The Effect of Ficin Immobilized on Carboxymethyl Chitosan on Biofilms of Oral Pathogens
Abstract
:1. Introduction
2. Results
2.1. Immobilization of Ficin on Carboxymethyl Chitosan
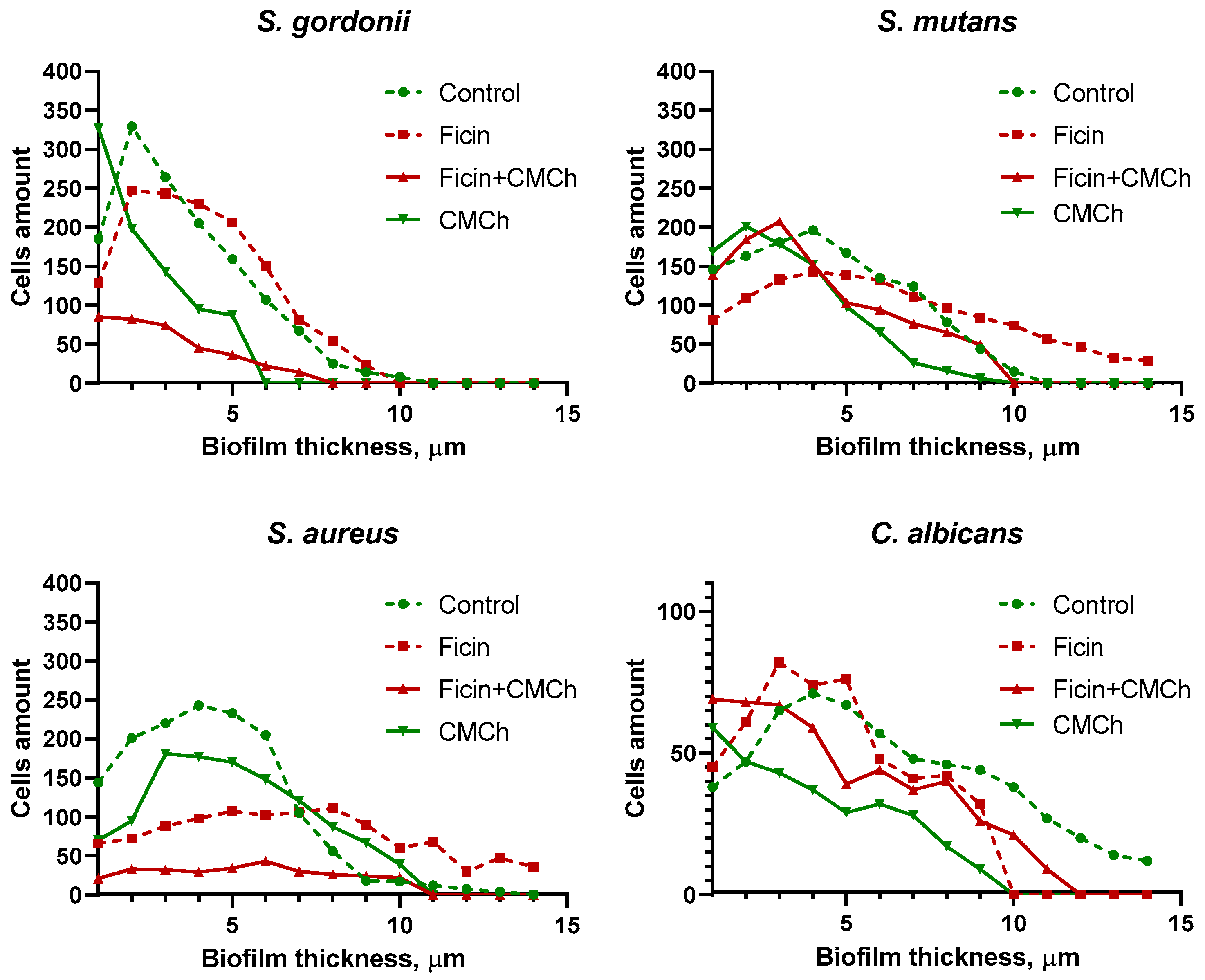
2.2. Anti-Biofilm Properties of Immobilized Ficin
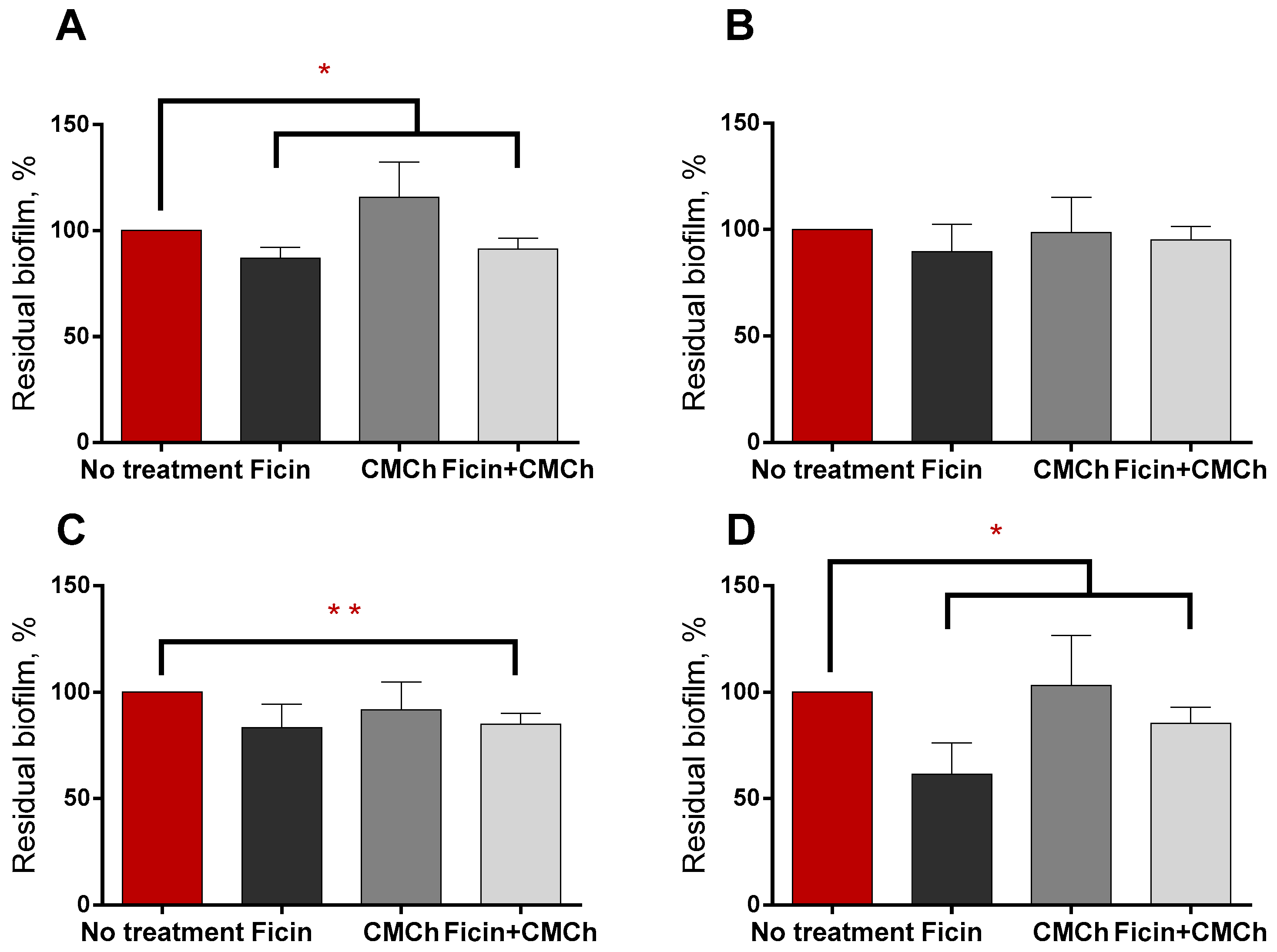
2.3. Anti-Biofilm Properties of Immobilized Ficin on Mixed Biofilms
2.4. Increasing the Efficiency of Antimicrobials against Staphylococcal Biofilms by Soluble and Chitosan-Immobilized Ficin
3. Discussion
4. Materials and Methods
4.1. Synthesis of Carboxymethyl Chitosan
4.2. Ficin Immobilization on Carboxymethyl Chitosan, Enzymatic Activity Measurements
4.3. Bacterial Strains and Growth Conditions
4.4. Biofilm Assays
4.5. Metagenomics
4.6. Scanning Electron Microscopy
4.7. Confocal Laser Scanning Microscopy
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deo, P.N.; Deshmukh, R. Oral microbiome: Unveiling the fundamentals. J. Oral Maxillofac. Pathol. 2019, 23, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Soares, J.C.M.; Luiz, M.T.; Oshiro Junior, J.A.; Besegato, J.F.; de Melo, P.B.G.; Rastelli, A.N.S.; Chorilli, M. Antimicrobial photodynamic therapy mediated by methylene blue-loaded polymeric micelles against Streptococcus mutans and Candida albicans biofilms. Photodiagn. Photodyn. Ther. 2023, 41, 103285. [Google Scholar] [CrossRef] [PubMed]
- Berger, D.; Rakhamimova, A.; Pollack, A.; Loewy, Z. Oral Biofilms: Development, Control, and Analysis. High Throughput 2018, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Mitterer, F.; Pombo, J.P.; Schild, S. Biofilms by bacterial human pathogens: Clinical relevance—Development, composition and regulation—Therapeutical strategies. Microb. Cell 2021, 8, 28–56. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Valen, H.; Scheie, A.A. Biofilms and their properties. Eur. J. Oral Sci. 2018, 126 (Suppl. S1), 13–18. [Google Scholar] [CrossRef]
- Abebe, G.M. Oral biofilm and its impact on oral health, psychological and social interaction. Int. J. Oral Dent. Health 2021, 7, 127–137. [Google Scholar]
- Qiu, W.; Zhou, Y.; Li, Z.; Huang, T.; Xiao, Y.; Cheng, L.; Peng, X.; Zhang, L.; Ren, B. Application of Antibiotics/Antimicrobial Agents on Dental Caries. BioMed Res. Int. 2020, 2020, 5658212. [Google Scholar] [CrossRef]
- Ostadhossein, F.; Moitra, P.; Altun, E.; Dutta, D.; Sar, D.; Tripathi, I.; Hsiao, S.H.; Kravchuk, V.; Nie, S.; Pan, D. Function-adaptive clustered nanoparticles reverse Streptococcus mutans dental biofilm and maintain microbiota balance. Commun. Biol. 2021, 4, 846. [Google Scholar] [CrossRef]
