Abstract
Non-small cell lung cancer (NSCLC) is one of the deadliest diseases worldwide. Tissue biopsy is the current gold standard for the diagnosis and molecular profiling of NSCLC. However, this approach presents some limitations due to inadequate tissue sampling, and intra- and intertumour heterogenicity. Liquid biopsy is a noninvasive method to determine cancer-related biomarkers in peripheral blood, and can be repeated at multiple timepoints. One of the most studied approaches to liquid biopsies is represented by circulating tumour cells (CTCs). Several studies have evaluated the prognostic and predictive role of CTCs in advanced NSCLC. Despite the limitations of these studies, the results of the majority of studies seem to be concordant regarding the correlation between high CTC count and poor prognosis in patients with NSCLC. Similarly, the decrease of CTC count during treatment may represent an important predictive marker of sensitivity to therapy in advanced NSCLC. Furthermore, molecular characterization of CTCs can be used to provide information on tumour biology, and on the mechanisms involved in resistance to targeted treatment. This review will discuss the current status of the clinical utility of CTCs in patients with advanced NSCLC, highlighting their potential application to prognosis and to treatment decision making.
Keywords:
CTCs; epithelial-to-mesenchymal transition (EMT); lung cancer; NSCLC; prognosis; treatment 1. Introduction
Lung cancer remains the leading cause of cancer-related deaths worldwide [1]. Non-small cell lung cancer (NSCLC) is a heterogeneous disease that represents about 85% of all lung malignancies [2]. More than 60% of NSCLC patients are diagnosed with advanced disease, and the 5-year survival rate is lower than 15% [1,3]. In the past twenty years, there has been a great improvement in understanding the molecular biology of oncogene-driven tumours, leading to significant advances in the treatment of these patients [4]. At the same time, the treatment of advanced non-oncogene addicted NSCLC has been revolutionized with the introduction of immune checkpoint inhibitors (ICIs) in clinical practice [5]. Unfortunately, not all patients respond, and only a subgroup of them benefit from immunotherapy. Hence, in the era of personalized medicine, there is an urgent need for new predictive biomarkers that may allow early evaluation of treatment response, selection of patients, and identification of drug-resistance.
Based on the current evidence, histopathological analysis and molecular profiling of all NSCLC tissue specimens is necessary before any therapeutic decision-making [6]. However, up to 30% of NSCLC patients do not undergo molecular profiling because of limited quantity or quality of tissue available [7], representing these two features as a strong limitation. Moreover, tissue biopsy is invasive and has low repeatability, thus limiting the possibility to define the intra- and intertumour heterogenicity in both spatial and temporal terms.
Liquid biopsy is a non-invasive method that may identify cancer-related biomarkers in peripheral blood, and can be repeated at multiple timepoints with the potential to define temporal and spatial tumour heterogeneity [8]. Circulating tumour cells (CTCs), due to recent advances in molecular knowledge, represent one of the most studied components of liquid biopsy as prognostic and predictive biomarkers in the management of NSCLC patients.
In this review, a search was conducted on the Pubmed database with the following strategy: “advanced non-small cell lung cancer” and “CTCs” in title in the last 10 years to describe the concept of CTCs and their clinical significance in the management of advanced NSCLC patients.
2. Circulating Tumour Cells
Circulating tumour cells (CTCs) are rare; one CTC per 106–107 white blood cells, found in the bloodstream of patients with solid tumours [9]. CTCs serve as a “liquid biopsy”, together with circulating metabolites, extracellular vesicles (EVs), and circulating nucleic acids such as cell-free DNA (cfDNA), microRNAs, and messenger RNA (mRNA) [9]. Liquid biopsy refers to the cytological and molecular analysis of cancer markers shed by the tumour into blood. It represents a minimally invasive and easily repeatable test, allowing for longitudinal studies. Collection and isolation of CTCs is more complex compared to ctDNA or cfDNA, but it allows protein, transcriptomic, or methylation analyses. In Figure 1, we report an overview of liquid biopsy approaches, including advantages and disadvantages (Figure 1).
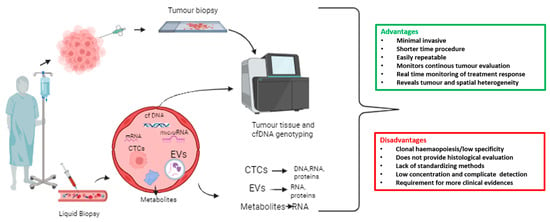
Figure 1.
Overview of liquid biopsy approach with advantages and disadvantages. A wide range of biomarker sources are included in the liquid biopsy, such as circulating tumour cells (CTCs), extracellular vesicles (EVs), messenger RNAs (mRNAs), microRNAs, cell-free DNA (cfDNA), and tumour-derived metabolites that are present in bodily fluids such as blood.
CTCs slough off of the primary tumour and, through a process known as intravasation, enter the circulating blood [10], initiating the metastatic process. They can migrate either as single cells (single CTCs), or as clusters of two or more CTCs (CTC clusters or circulating microemboli), held together by intercellular junctions [11,12]. Single CTCs and CTC clusters have a short half-life in the bloodstream (6–10 min for CTC clusters and 25–30 for single CTCs) [13], as they face several insults such as attack by immune cells, cell death by anoikis, and physical stress due to fluid shear, limiting the metastatic diffusion. However, CTC clusters are associated with poor clinical outcome in cancer patients, including lung cancer, and are characterized by a significant higher metastatic potential thanks to their stem features [11,14]. CTC clusters can be further distinguished in homotypic CTC clusters, composed of cancer cells only, and heterotypic CTC clusters, formed by cancer cells and other kind of cells, including nonmalignant immune and stromal cells [12]. Clustering of CTCs with neutrophils has been widely studied, as this interaction led to cell cycle progression in circulation and metastatic seeding acceleration [15,16]. The potential of CTCs as metastatic-initiating cells was proven in xenograft models [17], while in patients, a higher number of CTCs is correlated with increased tumour aggressiveness and decreased time to relapse in different cancer types, including NSCLC [18,19,20,21].
