The Association of Helicobacter pylori Biofilm with Enterovirus 71 Prolongs Viral Viability and Survival
Abstract
:1. Introduction
2. Results
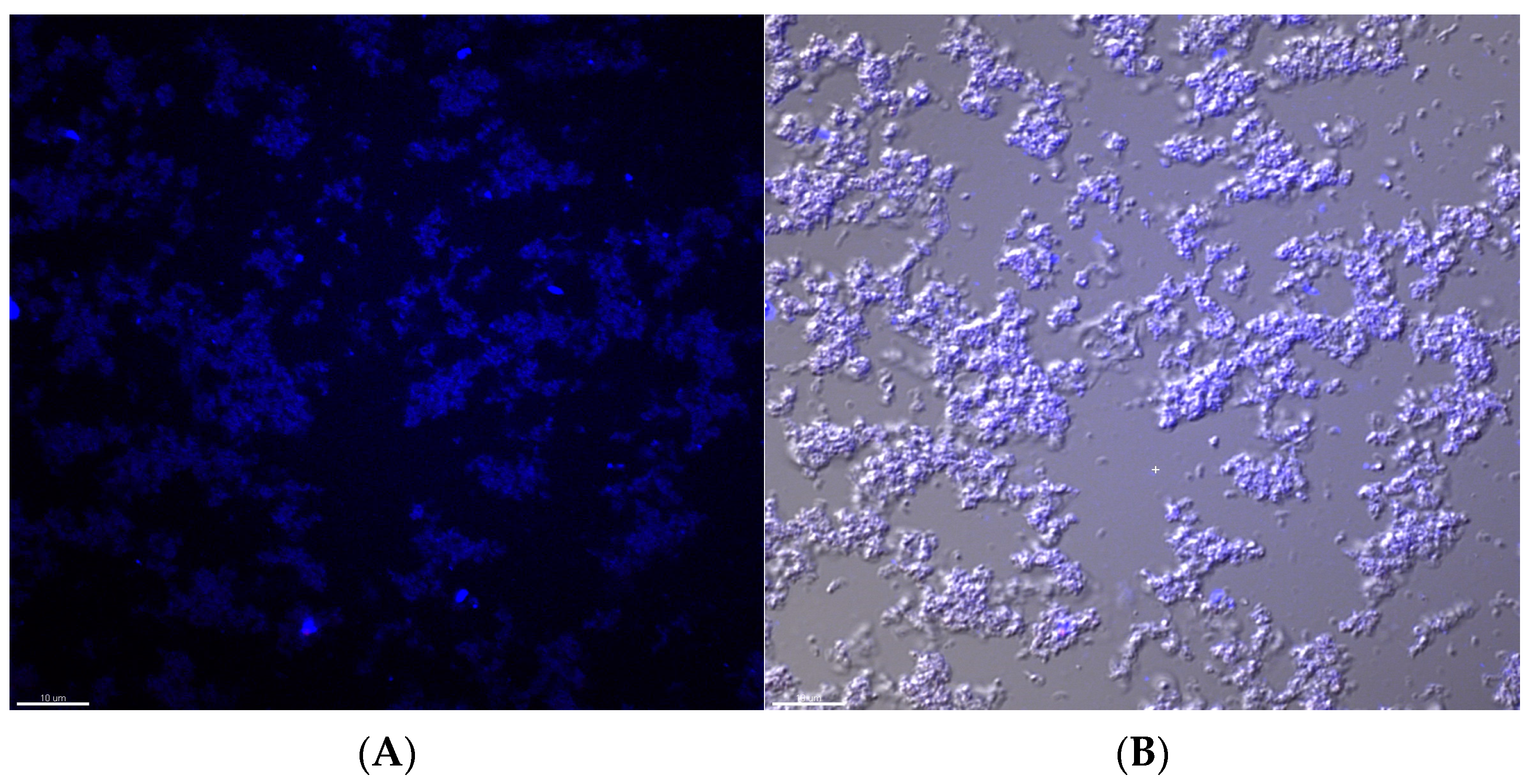
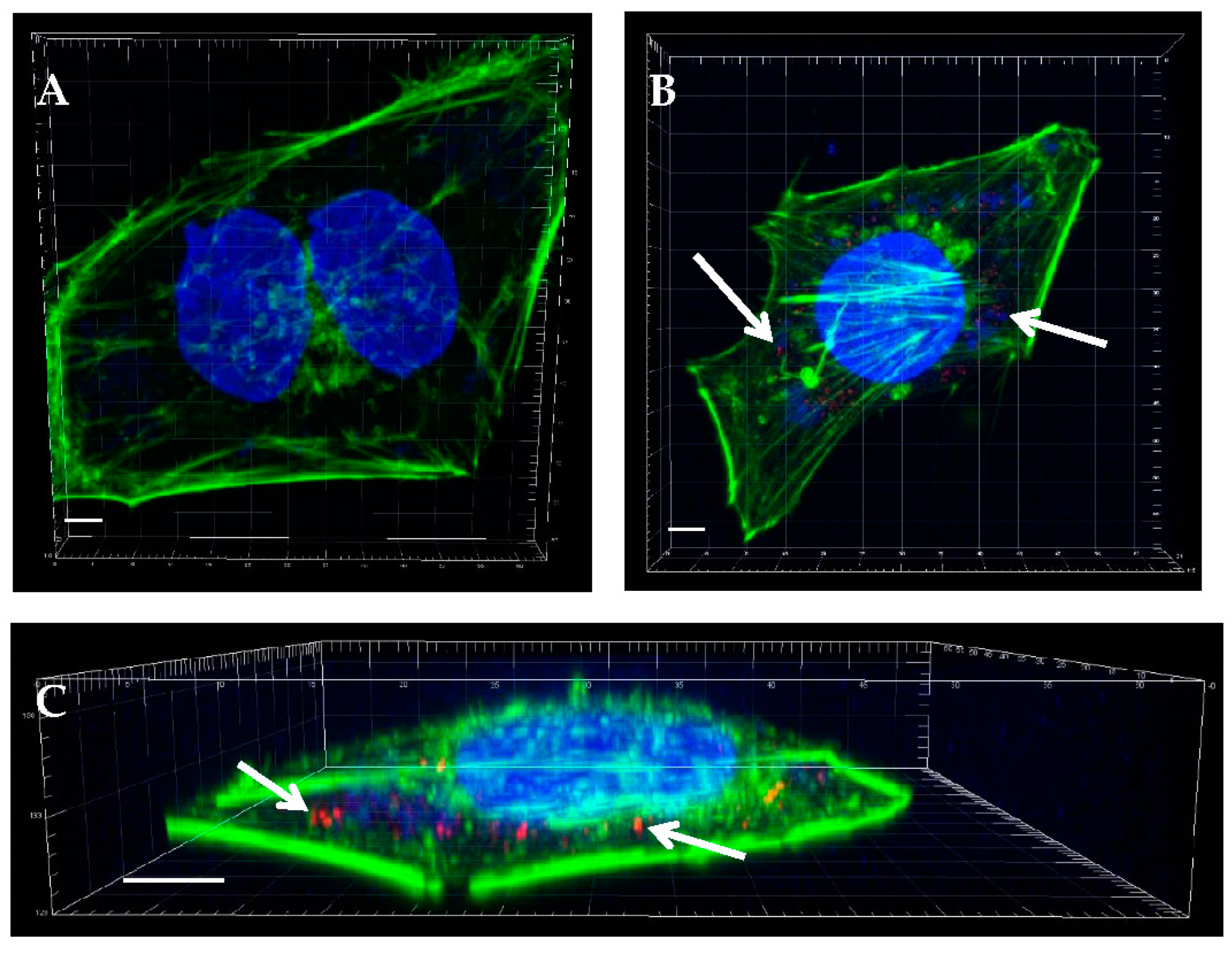
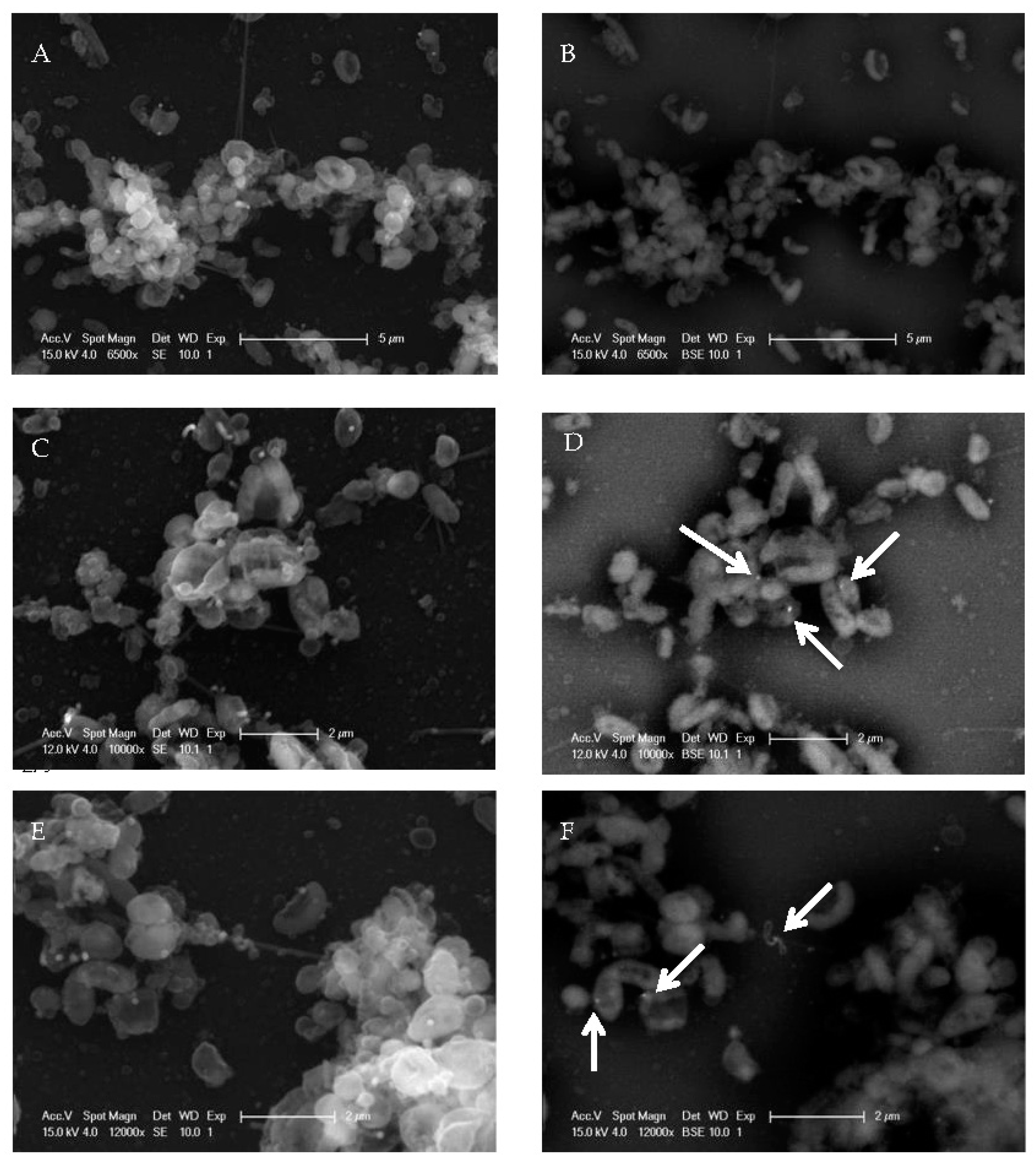
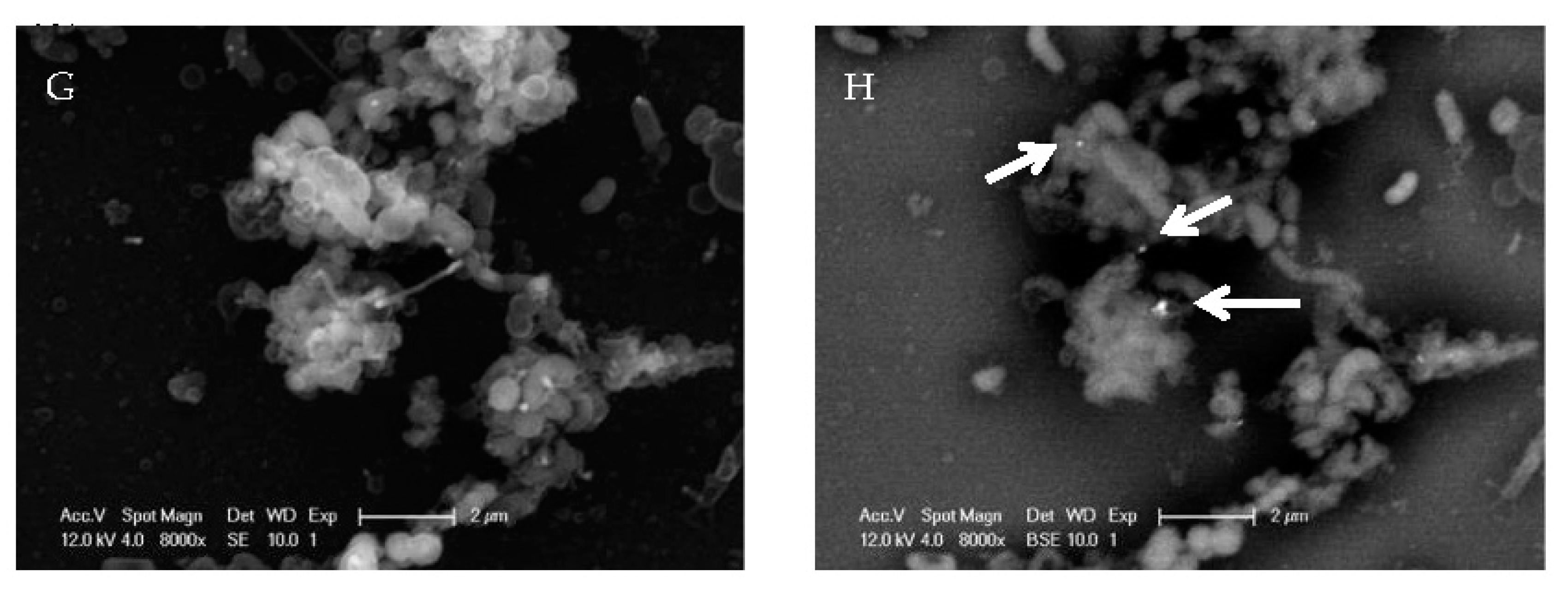
2.1. EV71 Particles Associate with the Preformed Biofilm of H. pylori
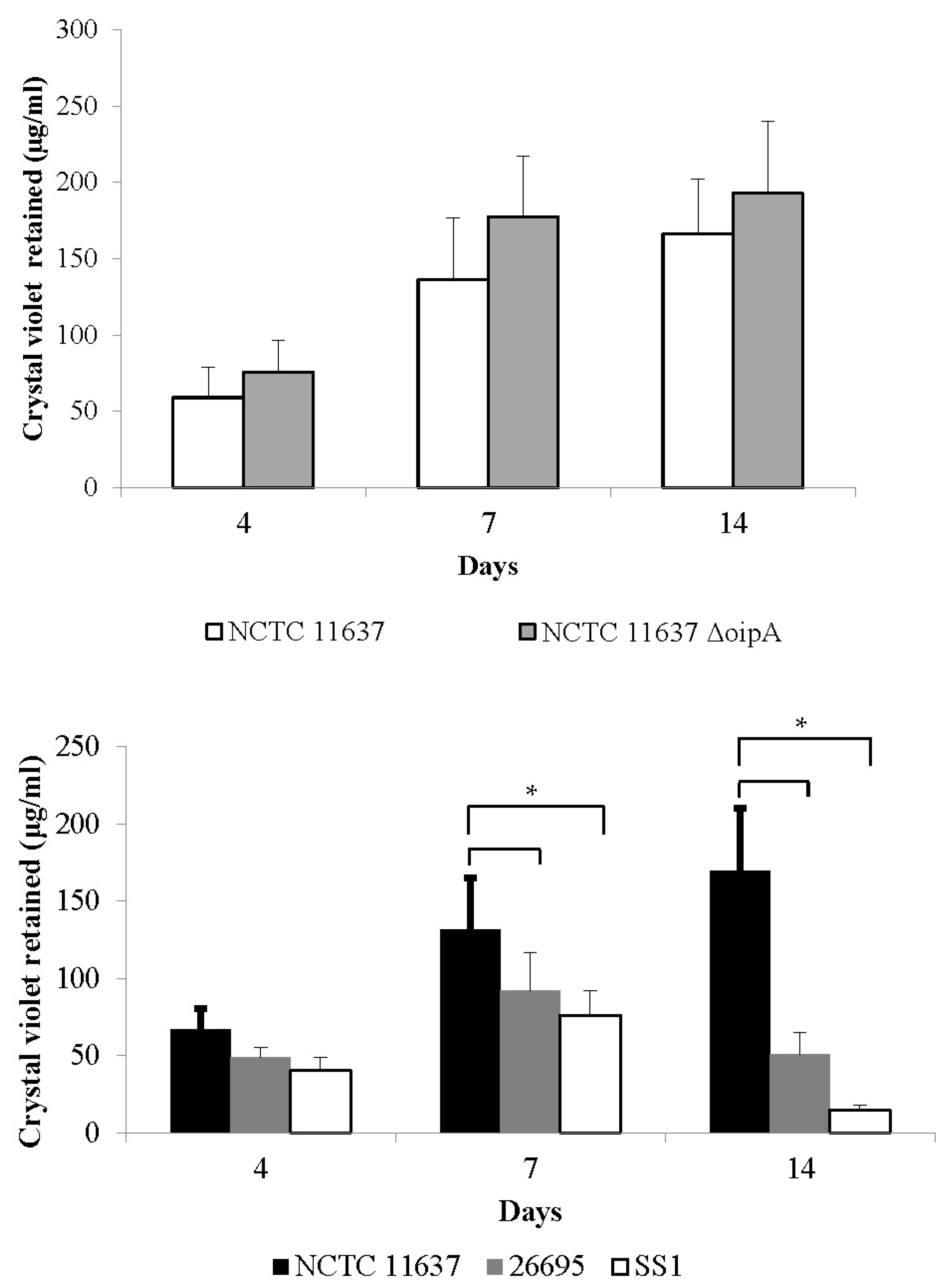
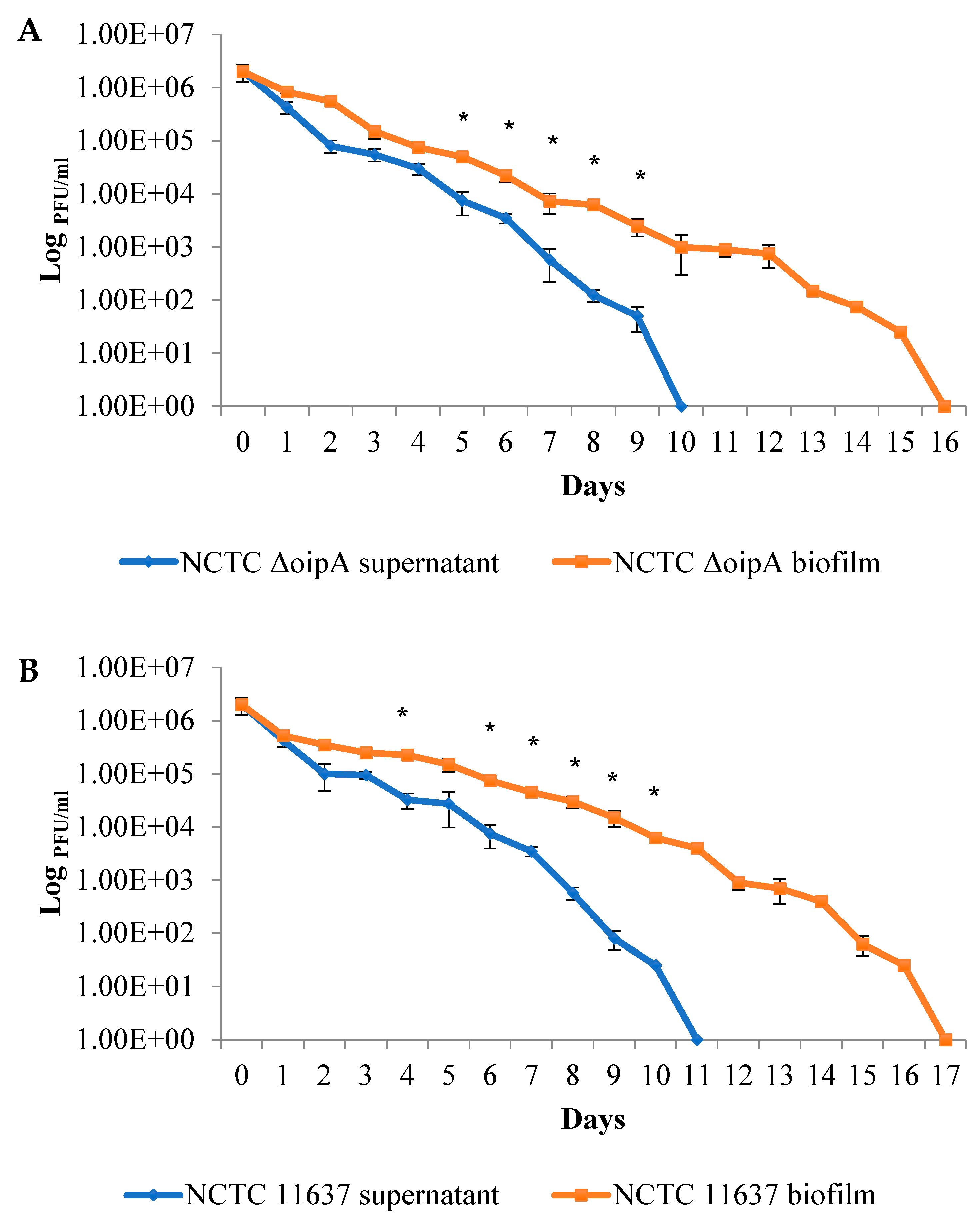
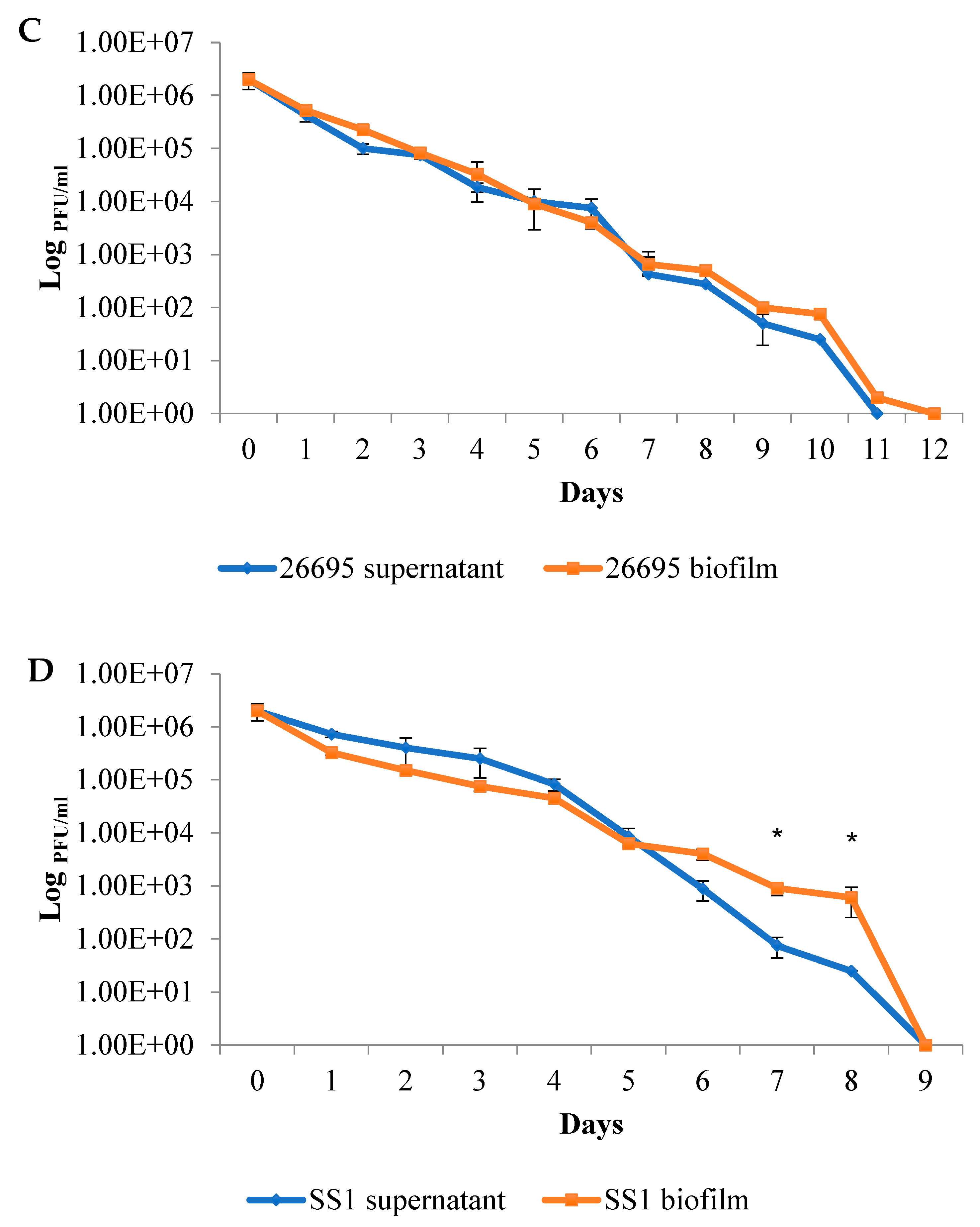
2.2. The Duration of Virus Viability Is Dependent on the Quantity of Biofilm Formation
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Preparation of Dehydrated Bacterial Biofilm and Quantification through Crystal Violet Staining
4.3. EV71 Strain, Cell Line, and Virus Plaque Formation Assay
4.4. Immunofluorescence Staining and Confocal Microscopy to Image the H. pylori Biofilm and EV71
4.5. Scanning Electron Microscopy and Gold Labeling of Virus Particles
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; van Hullebusch, E.D.; Neu, T.R.; Nielsen, P.H.; Seviour, T.; Stoodley, P.; Wingender, J.; Wuertz, S. The biofilm matrix: Multitasking in a shared space. Nat. Rev. Microbiol. 2023, 21, 70–86. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Kamiya, S. Biofilm formation by Helicobacter pylori and its involvement for antibiotic resistance. Biomed. Res. Int. 2015, 2015, 914791. [Google Scholar] [CrossRef] [PubMed]
- Krzyżek, P.; Grande, R.; Migdał, P.; Paluch, E.; Gościniak, G. Biofilm formation as a complex result of virulence and adaptive responses of Helicobacter pylori. Pathogens 2020, 9, 1062. [Google Scholar] [CrossRef] [PubMed]
- Klein, P.D.; Graham, D.Y.; Gaillour, A.; Opekun, A.R.; Smith, E.O. Water source as risk factor for Helicobacter pylori infection in Peruvian children. Lancet 1991, 337, 1503–1506. [Google Scholar] [CrossRef]
