrid between Brassica napus and B. rapa Reveals Heterosis for Oil Rape Improvement. Int. J. Genom. 2015, 230985. [Google Scholar]
- Itoh, J.; Nonomura, K.; Ikeda, K.; Yamaki, S.; Inukai, Y.; Yamagishi, H.; Kitano, H.; Nagato, Y. Rice plant development: From zygote to spikelet. Plant Cell Physiol. 2005, 46, 23–47. [Google Scholar] [CrossRef]
- Gallardo, K.; Le Signor, C.; Vandekerckhove, J.; Thompson, R.D.; Burstin, J. Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiol. 2003, 133, 664–682. [Google Scholar] [CrossRef]
- Zhang, D.; Pan, Q.; Cui, C.; Tan, C.; Ge, X.; Shao, Y.; Li, Z. Genome-specific differential gene expressions in resynthesized Brassica allotetraploids from pair-wise crosses of three cultivated diploids revealed by RNA-seq. Front. Plant Sci. 2015, 6, 957. [Google Scholar] [CrossRef]
- Jiang, J.; Shao, Y.; Du, K.; Ran, L.; Fang, X.; Wang, Y. Use of digital gene expression to discriminate gene expression differences in early generations of resynthesized Brassica napus and its diploid progenitors. BMC Genom. 2013, 14, 72. [Google Scholar] [CrossRef]
- Wei, Y.; Li, G.; Zhang, S.; Zhang, S.; Zhang, H.; Sun, R.; Zhang, R.; Li, F. Analysis of Transcriptional Changes in Different Brassica napus Synthetic Allopolyploids. Genes 2021, 12, 82. [Google Scholar] [CrossRef]
- Quan, C.; Li, Y.; Chen, G.; Tian, X.; Jia, Z.; Tu, J.; Shen, J.; Yi, B.; Fu, T.; Ma, C.; et al. The dynamics of lncRNAs transcription in interspecific F1 allotriploid hybrids between Brassica species. Genomics 2022, 114, 110505. [Google Scholar] [CrossRef]
- Smith, L.M.; Weigel, D. On epigenetics and epistasis: Hybrids and their non-additive interactions. EMBO J. 2012, 31, 249–250. [Google Scholar] [CrossRef] [PubMed]
- Sheikhi, S.; Ebrahimi, A.; Heidari, P.; Amerian, M.R.; Rashidi-Monfared, S.; Alipour, H. Exogenous 24-epibrassinolide ameliorates tolerance to high-temperature by adjusting the biosynthesis of pigments, enzymatic, non-enzymatic antioxidants, and diosgenin content in fenugreek. Sci. Rep. 2023, 13, 6661. [Google Scholar] [CrossRef] [PubMed]
- Mahati, K.; Padmasree, K. Brassinolide promotes interaction between chloroplasts and mitochondria during the optimization of photosynthesis by the mitochondrial electron transport chain in mesophyll cell protoplasts of Arabidopsis thaliana. Front. Plant Sci. 2023, 14, 1099474. [Google Scholar] [CrossRef]
- Li, G.; Yao, X.; Chen, Z.; Tian, X.; Lu, L. The Overexpression of Oryza sativa L. CYP85A1 Promotes Growth and Biomass Production in Transgenic Trees. Int. J. Mol. Sci. 2023, 24, 6480. [Google Scholar] [CrossRef] [PubMed]
- An, Y.-Q.; Qin, Z.-T.; Li, D.-D.; Zhao, R.-Q.; Bi, B.-S.; Wang, D.-W.; Ma, D.-J.; Xi, Z. The combined formulation of brassinolide and pyraclostrobin increases biomass and seed yield by improving photosynthetic capacity in Arabidopsis thaliana. Front. Plant Sci. 2023, 14, 1138563. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, L.; Cui, Y.; Yang, Y.; Wu, J.; Liang, J.; Li, X.; Zhang, X.; Zhang, Y.; Guo, Z.; et al. The lack of negative association between TE load and subgenome dominance in synthesized Brassica allotetraploids. Proc. Natl. Acad. Sci. USA 2023, 120, e2305208120. [Google Scholar] [CrossRef] [PubMed]
- de Jong, G.W.; Adams, K.L. Subgenome-dominant expression and alternative splicing in response to Sclerotinia infection in polyploid Brassica napus and progenitors. Plant J. 2023, 114, 142–158. [Google Scholar] [CrossRef] [PubMed]
- Orantes-Bonilla, M.; Wang, H.; Lee, H.T.; Golicz, A.A.A.; Hu, D.; Li, W.; Zou, J.; Snowdon, R.J.J. Transgressive and parental dominant gene expression and cytosine methylation during seed development in Brassica napus hybrids. Theor. Appl. Genet. 2023, 136, 113. [Google Scholar] [CrossRef]
- Zhu, W.; Xie, Z.; Chu, Z.; Ding, Y.; Shi, G.; Chen, W.; Wei, X.; Yuan, Y.; Wei, F.; Tian, B. The Chromatin Remodeling Factor BrCHR39 Targets DNA Methylation to Positively Regulate Apical Dominance in Brassica rapa. Plants 2023, 12, 1384. [Google Scholar] [CrossRef]
- Zhou, R.; Moshgabadi, N.; Adams, K.L. Extensive changes to alternative splicing patterns following allopolyploidy in natural and resynthesized polyploids. Proc. Natl. Acad. Sci. USA 2011, 108, 16122–16127. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhang, K.; Yan, S.; Tang, M.; Zhou, W.; Yin, Y.; Chen, C.; Li, M. Genome-scale targeted mutagenesis in Brassica napus using a pooled CRISPR library. Genome Res. 2023, 33, 798–809. [Google Scholar] [CrossRef]
