Roles of DJ41_1407 and DJ41_1408 in Acinetobacter baumannii ATCC19606 Virulence and Antibiotic Response
Abstract
1. Introduction
2. Results
2.1. Exploring the Potential Roles of DJ41_1407, and DJ41_1408 in A. baumannii ATCC19606
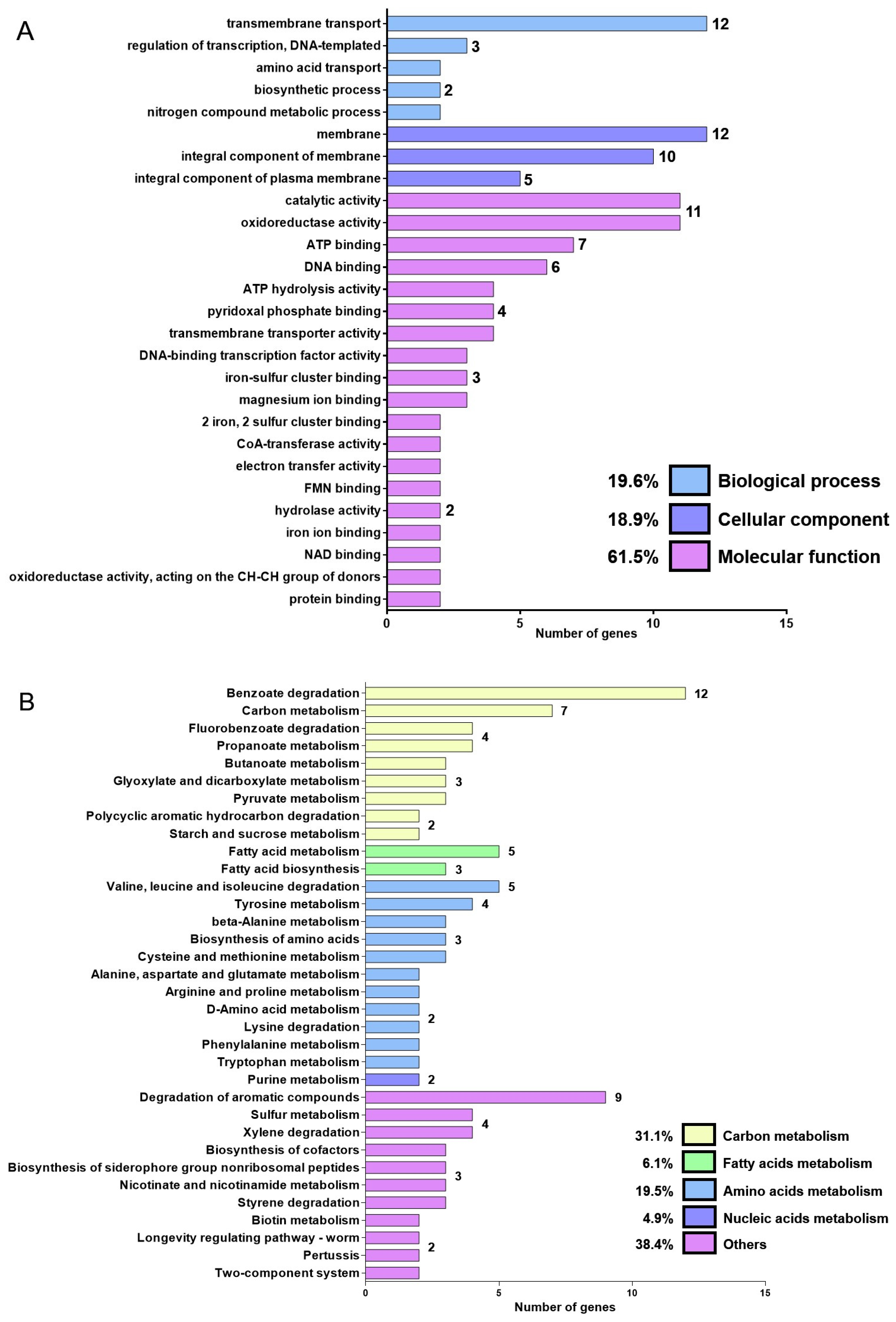
2.2. DJ41_1408 Transcriptome Analysis
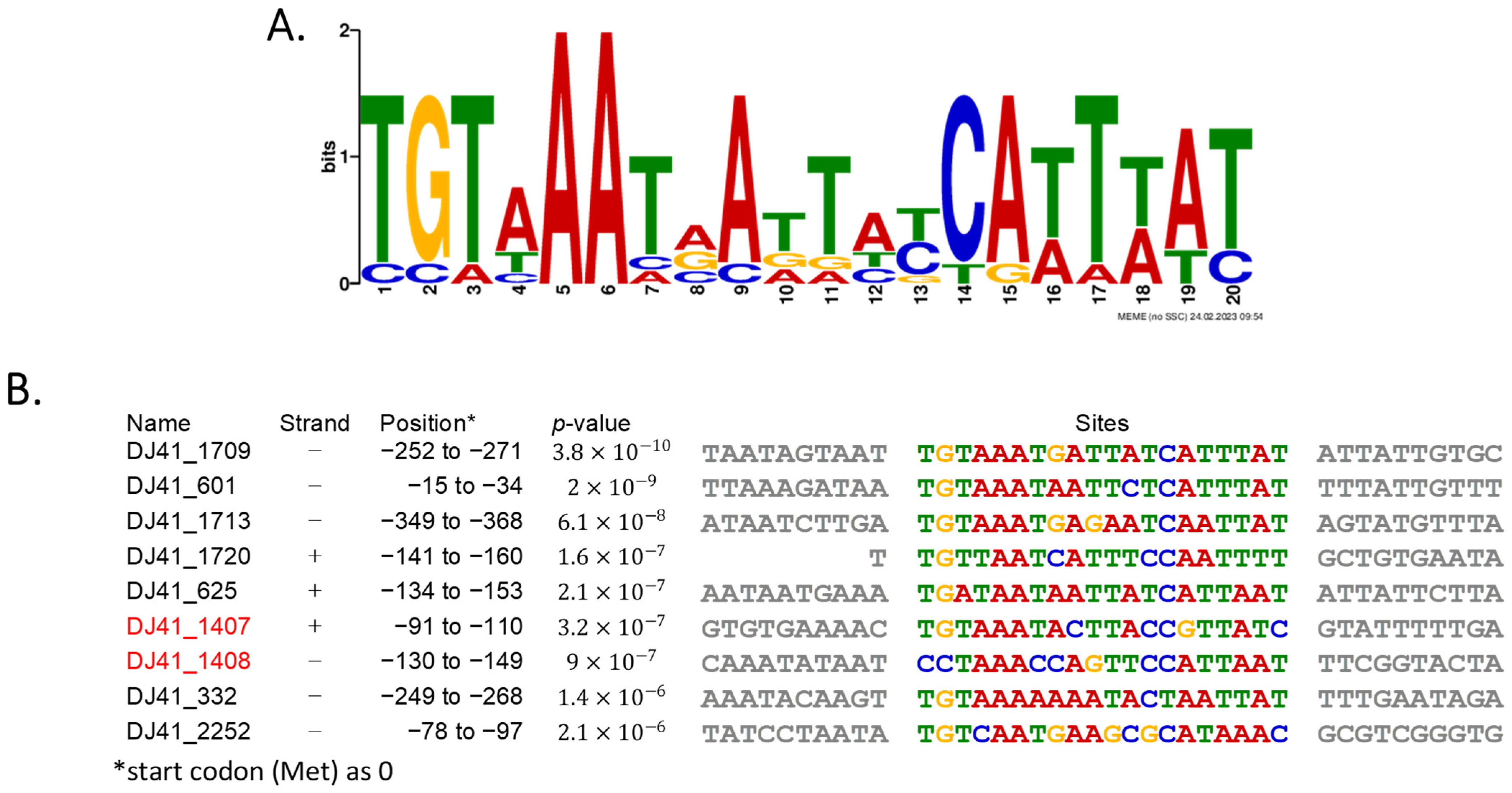
2.3. Possible Binding Region of DJ41_1408
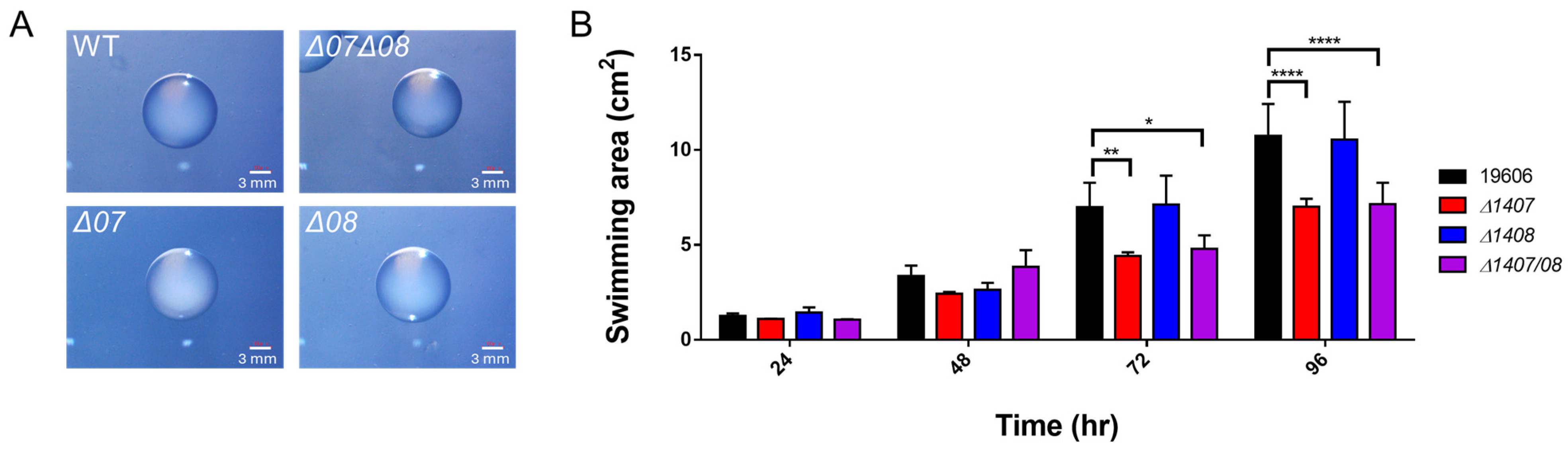
2.4. DJ41_1407 and DJ41_1408 Regulate Motility but Not Colony Size
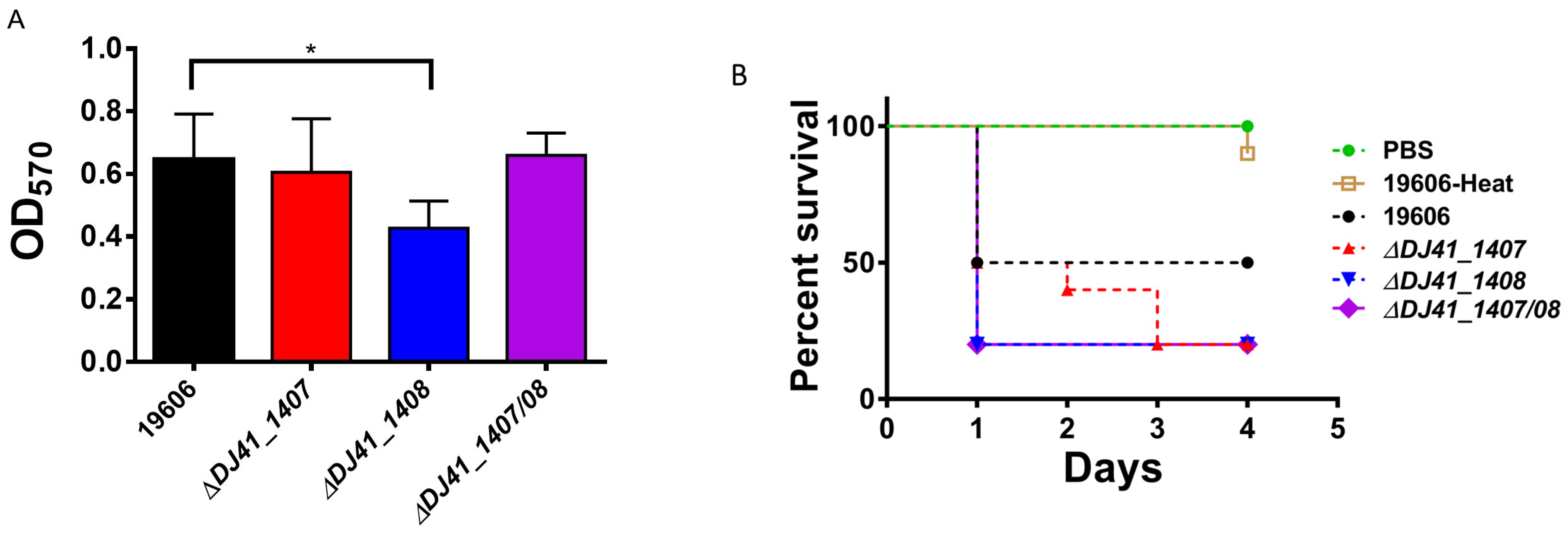
2.5. DJ41_1407 and DJ41_1408 Regulate Biofilm Formation and Virulence
2.6. DJ41_1408 Modulates Antibiotic Response
3. Discussion
4. Materials and Methods
4.1. Bacteria Strains, Plasmids, and Media
4.2. Markerless Mutation
4.3. Transcriptome Analysis Preparation
4.4. Galleria Mellonella Larvae Infection Assay
4.5. Biofilm Formation Assay
4.6. Motility Assay
4.7. MIC Test
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lastoria, L.C.; Caldeira, S.M.; Moreira, R.G.; Akazawa, R.T.; Maion, J.C.; Fortaleza, C.M. Ecological competition and the incidence of Acinetobacter baumannii bloodstream infections in a teaching hospital in Southeastern Brazil. Rev. Soc. Bras. Med. Trop. 2014, 47, 583–588. [Google Scholar] [CrossRef][Green Version]
- McConnell, M.J.; Actis, L.; Pachón, J. Acinetobacter baumannii: Human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 2013, 37, 130–155. [Google Scholar] [CrossRef]
- Roca, I.; Espinal, P.; Vila-Farrés, X.; Vila, J. The Acinetobacter baumannii Oxymoron: Commensal Hospital Dweller Turned Pan-Drug-Resistant Menace. Front. Microbiol. 2012, 3, 148. [Google Scholar] [CrossRef]
- Bergogne-Bérézin, E.; Towner, K.J. Acinetobacter spp. as nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 1996, 9, 148–165. [Google Scholar] [CrossRef]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef]