- Horiuchi, T.; Ohkusa, T.; Watanabe, M.; Kobayashi, D.; Miwa, H.; Eishi, Y. Helicobacter pylori DNA in drinking water in Japan. Microbiol. Immunol. 2001, 45, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, S.; Kato, S.; Watanabe, A. Water source as a Helicobacter pylori transmission route: A 3-year follow-up study of Japanese children living in a unique district. J. Med. Microbiol. 2008, 57, 909–910. [Google Scholar] [CrossRef]
- Mazari-Hiriart, M.; Lopez-Vidal, Y.; Calva, J.J. Helicobacter pylori in water systems for human use in Mexico City. Water Sci. Technol. 2001, 43, 93–98. [Google Scholar] [CrossRef]
- Lu, Y.Z.; Redlinger, T.E.; Avitia, R.; Galindo, A.; Goodman, K. Isolation and genotyping of Helicobacter pylori from untreated municipal wastewater. Appl. Environ. Microbiol. 2002, 68, 1436–1439. [Google Scholar] [CrossRef]
- Watson, C.L.; Owen, R.J.; Said, B.; Lai, S.; Lee, J.V.; Surman-Lee, S.; Nichols, G. Detection of Helicobacter pylori by PCR but not culture in water and biofilm samples from drinking water distribution systems in England. J. Appl. Microbiol. 2004, 97, 690–698. [Google Scholar] [CrossRef]
- Hulten, K.; Han, S.W.; Enroth, H.; Klein, P.D.; Opekun, A.R.; Gilman, R.H.; Evans, D.G.; Engstrand, L.; Graham, D.Y.; ElZaatari, F.A.K. Helicobacter pylori in the drinking water in Peru. Gastroenterology 1996, 110, 1031–1035. [Google Scholar] [CrossRef] [PubMed]
- Bunn, J.E.G.; MacKay, W.G.; Thomas, J.E.; Reid, D.C.; Weaver, L.T. Detection of Helicobacter pylori DNA in drinking water biofilms: Implications for transmission in early life. Lett. Appl. Microbiol. 2002, 34, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Park, S.R.; Mackay, W.G.; Reid, D.C. Helicobacter sp recovered from drinking water biofilm sampled from a water distribution system. Water Res. 2001, 35, 1624–1626. [Google Scholar] [CrossRef]