- Kassebaum, N.; Bernabé, E.; Dahiya, M.; Bhandari, B.; Murray, C.; Marcenes, W. Global burden of untreated caries: A systematic review and metaregression. J. Dent. Res. 2015, 94, 650–658. [Google Scholar] [CrossRef]
- Yu, O.Y.; Lam, W.Y.; Wong, A.W.; Duangthip, D.; Chu, C.H. Nonrestorative Management of Dental Caries. Dent. J. 2021, 9, 121. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Macleod, L.C.; Kitten, T.; Xu, P. Streptococcus sanguinis biofilm formation & interaction with oral pathogens. Future Microbiol. 2018, 13, 915–932. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Wang, L.L.; Lin, Y.Q.; Liang, H.M.; Zhou, S.Y.; Zheng, F.; Feng, X.L.; Rui, Y.Y.; Shao, L.Q. Nanoparticles for the Treatment of Oral Biofilms: Current State, Mechanisms, Influencing Factors, and Prospects. Adv. Healthc. Mater. 2019, 8, e1901301. [Google Scholar] [CrossRef] [PubMed]
- Mora-Martínez, A.; Murcia, L.; Rodríguez-Lozano, F.J. Oral Manifestations of Mucormycosis: A Systematic Review. J. Fungi 2023, 9, 935. [Google Scholar] [CrossRef]
- Mishra, S.; Gupta, A.; Upadhye, V.; Singh, S.C.; Sinha, R.P.; Häder, D.P. Therapeutic Strategies against Biofilm Infections. Life 2023, 13, 172. [Google Scholar] [CrossRef]
- Xiao, J.; Klein, M.I.; Falsetta, M.L.; Lu, B.; Delahunty, C.M.; Yates, J.R.; Heydorn, A.; Koo, H. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012, 8, e1002623. [Google Scholar] [CrossRef]
- Morel du Boil, P.; Wienese, S. Enzymic reduction of dextran in process-laboratory evaluation of dextranases. In Proceedings of the Annual Congress South African Sugar Technologists’ Association, Durban, South Africa, 30 July–2 August 2002; Volume 76, pp. 435–443. [Google Scholar]
- Ren, W.; Cai, R.; Yan, W.; Lyu, M.; Fang, Y.; Wang, S. Purification and Characterization of a Biofilm-Degradable Dextranase from a Marine Bacterium. Mar. Drugs 2018, 16, 51. [Google Scholar] [CrossRef]
- Karygianni, L.; Attin, T.; Thurnheer, T. Combined DNase and Proteinase Treatment Interferes with Composition and Structural Integrity of Multispecies Oral Biofilms. J. Clin. Med. 2020, 9, 983. [Google Scholar] [CrossRef]
- Lin, Y.; Zhou, X.; Li, Y. Strategies for Streptococcus mutans biofilm dispersal through extracellular polymeric substances disruption. Mol. Oral. Microbiol. 2022, 37, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hukić, M.; Seljmo, D.; Ramovic, A.; Ibrišimović, M.A.; Dogan, S.; Hukic, J.; Bojic, E.F. The Effect of Lysozyme on Reducing Biofilms by Staphylococcus aureus, Pseudomonas aeruginosa, and Gardnerella vaginalis: An In Vitro Examination. Microb. Drug Resist. 2018, 24, 353–358. [Google Scholar] [CrossRef]