- Espinal, P.; Martí, S.; Vila, J. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J. Hosp. Infect. 2012, 80, 56–60. [Google Scholar] [CrossRef]
- Roy, S.; Chowdhury, G.; Mukhopadhyay, A.K.; Dutta, S.; Basu, S. Convergence of Biofilm Formation and Antibiotic Resistance in Acinetobacter baumannii Infection. Front. Med. 2022, 9, 793615. [Google Scholar] [CrossRef]
- Shan, W.; Kan, J.; Cai, X.; Yin, M. Insights into mucoid Acinetobacter baumannii: A review of microbiological characteristics, virulence, and pathogenic mechanisms in a threatening nosocomial pathogen. Microbiol. Res. 2022, 261, 127057. [Google Scholar] [CrossRef]
- Chen, R.; Lv, R.; Xiao, L.; Wang, M.; Du, Z.; Tan, Y.; Cui, Y.; Yan, Y.; Luo, Y.; Yang, R.; et al. A1S_2811, a CheA/Y-like hybrid two-component regulator from Acinetobacter baumannii ATCC17978, is involved in surface motility and biofilm formation in this bacterium. Microbiologyopen 2017, 6, e00510. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, X.; Zhou, D.; He, J.; Chen, Q.; Xu, Q.; Wu, S.; Zhang, W.; Yao, Y.; Fu, Y.; et al. Alteration of adeS Contributes to Tigecycline Resistance and Collateral Sensitivity to Sulbactam in Acinetobacter baumannii. Microbiol. Spectr. 2023, 11, e0459422. [Google Scholar] [CrossRef]
- Liu, X.; Chang, Y.; Xu, Q.; Zhang, W.; Huang, Z.; Zhang, L.; Weng, S.; Leptihn, S.; Jiang, Y.; Yu, Y.; et al. Mutation in the two-component regulator BaeSR mediates cefiderocol resistance and enhances virulence in Acinetobacter baumannii. mSystems 2023, 8, e0129122. [Google Scholar] [CrossRef]