- Yang, F.L.; Hassanbhai, A.M.; Chen, H.Y.; Huang, Z.Y.; Lin, T.L.; Wu, S.H.; Ho, B. Proteomannans in biofilm of Helicobacter pylori ATCC 43504. Helicobacter 2011, 16, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.G.; Loke, M.F.; Goh, K.L.; Vadivelu, J.; Ho, B. Biofilm formation enhances Helicobacter pylori survivability in vegetables. Food Microbiol. 2017, 62, 68–76. [Google Scholar] [CrossRef]
- Leclerc, H.; Schwartzbrod, L.; Dei-Cas, E. Microbial agents associated with waterborne diseases. Crit. Rev. Microbiol. 2002, 28, 371–409. [Google Scholar] [CrossRef]
- Carter, M.J. Enterically infecting viruses: Pathogenicity, transmission and significance for food and waterborne infection. J. Appl. Microbiol. 2005, 98, 1354–1380. [Google Scholar] [CrossRef] [PubMed]
- Bible, J.M.; Pantelidis, P.; Chan, P.K.S.; Tong, C.Y.W. Genetic evolution of enterovirus 71: Epidemiological and pathological implications. Rev. Med. Virol. 2007, 17, 371–379. [Google Scholar] [CrossRef]
- Solomon, T.; Lewthwaite, P.; Perera, D.; Cardoso, M.J.; McMinn, P.; Ooi, M.H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 2010, 10, 778–790. [Google Scholar] [CrossRef]
- Wu, Y.; Yeo, A.; Phoon, M.C.; Tan, E.L.; Poh, C.L.; Quak, S.H.; Chow, V.T. The largest outbreak of hand; foot and mouth disease in Singapore in 2008: The role of enterovirus 71 and coxsackievirus A strains. Int. J. Infect. Dis. 2010, 14, e1076-81. [Google Scholar] [CrossRef]
- Weng, K.F.; Chen, L.L.; Huang, P.N.; Shih, S.R. Neural pathogenesis of enterovirus 71 infection. Microbes Infect. 2010, 12, 505–510. [Google Scholar] [CrossRef] [PubMed]
- McMinn, P.C. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol. Rev. 2002, 26, 91–107. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Sullivan, G.; Venosa, A.D. Low-temperature stability of viruses in sludges. Appl. Environ. Microbiol. 1988, 54, 839–841. [Google Scholar] [CrossRef]