- Krzyściak, W.; Jurczak, A.; Piątkowski, J.; Kościelniak, D.; Gregorczyk-Maga, I.; Kołodziej, I.; Papież, M.A.; Olczak-Kowalczyk, D. Effect of histatin-5 and lysozyme on the ability of Streptococcus mutans to form biofilms in in vitro conditions. Postępy Higieny I Medycyny Doświadczalnej 2015, 69, 1056–1066. [Google Scholar]
- Shukla, S.K.; Rao, T.S. Dispersal of Bap-mediated Staphylococcus aureus biofilm by proteinase K. J. Antibiot. 2013, 66, 55–60. [Google Scholar] [CrossRef] [PubMed]
- (24] Eladawy, M.; El-Mowafy, M.; El-Sokkary, M.M.A.; Barwa, R. Effects of lysozyme, proteinase K, and cephalosporins on biofilm formation by clinical isolates of Pseudomonas aeruginosa. Interdiscip. Perspect. Infect. Dis. 2020, 2020, 6156720. [Google Scholar] [CrossRef]
- Niazi, S.A.; Clark, D.; Do, T.; Gilbert, S.C.; Foschi, F.; Mannocci, F.; Beighton, D. The effectiveness of enzymic irrigation in removing a nutrient-stressed endodontic multispecies biofilm. Int. Endod. J. 2014, 47, 756–768. [Google Scholar] [CrossRef]
- Praveen, N.C.; Rajesh, A.; Madan, M.; Chaurasia, V.R.; Hiremath, N.V.; Sharma, A.M. In vitro Evaluation of Antibacterial Efficacy of Pineapple Extract (Bromelain) on Periodontal Pathogens. J. Int. Oral Health 2014, 6, 96–98. [Google Scholar] [PubMed]
- Yang, Y.; Shen, D.; Long, Y.; Xie, Z.; Zheng, H. Intrinsic Peroxidase-like Activity of Ficin. Sci. Rep. 2017, 7, 43141. [Google Scholar] [CrossRef]
- Baidamshina, D.R.; Trizna, E.Y.; Holyavka, M.G.; Bogachev, M.I.; Artyukhov, V.G.; Akhatova, F.S.; Rozhina, E.V.; Fakhrullin, R.F.; Kayumov, A.R. Targeting microbial biofilms using Ficin, a nonspecific plant protease. Sci. Rep. 2017, 7, 46068. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, W.; Zhang, M.; Zhang, L.; Shen, Y.; Huang, S.; Li, M.; Qiu, W.; Pan, Y.; Zhou, L.; et al. The Inhibitory Effects of Ficin on. BioMed Res. Int. 2021, 2021, 6692328. [Google Scholar] [CrossRef]
- Yu, J.; Wang, F.; Shen, Y.; Yu, F.; Qiu, L.; Zhang, L.; Chen, Y.; Yuan, Q.; Zhang, H.; Sun, Y.; et al. Inhibitory effect of ficin on Candida albicans biofilm formation and pre-formed biofilms. BMC Oral Health 2022, 22, 350. [Google Scholar] [CrossRef]
- Baidamshina, D.R.; Koroleva, V.A.; Olshannikova, S.S.; Trizna, E.Y.; Bogachev, M.I.; Artyukhov, V.G.; Holyavka, M.G.; Kayumov, A.R. Biochemical Properties and Anti-Biofilm Activity of Chitosan-Immobilized Papain. Mar. Drugs 2021, 19, 197. [Google Scholar] [CrossRef]