- Perez Mora, B.; Giordano, R.; Permingeat, V.; Calderone, M.; Arana, N.; Müller, G.; Rodríguez, R.E.; Krasauskas, R.; Mussi, M.A. BfmRS encodes a regulatory system involved in light signal transduction modulating motility and desiccation tolerance in the human pathogen Acinetobacter baumannii. Sci. Rep. 2023, 13, 175. [Google Scholar] [CrossRef]
- Massmig, M.; Reijerse, E.; Krausze, J.; Laurich, C.; Lubitz, W.; Jahn, D.; Moser, J. Carnitine metabolism in the human gut: Characterization of the two-component carnitine monooxygenase CntAB from Acinetobacter baumannii. J. Biol. Chem. 2020, 295, 13065–13078. [Google Scholar] [CrossRef]
- Shu, H.Y.; Huang, Y.W.; Tsai, P.Y.; Hsieh, K.S.; Lin, G.H. Role of EmaSR in Ethanol Metabolism by Acinetobacter baumannii. Int. J. Mol. Sci. 2022, 23, 12606. [Google Scholar] [CrossRef]
- Feng, Z.; Wang, L.; Guan, Q.; Chu, X.; Luo, Z.-Q. Acinetobacter baumannii coordinates central metabolism, plasmid dissemination, and virulence by sensing nutrient availability. mBio 2023, 14, e0227623. [Google Scholar] [CrossRef]
- Trebosc, V.; Lucchini, V.; Narwal, M.; Wicki, B.; Gartenmann, S.; Schellhorn, B.; Schill, J.; Bourotte, M.; Frey, D.; Grünberg, J.; et al. Targeting virulence regulation to disarm Acinetobacter baumannii pathogenesis. Virulence 2022, 13, 1868–1883. [Google Scholar] [CrossRef]
- Tipton, K.A.; Rather, P.N. An ompR-envZ Two-Component System Ortholog Regulates Phase Variation, Osmotic Tolerance, Motility, and Virulence in Acinetobacter baumannii Strain AB5075. J. Bacteriol. 2017, 199, e00705–e00716. [Google Scholar] [CrossRef]
- Park, J.; Kim, M.; Shin, B.; Kang, M.; Yang, J.; Lee, T.K.; Park, W. A novel decoy strategy for polymyxin resistance in Acinetobacter baumannii. eLife 2021, 10, e66988. [Google Scholar] [CrossRef] [PubMed]
- Giles, S.K.; Stroeher, U.H.; Papudeshi, B.; Edwards, R.A.; Carlson-Jones, J.A.; Roach, M.; Brown, M.H. The StkSR Two-Component System Influences Colistin Resistance in Acinetobacter baumannii. Microorganisms 2022, 10, 985. [Google Scholar] [CrossRef] [PubMed]