Therefore, due to the crucial role of CTCs in the metastatic cascade, many researchers support CTC investigation, as a tool to obtain further information concerning the tumour phenotype and genotype, and to predict disease progression and survival in early and advanced stage cancer patients.
2.1. Methods for CTC Isolation and Detection in NSCLC
As reported above, CTCs are rare, with a limited survival time in the bloodstream, making both their identification and isolation difficult and challenging [12]. Various methods have been developed for CTC enrichment and isolation, which can be performed either by positive selection [22,23] or negative selection [24,25,26], as reported in Table 1.

Table 1.
Current technologies for CTC enrichment and detection used in NSCLC. Abbreviations: EpCAM, epithelial cell adhesion molecule.
As the origin of most cancers is epithelial, several enrichment methods have been developed to select cells with positive expression of the epithelial cell adhesion molecule (EpCAM) antigen [34]. In this context, one of the most used platforms for CTC immune-magnetic enrichment, isolation, and enumeration relies on the EpCAM-dependent method CellSearch® technology (Veridex, Raritan, NJ). More specifically, CellSearch-based CTC enumeration is a helpful tool to stratify patients into favourable and unfavourable prognostic groups in advanced breast, colon, and prostate tumours [35,36,37,38], and represents the only FDA-approved CTC-based assay [39]. Concerning advanced NSCLC patients, the CellSearch detection of >5 CTCs/7.5 mL of blood was considered as a poor prognostic factor. However, CTC are not detectable in two-thirds of stage IV NSCLC patients, while in stage III NSCLC patients CTCs are detectable in less than 5% of patients [27,28]. Other EpCAM-dependent CTC detection methods based on immuno-affinity include the Adnatest assay (Qiagen) and the GILUPI Cell-collector [40].
However, the use of EpCAM as the main CTC marker may limit their detection, due to the variable expression of different markers. For instance, in the case of NSCLC using label-independent procedures, a higher number of CTCs were detected than using EpCAM-dependent methods, consistent with the low CTC-positivity rates at CellSearch [27,41]. The variable marker expression is due to epithelial–mesenchymal transition (EMT), which plays a significant role in the spread of systemic cancer. During EMT, cancer cells lose their epithelial features by a downregulation of epithelial genes, such as E-cadherin, EpCAM, occludins, claudins, and α- and β-catenin [42,43]. On the other hand, they acquire mesenchymal properties by an upregulation of mesenchymal genes such as N-cadherin, matrix metalloproteinases, integrins αv and β1, vimentin, and fibronectin [42,43]. In addition, some transcriptional factors such as Snail and Twist have a crucial role in survival of CTCs during EMT process [43]. By combining epithelial (EpCAM and CK8/18/19) and mesenchymal (vimentin and TWIST1) markers, three categories of CTCs were identified: epithelial CTCs (E-CTCs), mesenchymal CTCs (M-CTCs), and hybrid epithelial/mesenchymal phenotypes (E/M-CTCs) [43,44]. While E/M-CTCs were shown to be predictive in distinguishing patients with malignant NSCLC from those with benign disease, M-CTCs were associated with the presence of metastasis [43,44,45]. Interestingly, CTCs most frequently maintain a hybrid E/M phenotype, allowing them to retain their epithelial properties, but with an increase of the aggressive potential due to mesenchymal features [46,47].
Several studies demonstrated that all of these changes confer a higher metastatic potential and chemoresistance to cancer cells. Zhang et al. demonstrated the association between the EMT phenotype of CTCs from the peripheral blood and distant metastasis in patients with NSCLC [45]. A total of 110 patients were evaluated: 85 patients had NSCLC, of which 41 were with metastatic and 44 were with nonmetastatic disease, and 25 were with benign diseases. Overall, 80% of patients were characterized as CTC-positive with EMT markers (≥ 1 cell/5 mL blood) using a Canpatrol™ CTC assay; the CTC-positive rate was not related to the nonmetastatic and metastatic status. The total number and the mean numbers of CTCs of each subpopulation were statistically higher in NSCLC than in patients with benign pulmonary diseases. In particular, E/M- and M-CTCs rates were significantly superior in patients with NSCLC (75.3 and 44.7%, respectively) than in those with benign pulmonary diseases (0% and 0%, respectively; p < 0.05). Finally, M-CTCs were significantly higher in metastatic than in nonmetastatic patients (p < 0.001) [45]. Given these data, the EpCAM-negative CTC population may be not recognized, and detection approaches based on CTC epithelial markers may fail to detect those with mesenchymal characteristics [48].
Among the marker-independent approaches, isolation by size of epithelial tumour cells (ISET) is another method which can isolate CTCs [49]. In a study by Krebs et al., conducted on chemo-naive patients with stage IIIA to IV NSCLC, the CellSearch platform detected CTCs in 9 patients (40%), and while using ISET, 32 patients (80%) emerged as CTC-positive in a subpopulation of CTCs negative for the expression of epithelial markers [27]. Moreover, the CellSearch platform did not detect circulating tumour microemboli, while the ISET system detected them in 43% of patients [27]. However, this approach could underestimate very small CTCs [9]. In addition to their size, deformability is another property that can be exploited for CTC detection through the Parsortix microfluidic platform, which allows the recovery of CTCs and CTC clusters with preserved viability [50]. In NSCLC, the Parsortix platform provided the highest CTC-positivity compared to other technologies such as ISET and Ficoll. In addition, CTC detection using Parsortix was associated with disease progression, reduced PFS, and a high risk of relapse in NSCLC patients treated with anti-PD-1 agents [51]. In addition, microfluidic devices allow the continued collection of CTCs for further downstream analysis, and offer the possibility to integrate both isolation and detection of CTCs in a single device [52].
Other emerging approaches have been described to be suitable for CTC detection, such as spectroscopy-based techniques. Surface-enhanced Raman spectroscopy (SERS) allows for the highly sensitive and accurate detection of single cells by generating molecular fingerprint signals [53]. This method was found to be rapid and cost-effective, with high recognition rates (90%), and bio-probes with specific aptamers on parylene micropore membranes could distinguish cancer cells from white blood cells [54]. Recent findings have demonstrated that SERS can be functionalised and complemented with machine learning to detect metastasis-initiating cells directly in the blood of lung cancer patients, with a 100% diagnostic sensitivity [55].