- Gatina, A.; Trizna, E.; Kolesnikova, A.; Baidamshina, D.; Gorshkova, A.; Drucker, V.; Bogachev, M.; Kayumov, A. The Bovhyaluronidase Azoximer (Longidaza®) Disrupts Candida albicans and Candida albicans-Bacterial Mixed Biofilms and Increases the Efficacy of Antifungals. Medicina 2022, 58, 1710. [Google Scholar] [CrossRef] [PubMed]
- Elchinger, P.H.; Delattre, C.; Faure, S.; Roy, O.; Badel, S.; Bernardi, T.; Taillefumier, C.; Michaud, P. Immobilization of proteases on chitosan for the development of films with anti-biofilm properties. Int. J. Biol. Macromol. 2015, 72, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- Baidamshina, D.R.; Koroleva, V.A.; Trizna, E.Y.; Pankova, S.M.; Agafonova, M.N.; Chirkova, M.N.; Vasileva, O.S.; Akhmetov, N.; Shubina, V.V.; Porfiryev, A.G.; et al. Anti-biofilm and wound-healing activity of chitosan-immobilized Ficin. Int. J. Biol. Macromol. 2020, 164, 4205–4217. [Google Scholar] [CrossRef]
- Rostami, N.; Shields, R.C.; Serrage, H.J.; Lawler, C.; Brittan, J.L.; Yassin, S.; Ahmed, H.; Treumann, A.; Thompson, P.; Waldron, K.J.; et al. Interspecies competition in oral biofilms mediated by Streptococcus gordonii extracellular deoxyribonuclease SsnA. NPJ Biofilms Microbiomes 2022, 8, 96. [Google Scholar] [CrossRef] [PubMed]
- Donkor, E.S.; Kotey, F.C. Methicillin-Resistant Staphylococcus aureus in the Oral Cavity: Implications for Antibiotic Prophylaxis and Surveillance. Infect. Dis. 2020, 13, 1178633720976581. [Google Scholar] [CrossRef]
- Trizna, E.; Baidamshina, D.; Gorshkova, A.; Drucker, V.; Bogachev, M.; Tikhonov, A.; Kayumov, A. Improving the Efficacy of Antimicrobials against Biofilm-Embedded Bacteria Using Bovine Hyaluronidase Azoximer (Longidaza®). Pharmaceutics 2021, 13, 1740. [Google Scholar] [CrossRef]
- Fanaei Pirlar, R.; Emaneini, M.; Beigverdi, R.; Banar, M.; van Leeuwen, W.B.; Jabalameli, F. Combinatorial effects of antibiotics and enzymes against dual-species Staphylococcus aureus and Pseudomonas aeruginosa biofilms in the wound-like medium. PLoS ONE 2020, 15, e0235093. [Google Scholar] [CrossRef]
- Thallinger, B.; Prasetyo, E.N.; Nyanhongo, G.S.; Guebitz, G.M. Antimicrobial enzymes: An emerging strategy to fight microbes and microbial biofilms. Biotechnol. J. 2013, 8, 97–109. [Google Scholar] [CrossRef]
- Nahar, S.; Mizan, M.F.R.; Ha, A.J.W.; Ha, S.D. Advances and Future Prospects of Enzyme-Based Biofilm Prevention Approaches in the Food Industry. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1484–1502. [Google Scholar] [CrossRef]
- Harris, L.G.; Nigam, Y.; Sawyer, J.; Mack, D.; Pritchard, D.I. Lucilia sericata Chymotrypsin Disrupts Protein Adhesin-Mediated Staphylococcal Biofilm Formation. Appl. Environ. Microbiol. 2013, 79, 1393–1395. [Google Scholar] [CrossRef]