- Casella, L.G.; Torres, N.J.; Tomlinson, B.R.; Shepherd, M.; Shaw, L.N. The novel two-component system AmsSR governs alternative metabolic pathway usage in Acinetobacter baumannii. Front. Microbiol. 2023, 14, 1139253. [Google Scholar] [CrossRef]
- Marchand, I.; Damier-Piolle, L.; Courvalin, P.; Lambert, T. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 2004, 48, 3298–3304. [Google Scholar] [CrossRef]
- Lin, M.F.; Lin, Y.Y.; Yeh, H.W.; Lan, C.Y. Role of the BaeSR two-component system in the regulation of Acinetobacter baumannii adeAB genes and its correlation with tigecycline susceptibility. BMC Microbiol. 2014, 14, 119. [Google Scholar] [CrossRef]
- Lin, M.F.; Lin, Y.Y.; Lan, C.Y. The Role of the Two-Component System BaeSR in Disposing Chemicals through Regulating Transporter Systems in Acinetobacter baumannii. PLoS ONE 2015, 10, e0132843. [Google Scholar] [CrossRef]
- Adams, M.D.; Nickel, G.C.; Bajaksouzian, S.; Lavender, H.; Murthy, A.R.; Jacobs, M.R.; Bonomo, R.A. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 2009, 53, 3628–3634. [Google Scholar] [CrossRef]
- Arroyo, L.A.; Herrera, C.M.; Fernandez, L.; Hankins, J.V.; Trent, M.S.; Hancock, R.E. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob. Agents Chemother. 2011, 55, 3743–3751. [Google Scholar] [CrossRef]
- Tomaras, A.P.; Flagler, M.J.; Dorsey, C.W.; Gaddy, J.A.; Actis, L.A. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 2008, 154 Pt 11, 3398–3409. [Google Scholar] [CrossRef]
- Geisinger, E.; Isberg, R.R. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog. 2015, 11, e1004691. [Google Scholar] [CrossRef]