2.2. Molecular Evaluation of CTCs
2.2.1. Single Cell Analysis
Thanks to the efforts made in technology development, molecular evaluation of CTCs is now possible, even at the single-cell level, allowing for the genomic and transcriptomic characterization of individual CTCs [56].
Concerning single-cell genomics of CTCs, many studies have focused on the assessment of single nucleotide variants (SNVs). The first technologies available for this application consisted of polymerase chain reaction (PCR) genotyping, especially digital PCR (dPCR). This low-input technique allows highly sensitive and specific detection of mutations; indeed, dPCR targets specific sequences due to the presence of primers, which can be combined to allow target multiplexing. dPCR-based SNV detection at the single-cell level has been demonstrated to be a useful tool for monitoring patients throughout the course of a disease, in addition to unveiling their genetic heterogeneity [57,58,59]. In NSCLC, the mutational analysis of epidermal growth factor receptor (EGFR) through dPCR on single CTCs isolated prior treatment with tyrosine kinase inhibitors (TKIs), identified a low mutation rate at the single-CTC level in patients positive for EGFR mutations. At the same time this approach can detect rare EGFR mutations in those patients considered as wild-type [60]. However, the initial limitation of these techniques was the fact that they were able to detect only specific alterations, limiting the discovery of other variants. More recently, the development of panels for next generation sequencing (NGS), including oncology-relevant genes, was implemented. Pailler et al. investigated resistance mutations in 48 cancer-related genes in single CTCs from ALK-rearranged NSCLC patients treated with crizotinib or lorlatinib at disease progression. In crizotinib-treated patients, mutational heterogeneity in multiple genes involved in ALK-independent pathways (“off-target” mutations), such as the RTK-KRAS (EGFR, KRAS, BRAF genes) and TP53 pathways, were identified. CTCs from lorlatinib-treated patients harboured mutations affecting the ALK gene, with some variants not detected in the corresponding tumour biopsy [61]. Single-cell whole exome sequencing was also demonstrated to be critical for studying drug resistance in NSCLC, as well as to discover new drug targets in NSCLC. Chang et al. exploited this approach to explore the mutational landscape of CTCs compared to matched primary and progressive tumours in platinum-based treated NSCLC patients. In addition to the frequency of mutations in cancer-driver genes (i.e., EGFR and TP53), genes associated with cell-cycle and stem cell features were frequently altered in CTCs, most of which were derived from primary tumour samples, and played crucial roles in chemo-drug resistance and metastasis for NSCLCs [62].
Transcriptomic analysis of single CTCs through RNA sequencing is a technology that may help to better understand the molecular mechanisms underlying metastasis [63], although few studies are present in the literature for NSCLC. One of the most recent technologies relies on the single-cell 3′ RNA sequencing for gene expression analysis using the 10X Chromium platform [64]. In CTCs isolated from diagnostic leukapheresis of NSCLC patients, single-cell 10X Chromium was able to identify heterogeneous CTC phenotypes. The epithelial-like phenotype was characterized by the expression of epithelial markers, as well as Ki67 (highly proliferation) and IL-1B, and interferon-response pathways (immune responsive). The mesenchymal/invasive phenotype showed expression of vimentin, hypoxia, and glycolysis pathways, whereas CTCs from the mesenchymal/stem cell-like phenotype were enriched in genes including ALDH13, and genes associated with adipogenesis [65].
2.2.2. PD-L1 Determination of CTCs
Several studies have been conducted in order to evaluate the expression of the programmed death-ligand 1 (PD-L1) in CTCs from patients treated with immune checkpoint inhibitors, especially with immunofluorescence techniques. In advanced NSCLC patients, it has been shown that CTCs were more frequently PD-L1-positive compared to tumour tissue, therefore representing a potential tool to identify patients who would benefit from immunotherapy [66,67]. In 96 NSCLC patients treated with nivolumab, baseline PD-L1 immunofluorescent expression in CTCs (83%) was higher than tissue (41%), and a higher number of PD-L1 positive CTCs was associated with nonresponders (PFS < 6 months) (p = 0.04). All of the patients had PD-L1 positive CTCs at disease progression [67]. Similar data concerning the increased PD-L1 positivity in CTCs compared to tumour tissue (53% and 43.2%, respectively) in advanced NSCLC patients were reported by Zhou et al. [66].
Several techniques have been described for the assessment of PD-L1 in CTCs. Among them, immunofluorescence methods were described, with the use of fluorescent antibodies targeting PD-L1, following CTC enrichment [66,67,68]. Other studies have proposed the CellSearch system, by including an additional antibody targeting PD-L1 in the empty fluorescence channel (AlexaFluor488 or fluorescein) [69,70]. Other techniques for PD-L1 expression assessment in CTCs include the DEPArray platform (Menarini Silicon Biosystems). This technology allows one to visually inspect single cells marked with antibodies targeting specific antigens (i.e., PD-L1), and to recover the cells of interest at a single-cell resolution by exploiting the dielectrophoretic principle [71]. One of the main advantages of the DEPArray technology relies on the possibility to analyse the expression of PD-L1 at single-cell resolution, as already reported in advanced urothelial carcinoma [72].