- Donelli, G.; Francolini, I.; Romoli, D.; Guaglianone, E.; Piozzi, A.; Ragunath, C.; Kaplan, J.B. Synergistic activity of dispersin B and cefamandole nafate in inhibition of staphylococcal biofilm growth on polyurethanes. Antimicrob. Agents Chemother. 2007, 51, 2733–2740. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.B. Therapeutic potential of biofilm-dispersing enzymes. Int. J. Artif. Organs 2009, 32, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.K.; Bindhu, O.S. Plant Latex: A Rich Source of Haemostatic Proteases. In Herbal Medicine in India; Springer: Singapore, 2020; pp. 143–153. [Google Scholar]
- Yariswamy, M.; Shivaprasad, H.V.; Joshi, V.; Urs, A.N.N.; Nataraju, A.; Vishwanath, B.S. Topical application of serine proteases from Wrightia tinctoria R. Br. (Apocyanaceae) latex augments healing of experimentally induced excision wound in mice. J. Ethnopharmacol. 2013, 149, 377–383. [Google Scholar] [CrossRef]
- Holyavka, M.G.; Goncharova, S.S.; Sorokin, A.V.; Lavlinskaya, M.S.; Redko, Y.A.; Faizullin, D.A.; Baidamshina, D.R.; Zuev, Y.F.; Kondratyev, M.S.; Kayumov, A.R. Novel Biocatalysts Based on Bromelain Immobilized on Functionalized Chitosans and Research on Their Structural Features. Polymers 2022, 14, 5110. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Berenguer-Murcia, A.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of Different Enzyme Immobilization Strategies to Improve Enzyme Performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzym. Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Fernandez-Lafuente, R. Stabilization of multimeric enzymes: Strategies to prevent subunit dissociation. Enzym. Microb. Technol. 2009, 45, 405–418. [Google Scholar] [CrossRef]
- Cowan, D.A.; Fernandez-Lafuente, R. Enhancing the functional properties of thermophilic enzymes by chemical modification and immobilization. Enzym. Microb. Technol. 2011, 49, 326–346. [Google Scholar] [CrossRef]
- Palmer, S.R.; Ren, Z.; Hwang, G.; Liu, Y.; Combs, A.; Söderström, B.; Lara Vasquez, P.; Khosravi, Y.; Brady, L.J.; Koo, H.; et al. Streptococcus mutans yidC1 and yidC2 Impact Cell Envelope Biogenesis, the Biofilm Matrix, and Biofilm Biophysical Properties. J. Bacteriol. 2019, 201, 10–1128. [Google Scholar] [CrossRef]
- Hernández, P.; Sánchez, M.C.; Llama-Palacios, A.; Ciudad, M.J.; Collado, L. Strategies to Combat Caries by Maintaining the Integrity of Biofilm and Homeostasis during the Rapid Phase of Supragingival Plaque Formation. Antibiotics 2022, 11, 880. [Google Scholar] [CrossRef] [PubMed]