- Bhuiyan, M.S.; Ellett, F.; Murray, G.L.; Kostoulias, X.; Cerqueira, G.M.; Schulze, K.E.; Mahamad Maifiah, M.H.; Li, J.; Creek, D.J.; Lieschke, G.J.; et al. Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc. Natl. Acad. Sci. USA 2016, 113, 9599–9604. [Google Scholar] [CrossRef]
- Cerqueira, G.M.; Kostoulias, X.; Khoo, C.; Aibinu, I.; Qu, Y.; Traven, A.; Peleg, A.Y. A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J. Infect. Dis. 2014, 210, 46–55. [Google Scholar] [CrossRef]
- Weatherspoon-Griffin, N.; Yang, D.; Kong, W.; Hua, Z.; Shi, Y. The CpxR/CpxA two-component regulatory system up-regulates the multidrug resistance cascade to facilitate Escherichia coli resistance to a model antimicrobial peptide. J. Biol. Chem. 2014, 289, 32571–32582. [Google Scholar] [CrossRef]
- Teufel, R.; Mascaraque, V.; Ismail, W.; Voss, M.; Perera, J.; Eisenreich, W.; Haehnel, W.; Fuchs, G. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. USA 2010, 107, 14390–14395. [Google Scholar] [CrossRef]
- De la Cruz, M.A.; Pérez-Morales, D.; Palacios, I.J.; Fernández-Mora, M.; Calva, E.; Bustamante, V.H. The two-component system CpxR/A represses the expression of Salmonella virulence genes by affecting the stability of the transcriptional regulator HilD. Front. Microbiol. 2015, 6, 807. [Google Scholar] [CrossRef]
- Yarwood, J.M.; McCormick, J.K.; Schlievert, P.M. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 2001, 183, 1113–1123. [Google Scholar] [CrossRef]
- Xue, M.; Raheem, M.A.; Gu, Y.; Lu, H.; Song, X.; Tu, J.; Xue, T.; Qi, K. The KdpD/KdpE two-component system contributes to the motility and virulence of avian pathogenic Escherichia coli. Res. Vet. Sci. 2020, 131, 24–30. [Google Scholar] [CrossRef]
- Lv, M.; Ye, S.; Hu, M.; Xue, Y.; Liang, Z.; Zhou, X.; Zhang, L.; Zhou, J. Two-component system ArcBA modulates cell motility and biofilm formation in Dickeya oryzae. Front. Plant Sci. 2022, 13, 1033192. [Google Scholar] [CrossRef]