3. Prognostic Value of CTCs in Advanced NSCLC
The prognostic role of CTC count, evaluated at the baseline and after one cycle of chemotherapy by a semiautomated EpCAM–based immunomagnetic technique, has been evaluated for the first time in 101 chemotherapy-naive patients with stage III and IV NSCLC [28]. The number of CTCs in 7.5 mL of blood was higher in 60 patients with stage IV NSCLC (range, 0 to 146) than in 27 patients with stage IIIB (range, 0 to 3); in 14 IIIA patients, CTCs were not detected. In a univariate analysis, progression-free survival (PFS) was 6.8 vs. 2.4 months (p = 0.001), and overall survival (OS) was 8.1 vs. 4.3 months (p = 0.001) for patients with <5 CTCs per 7.5 mL of blood compared with ≥5 CTCs before chemotherapy, respectively. In a multivariate analysis, CTC number at baseline was the strongest predictor of OS, with a hazard ratio (HR) of 7.92 (95% CI, 2.85 to 22.01; p = 0.001) [28]. Similar results have been seen after a cycle of chemotherapy. Similarly, Nieva et al. evaluated a method using enrichment-free fluorescent labeling of CTCs, followed by automated digital microscopy in 28 patients with NSCLC [73]. CTCs were identified in 68% of analysed samples, with a median concentration of 1.6 per ml (range 0–182.6 CTCs/mL). Increased CTC detection seems to be related to progressive disease in individual patients; patients were grouped into those with ≥ 5 CTCs/mL and those with <5: the first group had a median survival of 244 days, while the second did not reach the median survival, with a median follow-up of 304 days. The HR for death was 4.0 in the group, with ≥5 CTCs/mL relative to those patients with a lower count, with a four-fold risk of dying at any given timepoint for patients with higher CTC counts (p = 0.0084) [73]. In another series of 43 advanced NSCLC, blood samples were obtained at the baseline, before the second and fifth cycles of chemotherapy. At baseline, 18 (41.9%) patients were positive for intact CTC counts, and 10 (23.2%) of them had ≥5 CTCs. The group of patients with CTC > 5 at baseline presented worse PFS and OS than those with <5 CTC (p = 0.034 and p = 0.008, respectively); during therapy, patients with a high amount of CTCs during the treatment demonstrated lower OS and PFS rates [74]. Moreover, they demonstrated that PFS and OS were shorter in patients with a progressive increase of CTC count from the baseline to final timepoint (p = 0.003 and p = 0.019, respectively) [74]. The prognostic role of CTCs has been evaluated in a pooled analysis of data from 550 patients with NSCLC enrolled in seven European centres. CTC counts of ≥ 2 and ≥ 5 per 7.5 mL were associated with reduced PFS (≥2 CTCs: HR = 1.72, p < 0.001; ≥ 5 CTCs: HR = 2.21, p < 0.001) and OS (≥2 CTCs: HR = 2.18, p < 0.001; ≥ 5 CTCs: HR = 2.75, p < 0.001), respectively [75]. In general, clinical trials performed with CellSearch methodology highlight the prognostic value of CTC count, even if negative studies have been reported in the literature [76,77]. Several methodologies, other than CellSearch, have been evaluated, including flow cytometry, the antigen-based platform Cytell, and ISET [49,78,79].
Despite interesting insights suggesting that these alternative methods may have higher sensitivity, the CellSearch approach remains the only clinically validated, FDA-cleared system for identification, isolation, and CTC counts.
Furthermore, interesting results regarding the prognostic role of CTCs in different molecular subtypes of advanced NSCLC have been reported in some studies. Tong et al. investigated the prognostic value of CTC count in 43 patients with EGFR -mutated or ALK -rearranged NSCLC at baseline and at progression of disease. Increased CTC count at baseline (≥8 CTCs/3.2 mL) was significantly associated with both shorter PFS and OS (11.6 vs. 8.5 months, p = 0.004 for PFS and 21 vs. 17.7 months, p = 0.013 for OS) [80]. Similar results were reported regarding the prognostic role of CTCs in terms of PFS in NSCLC patients treated with osimertinib [81]. According to CTC values at the baseline, patients were divided into favourable (CTCs < 5) and unfavourable (CTCs ≥ 5) prognostic groups; PFS was 9.3 in the former group and 6.5 months in the later one (HR 5.712, 95% CI: 3.781–9.577; p = 0.0002) [81]. The negative prognostic role of CTC count was also reported in a small cohort comprising 24 metastatic EGFR -positive NSCLC patients [58]. Patients with CTCs ≥ 1 in 7.5 mL had significantly shorted OS compared to patients without CTCs (3.0 months vs. not reached, p = 0.012, HR = 2.9, 95% CI 1.6–54.1 months) [82].
In Table 2, we report some recent studies evaluating the prognostic role of CTCs in advanced NSCLC.

Table 2.
Recent studies evaluating the prognostic role of CTCs in advanced NSCLC. Abbreviations: NSCLC, non-small cell lung cancer; CTC, circulating cancer cell; chemo, chemotherapy; FISH, fluorescence in situ hybridization; PFS, progression-free survival; OS, overall survival; ISET, isolation by size of tumour cells; PD-L1, programmed cell death ligand 1; WBCs, white blood cells; PD-1, programmed cell death protein 1; NA, not applicable; vs, versus; EML4, echinoderm microtubule-associated protein-like 4; ALK, anaplastic lymphoma kinase; PD, progressive disease; Nivo, nivolumab; TKI, tyrosine kinase inhibitor; EGFR, epidermal growth factor receptor; RR, response rate; mo., months.
Table 2.
Recent studies evaluating the prognostic role of CTCs in advanced NSCLC. Abbreviations: NSCLC, non-small cell lung cancer; CTC, circulating cancer cell; chemo, chemotherapy; FISH, fluorescence in situ hybridization; PFS, progression-free survival; OS, overall survival; ISET, isolation by size of tumour cells; PD-L1, programmed cell death ligand 1; WBCs, white blood cells; PD-1, programmed cell death protein 1; NA, not applicable; vs, versus; EML4, echinoderm microtubule-associated protein-like 4; ALK, anaplastic lymphoma kinase; PD, progressive disease; Nivo, nivolumab; TKI, tyrosine kinase inhibitor; EGFR, epidermal growth factor receptor; RR, response rate; mo., months.