- Radaic, A.; Kapila, Y.L. The oralome and its dysbiosis: New insights into oral microbiome-host interactions. Comput. Struct. Biotechnol. J. 2021, 19, 1335–1360. [Google Scholar] [CrossRef]
- Haffajee, A.D.; Teles, R.P.; Patel, M.R.; Song, X.; Veiga, N.; Socransky, S.S. Factors affecting human supragingival biofilm composition. I. Plaque mass. J. Periodontal Res. 2009, 44, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Hotz, P.; Guggenheim, B.; Schmid, R. Carbohydrates in pooled dental plaque. Caries Res. 1972, 6, 103–121. [Google Scholar] [CrossRef]
- Jakubovics, N.S.; Goodman, S.D.; Mashburn-Warren, L.; Stafford, G.P.; Cieplik, F. The dental plaque biofilm matrix. Periodontol 2000 2021, 86, 32–56. [Google Scholar] [CrossRef] [PubMed]
- Banas, J.A.; Vickerman, M.M. Glucan-binding proteins of the oral streptococci. Crit. Rev. Oral Biol. Med. 2003, 14, 89–99. [Google Scholar] [CrossRef]
- Mieher, J.L.; Larson, M.R.; Schormann, N.; Purushotham, S.; Wu, R.; Rajashankar, K.R.; Wu, H.; Deivanayagam, C. Glucan Binding Protein C of Streptococcus mutans Mediates both Sucrose-Independent and Sucrose-Dependent Adherence. Infect. Immun. 2018, 86, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Rozen, R.; Bachrach, G.; Bronshteyn, M.; Gedalia, I.; Steinberg, D. The role of fructans on dental biofilm formation by Streptococcus sobrinus, Streptococcus mutans, Streptococcus gordonii and Actinomyces viscosus. FEMS Microbiol. Lett. 2001, 195, 205–210. [Google Scholar] [CrossRef]
- Trizna, E.; Bogachev, M.I.; Kayumov, A. Degrading of the Pseudomonas Aeruginosa Biofilm by Extracellular Levanase SacC from Bacillus subtilis. Bionanoscience 2019, 9, 48–52. [Google Scholar] [CrossRef]
- Blanco-Cabra, N.; Paetzold, B.; Ferrar, T.; Mazzolini, R.; Torrents, E.; Serrano, L.; LLuch-Senar, M. Characterization of different alginate lyases for dissolving Pseudomonas aeruginosa biofilms. Sci. Rep. 2020, 10, 9390. [Google Scholar] [CrossRef]
- Lefebvre, E.; Vighetto, C.; Di Martino, P.; Garde, V.L.; Seyer, D. Synergistic antibiofilm efficacy of various commercial antiseptics, enzymes and EDTA: A study of Pseudomonas aeruginosa and Staphylococcus aureus biofilms. Int. J. Antimicrob. Agents 2016, 48, 181–188. [Google Scholar] [CrossRef]
- Chen, S.-C.; Wu, Y.-C.; Mi, F.-L.; Lin, Y.-H.; Yu, L.-C.; Sung, H.-W. A novel pH-sensitive hydrogel composed of N, O-carboxymethyl chitosan and alginate cross-linked by genipin for protein drug delivery. J. Control. Release 2004, 96, 285–300. [Google Scholar] [CrossRef] [PubMed]