- Jeong, G.J.; Khan, F.; Tabassum, N.; Kim, Y.M. Motility of Acinetobacter baumannii: Regulatory systems and controlling strategies. Appl. Microbiol. Biotechnol. 2024, 108, 3. [Google Scholar] [CrossRef]
- de Lorenzo, V.; Fernández, S.; Herrero, M.; Jakubzik, U.; Timmis, K.N. Engineering of alkyl- and haloaromatic-responsive gene expression with mini-transposons containing regulated promoters of biodegradative pathways of Pseudomonas. Gene 1993, 130, 41–46. [Google Scholar] [CrossRef]
- Chen, S.J.; Shu, H.Y.; Lin, G.H. Regulation of tert-Butyl Hydroperoxide Resistance by Chromosomal OhrR in A. baumannii ATCC 19606. Microorganisms 2021, 9, 629. [Google Scholar] [CrossRef]
- Bouvet, P.J.; Grimont, P.A. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Evol. Microbiol. 1986, 36, 228–240. [Google Scholar]
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Tsai, C.J.; Loh, J.M.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef]
- Lin, H.R.; Shu, H.Y.; Lin, G.H. Biological roles of indole-3-acetic acid in Acinetobacter baumannii. Microbiol. Res. 2018, 216, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Clinical & Laboratory Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard, 7th ed.; [M7-A7]; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2006. [Google Scholar]

| STRAIN | APR | GEN | KAN | TET | PMB | CST |
|---|---|---|---|---|---|---|
| Wild-type | 50 | 100 | 25 | 0.78 | 6.25 | 5 |
| ∆DJ41_1407 | 50 | 100 | 25 | 0.78 | 6.25 | 5 |
| ∆DJ41_1408 | 50 | 50 | 12.5 | 0.78 | 6.25 | 5 |
| ∆DJ41_1407∆DJ41_1408 | 50 | 50 | 12.5 | 0.78 | 6.25 | 5 |
| Bacteria | Description | References or Source |
|---|---|---|
| E. coli | ||
| DH5α | F−, supE44, hsdR17, recA1, gyrA96, endA1, thi-1, relA1, deoR, λ | ATCC53868 |