- Marshall, B.J.; Windsor, H.M. The relation of Helicobacter pylori to gastric adenocarcinoma and lymphoma: Pathophysiology, epidemiology, screening, clinical presentation, treatment, and prevention. Med. Clin. North Am. 2005, 89, 313–344. [Google Scholar] [CrossRef]
- Stark, R.M.; Gerwig, G.J.; Pitman, R.S.; Potts, L.F.; Williams, N.A.; Greenman, J.; Weinzweig, I.P.; Hirst, T.R.; Millar, M.R. Biofilm formation by Helicobacter pylori. Lett. Appl. Microbiol. 1999, 28, 121–126. [Google Scholar] [CrossRef] [PubMed]
- García, A.; Salas-Jara, M.J.; Herrera, C.; González, C. Biofilm and Helicobacter pylori: From environment to human host. World J. Gastroenterol. 2014, 20, 5632–5638. [Google Scholar] [CrossRef] [PubMed]
- Hathroubi, S.; Servetas, S.L.; Windham, I.; Merrell, D.S.; Ottemann, K.M. Helicobacter pylori biofilm formation and its potential role in pathogenesis. Microbiol. Mol. Biol. Rev. 2018, 82, e00001-18. [Google Scholar] [CrossRef]
- Krzyzek, P.; Migdal, P.; Grande, R.; Gosciniak, G. Biofilm formation of Helicobacter pylori in both static and microfluidic conditions is associated with resistance to clarithromycin. Front. Cell Infect. Microbiol. 2022, 12, 868905. [Google Scholar] [CrossRef]
- Al-Maleki, A.R.; Loke, M.F.; Lui, S.Y.; Ramli, N.S.K.; Khosravi, Y.; Ng, C.G.; Venkatraman, G.; Goh, K.L.; Ho, B.; Vadivelu, J. Helicobacter pylori outer inflammatory protein A (OipA) suppresses apoptosis of AGS gastric cells in vitro. Cell Microbiol. 2017, 19, e12771. [Google Scholar] [CrossRef]
- Quignon, F.; Sardin, M.; Kiene, L.; Schwartzbrod, L. Poliovirus-1 inactivation and interaction with biofilm: A pilot-scale study. Appl. Environ. Microbiol. 1997, 63, 978–982. [Google Scholar] [CrossRef]