- Malykhina, N.; Olshannikova, S.; Holyavka, M.; Sorokin, A.; Lavlinskaya, M.; Artyukhov, V.; Faizullin, D.; Zuev, Y.F. Preparation of Ficin Complexes with Carboxymethylchitosan and N-(2-Hydroxy) Propyl-3-Trimethylammoniumchitosan and Studies of Their Structural Features. Russ. J. Bioorg. Chem. 2022, 48 (Suppl. S1), S50–S60. [Google Scholar] [CrossRef]
- Sabirova, A.R.; Rudakova, N.L.; Balaban, N.P.; Ilyinskaya, O.N.; Demidyuk, I.V.; Kostrov, S.V.; Rudenskaya, G.N.; Sharipova, M.R. A novel secreted metzincin metalloproteinase from Bacillus intermedius. FEBS Lett. 2010, 584, 4419–4425. [Google Scholar] [CrossRef]
- Kayumov, A.R.; Khakimullina, E.N.; Sharafutdinov, I.S.; Trizna, E.Y.; Latypova, L.Z.; Lien, H.T.; Margulis, A.B.; Bogachev, M.I.; Kurbangalieva, A.R. Inhibition of biofilm formation in Bacillus subtilis by new halogenated furanones. J. Antibiot. 2015, 68, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Trizna, E.Y.; Khakimullina, E.N.; Latypova, L.Z.; Kurbangalieva, A.R.; Sharafutdinov, I.S.; Evtyugin, V.G.; Babynin, E.V.; Bogachev, M.I.; Kayumov, A.R. Thio Derivatives of 2(5H)-Furanone As Inhibitors against Bacillus subtilis Biofilms. Acta Nat. 2015, 7, 102–107. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A.; Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Bogachev, M.I.; Volkov, V.Y.; Markelov, O.A.; Trizna, E.Y.; Baydamshina, D.R.; Melnikov, V.; Murtazina, R.R.; Zelenikhin, P.V.; Sharafutdinov, I.S.; Kayumov, A.R. Fast and simple tool for the quantification of biofilm-embedded cells sub-populations from fluorescent microscopic images. PLoS ONE 2018, 13, e0193267. [Google Scholar] [CrossRef]

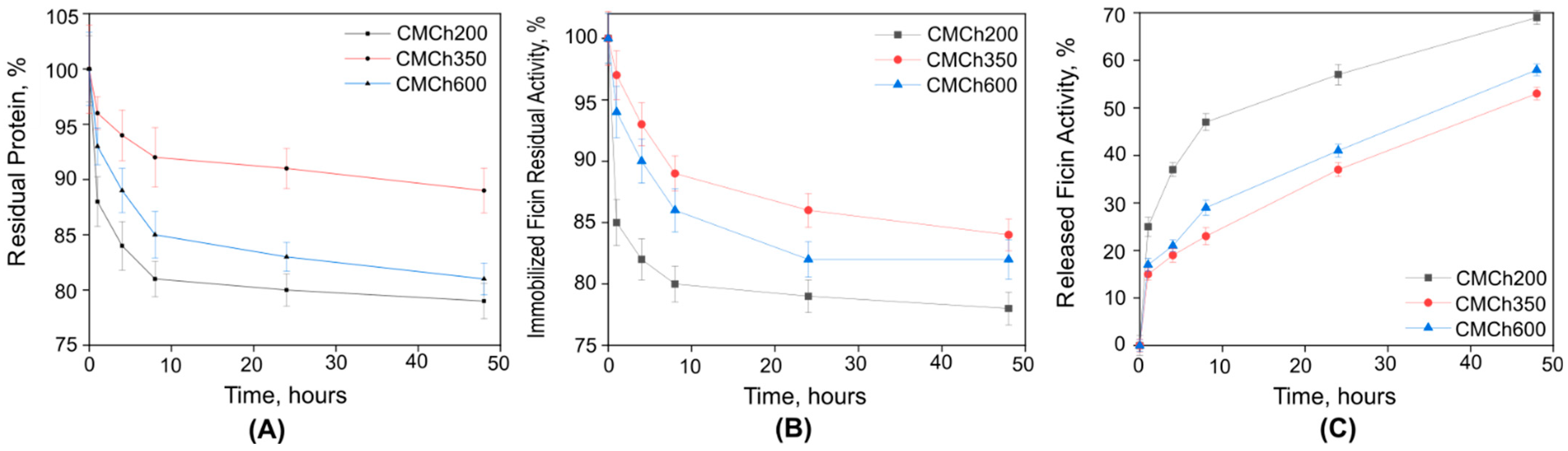
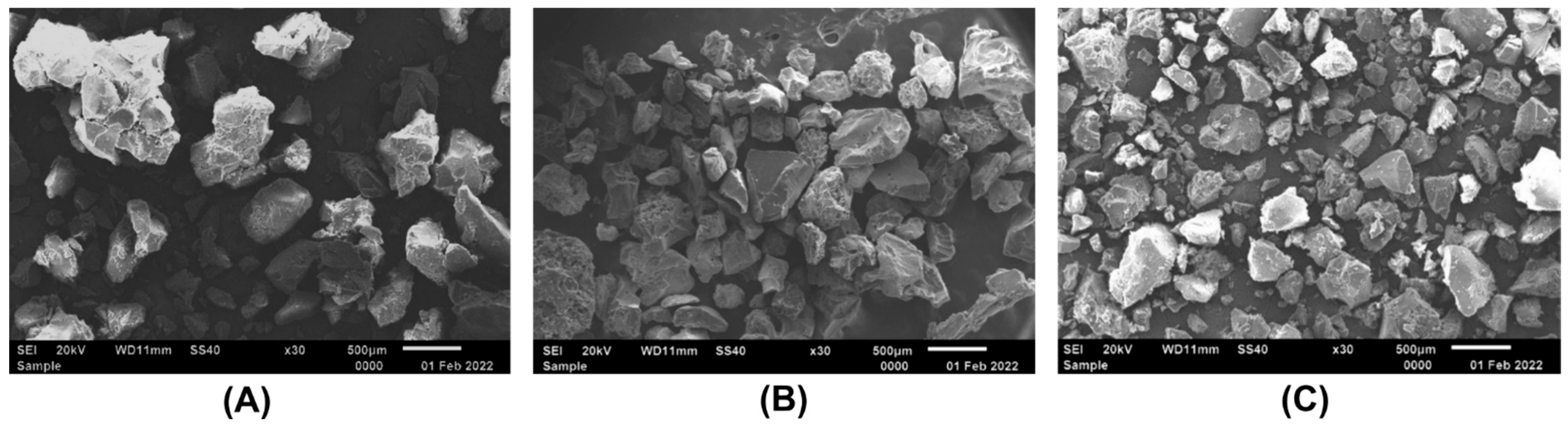
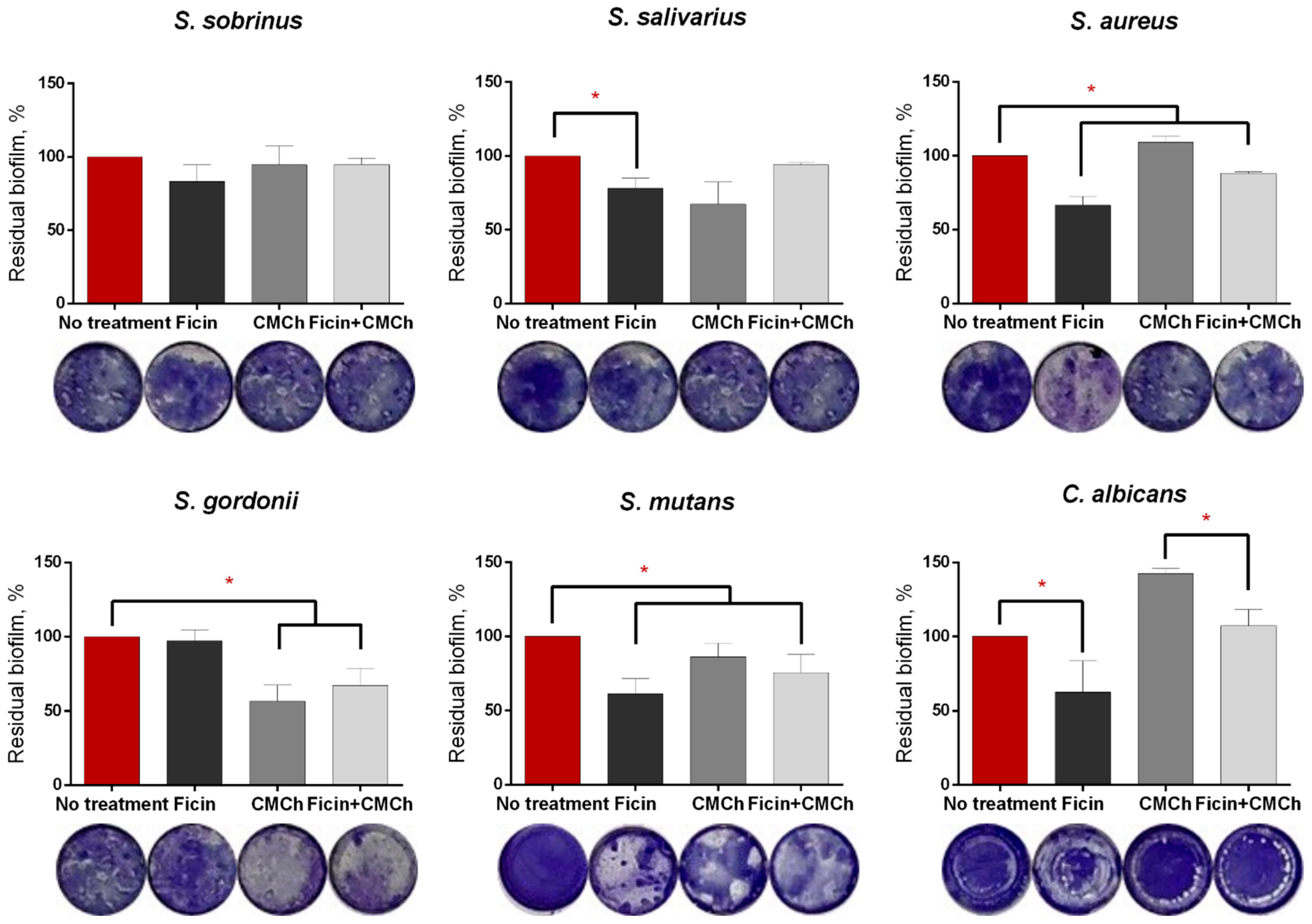
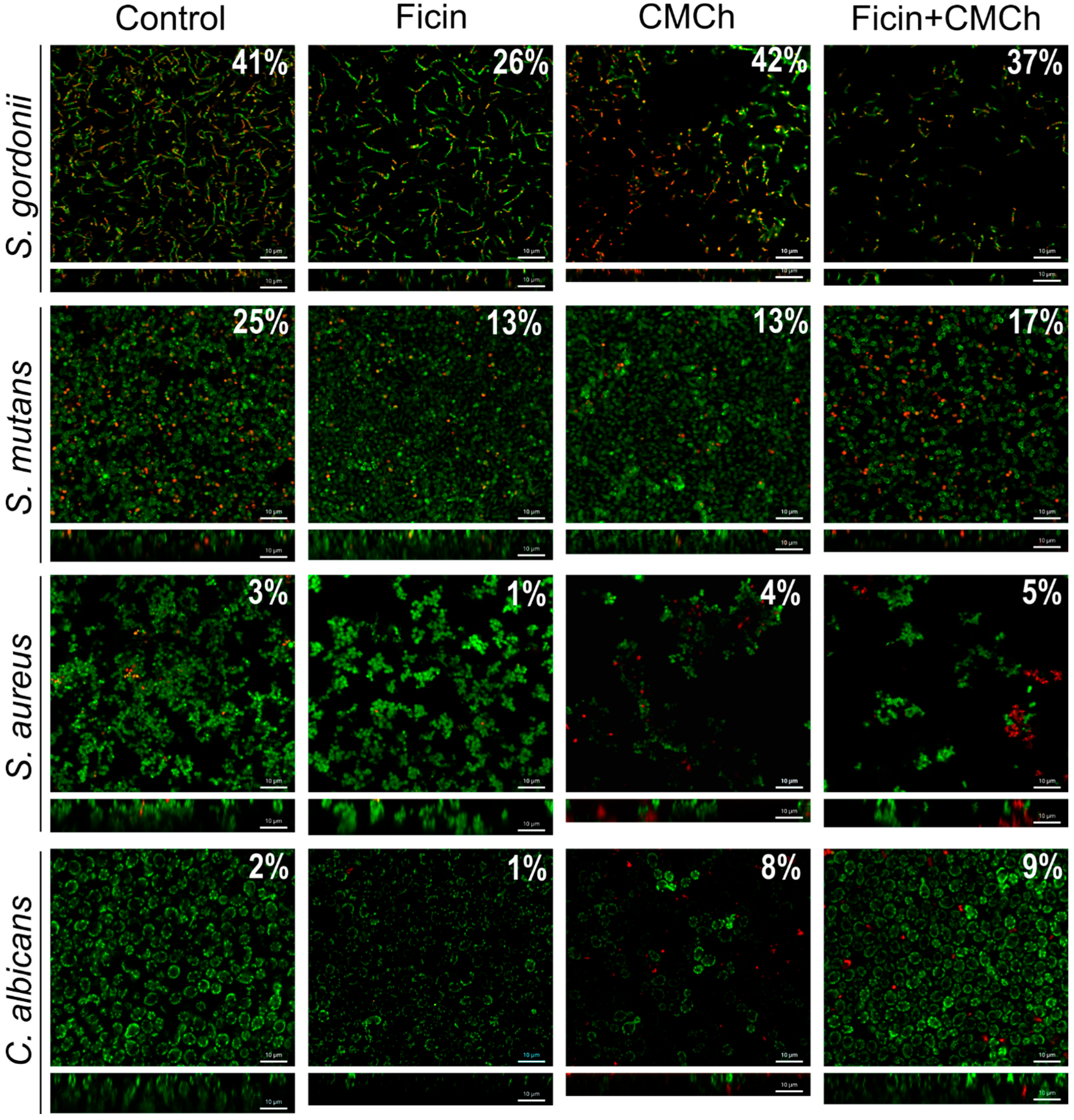


| Enzyme | Protein, mg/g of CMCh | Protein, % of Free Enzyme | Total Activity, U/mL | Total Activity, % of Free Enzyme | Optimal t, °C | Optimal pH |
|---|---|---|---|---|---|---|
| Soluble Ficin | 20.0 ± 0.1 | 100 | 96 ± 2.2 | 100 | 37–60 | 7.5 |
| Ficin on CMCh 200 kDa | 9.7 ± 0.2 | 49 ± 1.1 | 14.9 ± 1.7 | 15 | 37–60 | 6.5–7.5 |
| Ficin on CMCh 350 kDa | 3.9 ± 1.2 | 19 ± 1.3 | 31.6 ± 1.4 | 33 | 37–60 | 6.5–7.5 |
| Ficin on CMCh 600 kDa | 6.4 ± 2.4 | 32 ± 0.7 | 62.2 ± 3.9 | 65 | 37–60 | 6.5–7.5 |
| Enzyme Form | Km, µM | Vmax, µM mg−1 min−1 | kcat (min−1) | Vmax/Km | kcat/Km |
|---|---|---|---|---|---|
| Free Ficin | 20 ± 6.4 | 132 ± 20 | 6.3 ± 0.6 | 6.6 | 0.3 |
| Immobilized on CMCh200 | 16 ± 4.5 | 19 ± 3 | 0.9 ± 0.1 | 1.2 | 0.1 |
| Immobilized on CMCh350 | 17 ± 5.0 | 42 ± 5 | 2.0 ± 0.2 | 2.5 | 0.1 |
| Immobilized on CMCh600 | 13 ± 2.9 | 78 ± 8 | 3.7 ± 0.2 | 6.0 | 0.2 |
| Genus | Tooth Plaque | Biofilm Grown by Inoculation of Tooth Swab | Biofilm Grown by Inoculation of Saliva |