| DH5α/pK18-∆DJ41_1408 | Kanr, DH5α containing pK18_∆DJ41_1408 | This study |
| DH5α/pK18-∆DJ41_1407-08 | Kanr, DH5α containing pK18-∆DJ41_1407-08 | This study |
| S17-1λπ | thi-1, thr, leu, tonA, lacY, supE, recA, RP4-2 (Km::Tn7,Tc::Mu-1), Smr, lpir | [38] |
| S17-1λπ/pK18-∆DJ41_1407 | Kanr, S17-1λπ containing pK18_∆DJ41-1407 | [39] |
| S17-1λπ/pK18-∆DJ41_1408 | Kanr, S17-1λπ containing pK18_∆DJ41_1408 | This study |
| S17-1λπ/pK18-∆DJ41_1407-08 | Kanr, S17-1λπ containing pK18-∆DJ41_1407-08 | This study |
| A. baumannii | ||
| ATCC19606 | Ampr, clinical isolate, wild type | [40] |
| ∆DJ41_1407 | Ampr, deletion of DJ41_1407 | [39] |
| ∆DJ41_1408 | Ampr, deletion of DJ41_1408 | This study |
| ∆DJ41_1407∆DJ41_1408 | Ampr, deletion of DJ41_1407 and DJ41_1408 | This study |
| Primer | Sequence (5′-3′) | Application | Reference or Source |
|---|---|---|---|
| pK18-DJ41_1408UP_F | AATTCGAGCTCGGTACCCGGGATATTAACCGTTATATCTTA | construct and confirm mutant | This study |
| pK18-DJ41_1408DO_R | GTAAAACGACGGCCAGTGCCACGTCCGTGATAACTATGTCG | construct and confirm mutant | This study |
| DJ41_1408UP-DJ41_1408DO_F | AAATGGCAGAACAGCAACCGGAGACAGAACAGGAAGTA | construct mutant | This study |
| DJ41_1408DO-DJ41_1408UP_R | TACTTCCTGTTCTGTCTCCGGTTGCTGTTCTGCCATTT | construct mutant | This study |
| pK18-DJ41_1407UP_F | GAGCTCGGTACCCGGGGTGGTTGAGAACTGACGAAT | construct and confirm mutant | This study |
| DJ41_1408DO-DJ41_1407UP_R | TACTTCCTGTTCTGTCTCCGACGAAAAATCGTATGGGACA | construct mutant | This study |
| DJ41_1407UP-DJ41_1408DO_F | TGTCCCATACGATTTTTCGTCGGAGACAGAACAGGAAGTA | construct mutant | This study |
| 1406_up380-F | GAGCTCGGTACCCGGGACGGGTTATTGACGAGTTCT | confirm mutant | This study |
| 1408_do250-R | GTGCTTGGGTTATGGGTGAA | confirm mutant | This study |