- Sun, P.; Jiao, B.; Yang, Y.; Shan, L.; Li, T.; Li, X.; Xi, Z.; Wang, X.; Liu, J. WGDI: A user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. Cold Spring Harb. Lab. 2021, 15, 1841–1851. [Google Scholar] [CrossRef]
- Guo, L.; Chao, H.; Yin, Y.; Li, H.; Wang, H.; Zhao, W.; Hou, D.; Zhang, L.; Zhang, C.; Li, M. New insight into the genetic basis of oil content based on noninvasive three-dimensional phenotyping and tissue-specific transcriptome in Brassica napus. Biotechnol. Biofuels Bioprod. 2023, 16, 88. [Google Scholar] [CrossRef]
- Ding, Y.; Yu, S.; Wang, J.; Li, M.; Qu, C.; Li, J.; Liu, L. Comparative transcriptomic analysis of seed coats with high and low lignin contents reveals lignin and flavonoid biosynthesis in Brassica napus. BMC Plant Biol. 2021, 21, 246. [Google Scholar] [CrossRef]
- Chen, K.; Huang, Y.; Liu, C.; Liang, Y.; Li, M. Transcriptome Profile Analysis of Arabidopsis Reveals the Drought Stress-Induced Long Non-coding RNAs Associated With Photosynthesis, Chlorophyll Synthesis, Fatty Acid Synthesis and Degradation. Front. Plant Sci. 2021, 12, 643182. [Google Scholar] [CrossRef]
- Glover, N.M.; Redestig, H.; Dessimoz, C. Homoeologs: What Are They and How Do We Infer Them? Trends Plant Sci. 2016, 21, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Rapp, R.A.; Udall, J.A.; Wendel, J.F. Genomic expression dominance in allopolyploids. BMC Biol. 2009, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Hao, X.; He, S.; Hao, X.; Zhang, P.; Chen, X. Genome-wide identification, phylogeny and expression analysis of AP2/ERF transcription factors family in sweet potato. BMC Genom. 2021, 22, 748. [Google Scholar] [CrossRef] [PubMed]

| Additivity | Ar-Expression Level Dominance | An/Cn-Expression Level Dominance | Transgressive Up-Regulation or Transgressive Down-Regulation | |
|---|---|---|---|---|
| Ar-An | brassinosteroid mediated signaling pathway | translation | DNA replication | response to absence of light |
| translation | ubiquitin-dependent protein catabolic process | cellular response to water deprivation | polysaccharide biosynthetic process | |
| Golgi apparatus | chloroplast | cellular response to salt stress | regulation of plant organ formation | |
| mitochondrial matrix | mitochondrial matrix | lignan biosynthetic process | drug transport | |
| origin recognition complex | GTP binding | plant seed peroxidase activity | regulation of flower development | |
| catalytic activity | ligase activity | transferase activity | alcohol dehydrogenase (NAD) activity | |
| Ar-Cn | translation | photosynthesis | small GTPase mediated signal transduction | cellular amino acid biosynthetic process |
| cellular amino acid biosynthetic process | phosphorylation | DNA replication | methionine biosynthetic process | |
| Golgi apparatus | negative regulation of cell death | protein transport | glycogen metabolic process | |
| cytosol | brassinosteroid homeostasis | DNA replication initiation | microtubule-based process | |
| RNA binding | chloroplast | protein folding | chloroplast stroma | |
| endopeptidase activity | ATP binding | chloroplast | plant-type cell wall |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, W.; Zhang, L.; He, J.; Chen, W.; Zhao, F.; Fu, C.; Li, M. Transcriptome Shock in Developing Embryos of a Brassica napus and Brassica rapa Hybrid. Int. J. Mol. Sci. 2023, 24, 16238. https://doi.org/10.3390/ijms242216238
Zhou W, Zhang L, He J, Chen W, Zhao F, Fu C, Li M. Transcriptome Shock in Developing Embryos of a Brassica napus and Brassica rapa Hybrid. International Journal of Molecular Sciences. 2023; 24(22):16238. https://doi.org/10.3390/ijms242216238
Chicago/Turabian StyleZhou, Weixian, Libin Zhang, Jianjie He, Wang Chen, Feifan Zhao, Chunhua Fu, and Maoteng Li. 2023. "Transcriptome Shock in Developing Embryos of a Brassica napus and Brassica rapa Hybrid" International Journal of Molecular Sciences 24, no. 22: 16238. https://doi.org/10.3390/ijms242216238
APA StyleZhou, W., Zhang, L., He, J., Chen, W., Zhao, F., Fu, C., & Li, M. (2023). Transcriptome Shock in Developing Embryos of a Brassica napus and Brassica rapa Hybrid. International Journal of Molecular Sciences, 24(22), 16238. https://doi.org/10.3390/ijms242216238