|---|---|---|---|
| Actinomyces | 16.9% | 0.2% | 0.0% |
| Corynebacterium | 4.7% | 0.0% | 0.0% |
| Staphylococcus | 0.0% | 0.0% | 29.0% |
| Granulicatella | 1.0% | 29.9% | 0.6% |
| Streptococcus | 24.6% | 57.6% | 65.1% |
| Veillonella | 3.9% | 0.0% | 0.0% |
| Fusobacterium | 1.8% | 0.0% | 0.0% |
| Leptotrichia | 9.7% | 0.2% | 0.0% |
| Neisseria | 1.6% | 0.0% | 0.0% |
| Indices | Tooth Plaque | Biofilm Grown by Inoculation of Tooth Swab | Biofilm Grown by Inoculation of Saliva |
|---|---|---|---|
| PD_whole_tree | 17.7 | 10.3 | 11.1 |
| Chao1 | 644 | 325 | 257 |
| Shennon | 6.35 | 4.29 | 3.06 |
| Simpson | 0.96 | 0.84 | 0.73 |
| Species | Ficin Treatment | CMCh Treatment | CMCh-Ficin Treatment |
|---|---|---|---|
| S. aureus | 0.72 | 0.76 | 0.94 |
| C. albicans | 1.02 | 0.59 | 0.58 |
| S. gordonii | 0.84 | 1.35 | 1.14 |
| S. salivarius | 0.81 | 1.17 | 0.70 |
| S. mutans | 0.94 | 0.92 | 0.84 |
| S. sobrinus | 1.19 | 1.13 | 1.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baidamshina, D.R.; Trizna, E.Y.; Goncharova, S.S.; Sorokin, A.V.; Lavlinskaya, M.S.; Melnik, A.P.; Gafarova, L.F.; Kharitonova, M.A.; Ostolopovskaya, O.V.; Artyukhov, V.G.; et al. The Effect of Ficin Immobilized on Carboxymethyl Chitosan on Biofilms of Oral Pathogens. Int. J. Mol. Sci. 2023, 24, 16090. https://doi.org/10.3390/ijms242216090
Baidamshina DR, Trizna EY, Goncharova SS, Sorokin AV, Lavlinskaya MS, Melnik AP, Gafarova LF, Kharitonova MA, Ostolopovskaya OV, Artyukhov VG, et al. The Effect of Ficin Immobilized on Carboxymethyl Chitosan on Biofilms of Oral Pathogens. International Journal of Molecular Sciences. 2023; 24(22):16090. https://doi.org/10.3390/ijms242216090
Chicago/Turabian StyleBaidamshina, Diana R., Elena Yu. Trizna, Svetlana S. Goncharova, Andrey V. Sorokin, Maria S. Lavlinskaya, Anastasia P. Melnik, Leysan F. Gafarova, Maya A. Kharitonova, Olga V. Ostolopovskaya, Valeriy G. Artyukhov, and et al. 2023. "The Effect of Ficin Immobilized on Carboxymethyl Chitosan on Biofilms of Oral Pathogens" International Journal of Molecular Sciences 24, no. 22: 16090. https://doi.org/10.3390/ijms242216090
APA StyleBaidamshina, D. R., Trizna, E. Y., Goncharova, S. S., Sorokin, A. V., Lavlinskaya, M. S., Melnik, A. P., Gafarova, L. F., Kharitonova, M. A., Ostolopovskaya, O. V., Artyukhov, V. G., Sokolova, E. A., Holyavka, M. G., Bogachev, M. I., Kayumov, A. R., & Zelenikhin, P. V. (2023). The Effect of Ficin Immobilized on Carboxymethyl Chitosan on Biofilms of Oral Pathogens. International Journal of Molecular Sciences, 24(22), 16090. https://doi.org/10.3390/ijms242216090