- Storey, M.V.; Ashbolt, N.J. Persistence of two model enteric viruses (B40-8 and MS-2 bacteriophages) in water distribution pipe biofilms. Water Sci. Technol. 2001, 43, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Storey, M.V.; Ashbolt, N.J. Enteric virions and microbial biofilms–A secondary source of public health concern? Water Sci. Technol. 2003, 48, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.W.; Poh, C.L.; Sam, I.C.; Chan, Y.F. Enterovirus 71 uses cell surface heparan sulfate glycosaminoglycan as an attachment receptor. J. Virol. 2013, 87, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Lin, S.T.; Chen, C.Y.; Wu, S.C. Enterovirus 71 adsorption on metal ion-composite chitosan beads. Biotechnol. Prog. 2012, 28, 206–214. [Google Scholar] [CrossRef]
- Myint, S.; Manley, R.; Cubitt, D. Viruses in bathing waters. Lancet 1994, 343, 1640–1641. [Google Scholar] [CrossRef]
- Mackay, W.G.; Gribbon, L.T.; Barer, M.R.; Reid, D.C. Biofilms in drinking water systems: A possible reservoir for Helicobacter pylori. J. Appl. Microbiol. 1999, 85, 52s–59s. [Google Scholar] [CrossRef]
- Ali, M.A.; Al-Herrawy, A.Z.; El-Hawaary, S.E. Detection of enteric viruses, Giardia and Cryptosporidium in two different types of drinking water treatment facilities. Water Res. 2004, 38, 3931–3939. [Google Scholar] [CrossRef]
- Vivier, J.C.; Ehlers, M.M.; Grabow, W.O. Detection of enteroviruses in treated drinking water. Water Res. 2004, 38, 2699–2705. [Google Scholar] [CrossRef]
- Ehlers, M.M.; Grabow, W.O.; Pavlov, D.N. Detection of enteroviruses in untreated and treated drinking water supplies in South Africa. Water Res. 2005, 39, 2253–2258. [Google Scholar] [CrossRef]
- Ranganathan, S.; Singh, S.; Poh, C.L.; Chow, V.T. The hand, foot and mouth disease virus capsid: Sequence analysis and prediction of antigenic sites from homology modelling. Appl. Bioinform. 2002, 1, 43–52. [Google Scholar]