| CTC Biomarkers | Study | Analysis Time-Point | Patients (n) | Detection Method | Therapeutic Approach | Main Findings |
|---|---|---|---|---|---|---|
| NA | J Li et al. [83] | At baseline, before 2 and 4 cycles of chemo, or after 1 and 2 mo. of targeted therapy | 100 | Negative enrichment of immune magnetic beads and FISH | Chemo /EGFR TKIs | Patients were divided into low CTC levels (<4 CTCs, LL) and high CTC levels (≥4 CTCs, HL). PFS: 5.6 mo. in LL group vs. 4.2 mo. in HL group, (p < 0.001). |
| Alama et al. [84] | At baseline, after 4 and 8 cycles | 89 | ScreenCell CYTO | Nivolumab | OS in patients with ≤2 CTCs was 8.8 mo. vs. 6.2 mo. in patients with ≥2 CTCs (p = 0.05). | |
| Zhou et al. [85] | At the 1st and the 3rd cycle | 59 | CellSearch | Chemo | Pts with CTC ≥ 2 had a significant shorter PFS and OS (4.3 vs. 4.9 mo. and 8.3 vs. 11.2 mo., respectively) | |
| PD-L1/PD-1 | Ilié et al. [86] | At baseline | 106 | ISET® platform; Rarecells | Chemo | Trend for worse OS in patients receiving first-line cisplatin-based chemotherapy, whose tumours express PD-L1 in CTCs or immune cells |
| Kallergi et al. [87] | At baseline and after 3 cycles | 30 | ISET® platform; Rarecells | Chemo | CTCs were detected in 93.3% and 81.8% of patients at baseline, and after the third chemotherapy cycle, respectively. The presence of >3 PD-1 + CTCs before treatment is associated with shorter PFS (0.5 vs. 3.9 months, p = 0.022) | |
| Cheng et al. [88] | At baseline | 66 | ISET® platform | NA | PFS time of initial treated patients with positive PD-L1 expression was shorter than that of those with negative PD-L1 expression in CTCs or tumour tissue (p > 0.05). | |
| Sinoquet et al. [89] | At baseline, or later, at progression | 54 | CellSearch | NA | CTCs and PD-L1(+) CTCs were detected in 43.4% and 9.4% of patients with NSCLC, respectively. PD-L1 expression concordance between tumour tissue and CTCs was low (54%). CTCs and PD-L1(+) CTCs were associated with worse OS, whereas PD-L1 expression in tumour tissue was not. Survival was worse in patients with PD-L1(+) CTCs | |
| EGFR | Yang et al. [81] | At baseline and at day 28 | 68 | CellSearch | Osimertinib | Patients in the favourable group (<5 CTCs) at baseline exhibited significantly longer PFS compared with patients in the unfavourable group (≥5 CTCs) (9.3 vs. 6.5 mo.; p = 0.0002). |
| ALK | Rossi et al. [90] | At baseline, at the end of first cycle of therapy (T1), and at the first and subsequent radiological assessments and/or at PD | 199 | CellSearch Expanded cytokeratins profile (EA) | NA | Pts with ≥4 CTCs had a significant lower PFS (0.29 vs. 0.66 years; p = 0.004) and OS (0.59 vs. 1.29 years, p = 0.04). Similar results using 5 CTCs as cut-off value. EML4-ALK(+) CTCs were associated with shorter PFS compared to NSCLC that did not express EML4-ALK in CTCs (0.57 years vs. 0.94 years, p = 0.017). |
| PD-L1, ALK, EGFR | Kulasinghe et al. [91] | At baseline | 33 | ClearCell FX | Nivo Chemo TKI | PFS was not found to be associated with CTCs prior to therapy (p = 0.0632), nor the presence of PD-L1 expression (p = 0.4023). |
| 11 clinically relevant genes, including EGFR and ALK | Tamminga et al. [92] | At baseline | 86 | CellSearch | Chemo TKI | RR of patients with CTC were lower than in patients without CTC (OR = 0.22, p < 0.01). In both treatment groups, the difference in RR between patients with and without CTC was similar (interaction p = 0.17). No significant interaction between CTC presence and therapy was observed (p = 0.42 for PFS and p = 0.83 for OS). |
4. Predictive Role and Clinical Implications of CTCs in Advanced NSCLC
Several studies have investigated the potential predictive role of CTCs as a biomarker during different treatment strategies in advanced NSCLC patients. Specifically, variations of CTC counts and dynamic evolution of actionable genomic mutations during therapy have been investigated as biomarkers for treatment response monitoring. Moreover, interest regarding the role of PD-L1 expression on CTCs has increased in recent years.
4.1. Predictive Role during Chemotherapy
Several studies investigated the role of CTC count as a potential biomarker predicting chemotherapy response in advanced NSCLC patients using the CellSearch System (Table 3).
As previously mentioned, in the pivotal clinical trial by Krebs and colleagues, using a positive CTC cut-off a value of ≥2 in 7.5 mL, patients with a reduction of CTC numbers after the first cycle of treatment had a significantly longer PFS (5.4 versus 1.9 months; p < 0.001) and OS (8.3 versus 3.3 months; p < 0.009) compared with those with an increased or stable number of CTCs [28]. In another study, the association between changes in CTC count and treatment outcome in untreated advanced NSCLC patients became statistically significant only after the second cycle of chemotherapy [93]. A multicentre prospective study evaluated dynamic modification of CTCs during first-line platinum-based chemotherapy in 150 advanced NSCLC patients, monitoring their count at cycle 1, at cycle 4 of therapy, and at disease progression: a persistent presence of CTCs during treatment is correlated with shorter PFS (3 vs. 5.7 months) and OS (5.6 vs. 13.1 months) compared to patients negative for CTCs [94]. Nevertheless, other studies failed to reveal the association of CTC counts and treatment response [76,85].
Moreover, alternative methods, other than CellSearch, have been used to evaluate the validity of the predictive value of CTCs in NSCLC patients treated with chemotherapy (Table 3). The research group of Gorges, using the GILUPI CellCollector, showed that the treatment response during therapy was associated with significant decreases in CTC counts (p = 0.001) in 50 advanced NSCLC patients [95]. Another study evaluated the Survivin mRNA-CTCs in 78 patients with advanced NSCLC before, after 1 cycle, and 3 cycles of first-line chemotherapy, showing that persistence of Survivin mRNA-positive CTCs after 1 and 3 cycles was significantly associated with shorter PFS and OS [96].

Table 3.