| AbEraS+R_intergenic region-qF | GCGATTCGTTAC GGTTTGAT | amplify intergenic region | [39] |
| AbEraS+R_intergenic region-qR | AGGAATATAAGGCAGGTTGCTG | amplify intergenic region | [39] |
| 1407/08-IGR-qF | GGGGCATGCTTCAGAATAGA | amplify intergenic region | This study |
| 1407/08-IGR-qR | TAATCCCCATTCGTGCAAGT | amplify intergenic region | This study |
| Plasmids | Description | References or Source |
|---|---|---|
| pK18mobsacB | Kanr, mobilizable suicide vector, sacB, oriT | [41] |
| pK18-∆DJ41_1407 | Kanr, pK18mobsacB containing DJ41_1407 upstream and downstream 1 kb fragments | [39] |
| pK18-∆DJ41_1408 | Kanr, pK18mobsacB containing DJ41_1408 upstream and downstream 1 kb fragments | This study |
| pK18-∆DJ41_1407-08 | Kanr, pK18mobsacB containing DJ41_1407 and DJ41_1408 upstream and downstream 1 kb fragments | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toh, Y.-H.; Lin, G.-H. Roles of DJ41_1407 and DJ41_1408 in Acinetobacter baumannii ATCC19606 Virulence and Antibiotic Response. Int. J. Mol. Sci. 2024, 25, 3862. https://doi.org/10.3390/ijms25073862
Toh Y-H, Lin G-H. Roles of DJ41_1407 and DJ41_1408 in Acinetobacter baumannii ATCC19606 Virulence and Antibiotic Response. International Journal of Molecular Sciences. 2024; 25(7):3862. https://doi.org/10.3390/ijms25073862
Chicago/Turabian StyleToh, Yee-Huan, and Guang-Huey Lin. 2024. "Roles of DJ41_1407 and DJ41_1408 in Acinetobacter baumannii ATCC19606 Virulence and Antibiotic Response" International Journal of Molecular Sciences 25, no. 7: 3862. https://doi.org/10.3390/ijms25073862
APA StyleToh, Y.-H., & Lin, G.-H. (2024). Roles of DJ41_1407 and DJ41_1408 in Acinetobacter baumannii ATCC19606 Virulence and Antibiotic Response. International Journal of Molecular Sciences, 25(7), 3862. https://doi.org/10.3390/ijms25073862