- Lal, S.K.; Kumar, P.; Yeo, W.M.; Kar-Roy, A.; Chow, V.T. The VP1 protein of human enterovirus 71 self-associates via an interaction domain spanning amino acids 66-297. J. Med. Virol. 2006, 78, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, R.G.; Jones, E.L.; Gerba, C.P. Viruses in recreational water-borne disease outbreaks: A review. J. Appl. Microbiol. 2009, 107, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Love, D.C.; Rodriguez, R.A.; Gibbons, C.D.; Griffith, J.F.; Yu, Q.L.; Stewart, J.R.; Sobsey, M.D. Human viruses and viral indicators in marine water at two recreational beaches in Southern California, USA. J. Water Health 2014, 12, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Cabelli, V.J.; Dufour, A.P.; Mccabe, L.J.; Levin, M.A. Swimming-associated gastroenteritis and water quality. Am. J. Epidemiol. 1982, 115, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, P.L.; Tobin, R.S.; Brown, N.E.; Ness, P.F. A prospective study of swimming-related illness. I. Swimming-associated health risk. Am. J. Public Health 1985, 75, 108–1070. [Google Scholar] [CrossRef]
- Craun, G.F.; Calderon, R.L.; Craun, M.F. Outbreaks associated with recreational water in the United States. Int J. Environ. Health Res. 2005, 15, 243–262. [Google Scholar] [CrossRef]
- Cheung, W.H.; Chang, K.C.; Hung, R.P.; Kleevens, J.W. Health effects of beach water pollution in Hong Kong. Epidemiol. Infect. 1990, 105, 139–162. [Google Scholar] [CrossRef]
- Hughes, M.S.; Coyle, P.V.; Connolly, J.H. Enteroviruses in recreational waters of Northern Ireland. Epidemiol. Infect. 1992, 108, 529–536. [Google Scholar] [CrossRef]
- Corbett, S.J.; Rubin, G.L.; Curry, G.K.; Kleinbaum, D.G. The health effects of swimming at Sydney beaches. The Sydney Beach Users Study Advisory Group. Am. J. Public Health 1993, 83, 1701–1706. [Google Scholar] [CrossRef]
- Lee, C.S.; Lee, C.; Marion, J.; Wang, Q.; Saif, L.; Lee, J. Occurrence of human enteric viruses at freshwater beaches during swimming season and its link to water inflow. Sci. Total Environ. 2014, 472, 757–766. [Google Scholar] [CrossRef]