Selected studies investigating the predictive role of CTCs in advanced NSCLC patients treated with chemotherapy. Abbreviations: NSCLC, non-small cell lung cancer; CTC, circulating cancer cell; Chemo, chemotherapy; Pts, patients; FISH, fluorescence in situ hybridization; PFS, progression-free survival; OS, overall survival; PD, progressive disease; TKI, tyrosine kinase inhibitor; RR, response rate; mo., months; vs., versus.
Table 3.
Selected studies investigating the predictive role of CTCs in advanced NSCLC patients treated with chemotherapy. Abbreviations: NSCLC, non-small cell lung cancer; CTC, circulating cancer cell; Chemo, chemotherapy; Pts, patients; FISH, fluorescence in situ hybridization; PFS, progression-free survival; OS, overall survival; PD, progressive disease; TKI, tyrosine kinase inhibitor; RR, response rate; mo., months; vs., versus.
| Study | Analysis Time-Point | Patients (n) | Therapeutic Approach | Detection Method | Predictive Relevance |
|---|---|---|---|---|---|
| Krebs et al. [28] | At baseline and after 1st cycle | 101 | Chemo | CellSearch | Pts with <5 CTCs count at the second time point in comparison with those with ≥5 CTCs: PFS: 6.9 vs. 2.4 mo., p = 0.012 OS: 8.9 vs. 3.9 mo., p = 0.003 |
| Xu et al. [31] | Before the 1st, 2nd, and 3rd cycle | 66 | Chemo | CellSearch | A statistically significant difference between CTC value, a modification after 2 courses and PD was observed (p = 0.028) |
| Muinelo-Romay et al. [74] | Before the 1st, 2nd, and 5th cycles of chemo | 43 | Chemo | CellSearch | Pts with ≥2 CTCs at 2nd cycle had superior rates of PD and poorer PFS (4.2 mo. vs. 8.5 mo., p = 0.016) |
| Gorges et al. [95] | At baseline and every 12 weeks | 50 | Chemo | GILUPI CellCollector | Treatment response during therapy was associated with significant decreases in CTC counts (p = 0.001) |
| J Li et al. [83] | At baseline, before 2 and 4 cycle of chemo or after 1 and 2 months of targeted therapy | 100 | Chemo Targeted therapy (anti-EGFR) | Negative enrichment of immune magnetic beads and FISH | The changes in CTC levels before and after the 2nd cycle of chemo were correlated with the DCR: 38% in CTC-positive pts vs. 62% in CTC-negative pts (p < 0.001) |
| Tamminga et al. [92] | At baseline | 86 | Chemo (n = 52) TKI (n = 34) | CellSearch | Pts with CTCs vs. pts without CTCs: RR: (a) TKI: 25% vs. 73% (p = 0.02) (b) chemo: 35% vs. 51% (p = 0.05) PFS: 3.3 mo. vs. 8 mo., p = 0.01 OS: 5.2 mo. vs. 12.1 mo., p = 0.03 |
| Wang et al. [94] | At baseline, at cycle 1 and cycle 4, and at PD | 150 | Chemo | CellSearch | Pts with persistent negative CTCs had longer PFS (5.7 mo. vs. 3.0 mo.) and OS (13.1 mo. vs. 5.6 mo.) compared with pts with persistent positive CTCs, which was not impacted by chemotherapy. Chemotherapy decreased CTC from 36.0% to 13.7% |
| Zhou et al. [85] | Before the 1st and the 3rd cycle | 59 | Chemo | CellSearch | No correlation between the change of CTC counts and response to chemotherapy was observed |
| Hirose et al. [76] | At baseline | 33 | Chemo | CellSearch | No differences in response rates to cytotoxic chemotherapy between CTC-positive patients and CTC-negative patients. |
4.2. Predictive Role in Oncogene-Addicted NSCLC during Targeted Therapy
The predictive value of CTCs has been investigated in oncogene-addicted NSCLC patients receiving targeted therapy, especially in those with EGFR mutations and ALK rearrangements.
A phase II trial investigated the role of CTCs in 41 unselected patients with relapsed or refractory metastatic NSCLC treated with pertuzumab and erlotinib. CTC count was analysed using CellSearch at baseline and during the course of treatment. CTC value decreased in a statistically significant manner from day 56 of treatment, with a significant prolongation of longer PFS (p = 0.050) [97]. In the same line, another phase II trial using flow cytometry evaluated the correlation between the change of CTC values and response to first-generation EGFR TKIs in 66 advanced, EGFR-mutant NSCLC patients. No significant differences were observed in CTC counts between baseline and after treatment in patients with progressive disease (n = 26), whereas the CTC values were significant reduced during treatment in 40 patients with clinical benefit (p = 0.009) [98]. Moreover, PFS and 1-year and 3-year OS rates were longer for patients with low CTC counts than those with high CTC counts [98].
Finally, Breitenbuecher et al. evaluated the predictive role of CTCs in a small sample of treatment-naive patients with advanced EGFR DelEx19-mutated NSCLC [99]. Fifty percent of patients with EGFR-mutant positive CTC samples at baseline became negative within a median of 34 days of treatment, and CTC negativity lasted for a median of 109 days. The negativization of EGFR-mutant CTCs was associated with radiological and clinical response to treatment. Moreover, an exploratory analysis showed a significant statistical correlation between the duration of CTC negativity and the time to treatment failure [99].
In a similar way, a limited number of studies reported the ability to detect ALK-rearrangement in CTCs from ALK-rearranged NSCLC patients [100,101]. Interestingly, the correlation between dynamic modification of CTC counts during treatment and treatment response or disease progression in patients with ALK-rearranged NSCLC was confirmed in more studies [102,103]. The results of some studies which investigated the predictive value of CTCs in this setting of patients are shown in Table 4.

Table 4.
Selective recent studies investigating the predictive value of CTCs in NSCLC patients treated with TKI. Abbreviations: NSCLC, non-small cell lung cancer; CTC, circulating cancer cell; TKI, tyrosine kinase inhibitor; ISET, isolation by size of tumour cells; CK, cytokeratin; EMT, epithelial–mesenchymal transition; VIM, vimentin; PD-L1, programmed cell death ligand 1; PFS, progression-free survival; PFS 12 m: one-year progression-free survival; RR, response rate; EGFR, epidermal growth factor receptor; ALK, anaplastic lymphoma kinase; OS, overall survival; CNG, copy number gain; pts, patients.
4.3. Identification of Mechanisms of Resistance in Oncogene Addicted NSCLC during Treatment
In an attempt to identify drug resistance mechanisms, investigators explored the impact of molecular profiling of CTCs in NSCLC patients.
Maheswaran et al. reported a correlation between the emergence of T790M mutations in CTCs and acquisition of drug resistance in patients affected by advanced NSCLC treated with gefitinib [109]. Recently, a Japanese research group investigated the role of BIM-γ mRNA expression on CTCs in 30 EGFR-mutated NSCLC patients treated with osimertinib [107]. No significant different was found in terms of objective response rate (ORR) or PFS for osimertinib between CTC-positive patients with ≥10 CTCs per 7.5 mL and those with <10 CTCs per 7.5 mL. The ORR to osimertinib was inferior in patients with high expression of BIM-γ mRNA compared with those with low BIM-γ mRNA expression (26.6% vs. 73.3%, respectively; p = 0.011). However, there was no significant difference in terms of disease control rate (DCR), PFS, and OS between the groups. The authors concluded that BIM-γ mRNA overexpression in CTCs in this subset of patients could be a potential biomarker for inadequate response to osimertinib [107].
Moreover, a Greek study group studied changes in gene expression in CTC-enriched fractions of 30 EGFR-mutant NSCLC patients under osimertinib [104]. RNA-based molecular characterization of CTCs was performed before, after 1 cycle of treatment, and at progression of disease. They observed a dynamic role of EMT as a resistance mechanism during osimertinib treatment in these patients. Indeed, at PD, they compared the different CTC profiles, observing significant correlations between the epithelial CTC profile and ALDH-1 (p = 0.043), and the mesenchymal/EMT profile and ALDH-1 (p = 0.014). Interestingly, there was also a significant increase in the expression levels of PD-L1 in CTCs between baseline and disease progression (p = 0.016). Moreover, a correlation was found between PD-L1 status and the mesenchymal subtype (p = 0.036), and more precisely with VIM overexpression (p = 0.011) at PD. Therefore, based on these observations, the authors suggested a potential role of ICIs in this subpopulation of patients [104].
Recently, Chen et al. investigated drug-resistant gene function and dynamic change of CTCs in 20 EGFR-positive NSCLC patients before and after treatment with osimertinib. They found that osimertinib increased the expression of HER-2 on the cell surface of NSCLC cell lines as a drug resistance mechanism [110].
More recently, scientific interest has been focused on CTC profiling in patients with ALK-rearranged NSCLC. In a study by Pailler et al., the presence of resistance mutations in CTCs of 17 ALK-rearranged NSCLC patients progressing on Crizotinib (n = 14) or Lorlatinib (n = 3) was investigated. In spite of the limitations of this study, they highlighted the high genetic heterogeneity in both ALK-independent and ALK-dependent pathways, and potential clinical importance of CTCs to identify therapeutic resistance mutations in this population [103].
As a continuation of this study [61], Oulhen et al. [111] analysed 6 of these 17 ALK-rearranged patients with ≥10 CTCs/20 mL blood at resistance to 1st and 3rd ALK-TKIs. They described the copy number alteration (CNA) heterogeneity based on epithelial and/or ALK expression features. Phenotypic analysis showed similar CTC features for each patient that were either ALK−/CK+ or ALK+/CK−. CNA analysis performed on 43 CTC samples showed a significant CNA heterogeneity and high CIN at treatment resistance. RTK/RAS was found to be the predominant mutated ALK-independent pathway revealed by CTCs, followed by multiple gains in EGFR, FGFR1, ERBB2, and NTRK. Based on these results, authors suggested a potential utility of CNA evaluation on CTCs for better defining the different resistance mechanisms to ALK-TKIs, and identifying potential alternative targets bypassing these drug resistance pathways [111].
4.4. PD-L1 Expression in CTCs during Treatment with Immune Checkpoint Inhibitors
Programmed death ligand 1 (PD-L1) represents the tumour tissue biomarker in order to predict the responsivity to treatment with ICIs in NSCLC. The reliability to predict the response to ICIs of PD-L1 can be related to its variable expression within and between tumour sites and in different time points of tumour evolution [112]. Given that, the use of CTCs may overcome tumour spatiotemporal heterogeneity, and predict responses to ICIs in order to investigate PD-L1 expression on them [113]. In a study by Nicolazzo et al., 24 advanced NSCLC patients treated with nivolumab have been evaluated for PD-L1 expressing CTCs at three predefined timepoints (after 1, 3, and 6 months after starting treatment). Interestingly, the persistence of PD-L1-positive CTCs after 6 months of treatment was correlated with a lack of treatment response [114]. Similarly, Guibert et al. confirmed the prognostic role of PD-L1-positive CTCs in 96 advanced NSCLC patients treated with nivolumab using the ISET method [67]. However, PD-L1-positive CTCs have not shown a clear predictive effect. Moreover, as reported in most studies, they confirmed the lack of correlation between PD-L1 positivity in tumour tissue and PD-L1 positivity on CTCs [67].
Only one study suggested that PD-L1 expression on CTCs could be used as a valid alternative to tumour tissue PD-L1 evaluation for monitoring ICI treatment response, based on their results that demonstrated a correlation of PD-L1 status in CTCs and tumour tissue [86]. Furthermore, Raimondi et al. showed that the presence of PD-L1-positive CTCs isolated from a population of NSCLC patients were correlated with a partial EMT phenotype, suggesting that coexistence of both of these markers represents a possible immune escape mechanism [115]. To the contrary, Kulasinghe et al. reported no association between PFS and presence of PD-L1-positive CTCs in advanced NSCLC patients [91]. Spiliotaki et al. explored PD-L1 and Ki-67 in CTCs of 47 advanced NSCLC patients receiving pembrolizumab [116]. Blood samples were analysed at baseline, post-first cycle, post-third, and primary resistance (PMR). Patients in PD presented a higher number of total and PD-L1-low CTCs after the first cycle compared to patients with clinical benefit (83% vs. 37% and 67% vs. 8%, respectively). Moreover, patients with a reduction of total and PD-L1-low CTCs after the first cycle of pembrolizumab presented a longer PFS. To the contrary, PD-L1-positive patients with a Ki67 index > 30% on baseline presented shorter PFS and OS compared to those harbouring Ki67 ≤ 30% [116].