- Skraber, S.; Schijven, J.; Gantzer, C.; de Roda Husman, A.M. Pathogenic viruses in drinking-water biofilms: A public health risk? Biofilms 2005, 2, 105–117. [Google Scholar] [CrossRef]
- Lodder, W.J.; Vinje, J.; van De Heide, R.; de Roda Husman, A.M.; Leenen, E.J.; Koopmans, M.P. Molecular detection of Norwalk-like caliciviruses in sewage. Appl. Environ. Microbiol. 1999, 65, 5624–5627. [Google Scholar] [CrossRef]
- Skraber, S.; Ogorzaly, L.; Helmi, K.; Maul, A.; Hoffmann, L.; Cauchie, H.M.; Gantzer, C. Occurrence and persistence of enteroviruses, noroviruses and F-specific RNA phages in natural wastewater biofilms. Water Res. 2009, 43, 4780–4789. [Google Scholar] [CrossRef]
- Smith, E.M.; Gerba, C.P.; Melnick, J.L. Role of sediment in the persistence of enteroviruses in the estuarine environment. Appl. Environ. Microbiol. 1978, 35, 685–689. [Google Scholar] [CrossRef]
- Sakoda, A.; Sakai, Y.; Hayakawa, K.; Suzuki, M. Adsorption of viruses in water environment onto solid surfaces. Water Sci. Technol. 1997, 35, 107–114. [Google Scholar] [CrossRef]
- Karim, M.R.; Manshadi, F.D.; Karpiscak, M.M.; Gerba, C.P. The persistence and removal of enteric pathogens in constructed wetlands. Water Res. 2004, 38, 1831–1837. [Google Scholar] [CrossRef] [PubMed]
- Helmi, K.; Skraber, S.; Gantzer, C.; Willame, R.; Hoffmann, L.; Cauchie, H.M. Interactions of Cryptosporidium parvum, Giardia lamblia, vaccinal poliovirus type 1, and bacteriophages phiX174 and MS2 with a drinking water biofilm and a wastewater biofilm. Appl. Environ. Microbiol. 2008, 74, 2079–2088. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A.; Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef]
- Singh, S.; Chow, V.T.; Phoon, M.C.; Chan, K.P.; Poh, C.L. Direct detection of enterovirus 71 (EV71) in clinical specimens from a hand, foot, and mouth disease outbreak in Singapore by reverse transcription-PCR with universal enterovirus and EV71-specific primers. J. Clin. Microbiol. 2002, 40, 2823–2827. [Google Scholar] [CrossRef]
- Singh, S.; Poh, C.L.; Chow, V.T. Complete sequence analyses of enterovirus 71 strains from fatal and non-fatal cases of the hand, foot and mouth disease outbreak in Singapore (2000). Microbiol. Immunol. 2002, 46, 801–808. [Google Scholar] [CrossRef]
- Yeo, H.; Chong, C.W.H.; Chen, E.W.; Lim, Z.Q.; Ng, Q.Y.; Yan, B.; Chu, J.J.H.; Chow, V.T.K.; Alonso, S. A single amino acid substitution in structural protein VP2 abrogates the neurotropism of enterovirus A-71 in mice. Front. Microbiol. 2022, 13, 821976. [Google Scholar] [CrossRef] [PubMed]
- Hassanbhai, A.M. Characterization of Biofilm Formation by Helicobacter Pylori. PhD Thesis, National University of Singapore, Singapore, 2012. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassanbhai, A.M.; Phoon, M.C.; Chow, V.T.; Ho, B. The Association of Helicobacter pylori Biofilm with Enterovirus 71 Prolongs Viral Viability and Survival. Int. J. Mol. Sci. 2023, 24, 14500. https://doi.org/10.3390/ijms241914500
Hassanbhai AM, Phoon MC, Chow VT, Ho B. The Association of Helicobacter pylori Biofilm with Enterovirus 71 Prolongs Viral Viability and Survival. International Journal of Molecular Sciences. 2023; 24(19):14500. https://doi.org/10.3390/ijms241914500
Chicago/Turabian StyleHassanbhai, Ammar M., Meng Chee Phoon, Vincent T. Chow, and Bow Ho. 2023. "The Association of Helicobacter pylori Biofilm with Enterovirus 71 Prolongs Viral Viability and Survival" International Journal of Molecular Sciences 24, no. 19: 14500. https://doi.org/10.3390/ijms241914500
APA StyleHassanbhai, A. M., Phoon, M. C., Chow, V. T., & Ho, B. (2023). The Association of Helicobacter pylori Biofilm with Enterovirus 71 Prolongs Viral Viability and Survival. International Journal of Molecular Sciences, 24(19), 14500. https://doi.org/10.3390/ijms241914500