A summary of the recent studies evaluating PD-L1 expression on CTCs in advanced NSCLC patients is reported in Table 5.

Table 5.
Selective studies investigating the predictive role of PD-L1-positive CTCs in advanced NSCLC patients in treatment with ICIs. Abbreviations: NSCLC, non-small cell lung cancer; CTC, circulating cancer cell; ICI, immune checkpoint inhibitor; PD-L1, programmed cell death ligand 1; PD, progressive disease; ISET, isolation by size of tumour cells; PFS, progression-free survival; OS, overall survival; pts, patients; Nivo, nivolumab; EMT, epithelial–mesenchymal transition; MTV, metabolic tumour volume; MCA, automated microcavity array; PR, partial response; NR, not reached; Pembro, pembrolizumab.
5. Future Perspectives: CTCs vs. cfDNA
One of the most interesting fields in CTC research is the possibility to study their molecular profiling in order to identify biomarkers predictive of response to targeted therapy.
Indeed, several studies analysed the predictive role of some actionable genomic mutations or rearrangements, such as ALK, EGFR, and ROS-1 in CTCs of metastatic NSCLC patients, trying to identify drug resistance mechanisms and potentially leading to the development of new target-based agents [61,103,107,109]. However, to date, the field of molecular characterization of NSCLC is dominated by the use of circulating tumour DNA (cfDNA) that is already commonly integrated into clinical practice of NSCLC management, differently from CTCs. This is due to the fact that cfDNA analysis is easier and less expensive compared to CTC analysis [120]. In addition, the number of CTCs isolated in patients with NSCLC is too limited to consider molecular analysis.
However, CTC evaluation has some advantages over cfDNA, and enables one to obtain different information. First of all, CTC analysis allows the detection of protein expression, as PD-L1 can be a potential predictive marker in NSCLC patients treated with ICIs [67,114]. Second, on CTCs, it is possible to analyse morphological features. Finally, molecular profiling of CTCs depicts better tumour heterogeneity in spatiotemporal terms compared to cfDNA [121]. Recent studies have also demonstrated the significant role of CTCs in ex vivo models, including mouse xenografts [17,122], investigating the process of metastatization and of drug resistance mechanisms, leading to new therapeutic opportunities.
In the near future, we expect interesting progresses in this research field because of a progressive improvement of platforms for single-cell analysis of CTCs. Moreover, an alliance between CTC and cfDNA analysis will improve the clinical application of liquid biopsy, leading to an earlier identification of nonresponders who may benefit from an early switch to a more effective alternative treatment.
6. Conclusions
Despite a significant heterogeneity in the studies investigating the role of CTCs in terms of patient characteristics, CTC cut-off value, and methods of CTC analysis, the results of the majority of studies seem to be concordant regarding the correlation between CTC count and survival endpoints in advanced NSCLC. Moreover, several studies have confirmed the predictive role of CTCs during different types of therapies in terms of treatment response or drug resistance. Nevertheless, while the use of CTCs in NSCLC seems to be promising, their use was not implemented in routine clinical practice, and some important obstacles need to be overcome.
Development of well-standardized platforms with higher sensitivity and specificity for the detection and molecular analysis of CTCs, and subsequent validation of their role in large cohorts of patients, are unquestionably necessary. Moreover, large-scale prospective randomized trials with homogeneous patient features and robust endpoints should be conducted for validation of CTCs in NSCLC patients and their subsequent implementation in clinical practice. Furthermore, based on limited data regarding the prognostic and predictive role of CTCs in specific molecular subgroups of NSCLC patients eligible for targeted therapies or in patients treated with immune checkpoint inhibitors, additional research in these groups is absolutely necessary.
In conclusion, in the era of precision oncology, CTC analysis could be a promising tool for prognosis, selection of patients most likely to benefit from personalized treatment, and improving of our knowledge of the molecular evolution of this disease.
Author Contributions
K.A. and T.R. wrote the original draft. P.C., A.V., M.A.B., I.P. and S.B. revised the manuscript. L.C., A.D. and P.U. revised the manuscript and gave the final approval. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partly supported thanks to the contribution of Ricerca Corrente by the Italian Ministry of Health within the research line “Precision, gender and ethnicity-based medicine and geroscience: genetic-molecular mechanisms in the development, characterization, and treatment of tumors”.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Data sharing not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Luo, G.; Etxeberria, J.; Hao, Y. Global Patterns and Trends in Lung Cancer Incidence: A Population-Based Study. J. Thorac. Oncol. 2021, 16, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Chen, B.H.; Chou, W.C.; Yang, C.T.; Chang, J.W. Factors associated with the prognosis and long-term survival of patients with metastatic lung adenocarcinoma: A retrospective analysis. J. Thorac. Dis. 2018, 10, 2070–2078, Erratum in: J. Thorac. Dis. 2018, 10, E604. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.F.; Mardis, E.R. The emerging clinical relevance of genomics in cancer medicine. Nat. Rev. Clin. Oncol. 2018, 15, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Onoi, K.; Chihara, Y.; Uchino, J.; Shimamoto, T.; Morimoto, Y.; Iwasaku, M.; Kaneko, Y.; Yamada, T.; Takayama, K. Immune Checkpoint Inhibitors for Lung Cancer Treatment: A Review. J. Clin. Med. 2020, 9, 1362. [Google Scholar] [CrossRef]
- European Society for Medical Oncology. Metastatic Non-Small-Cell Lung Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Available online: https://www.esmo.org/content/download/347819/6934778/1/ESMO-CPG-mNSCLC-15SEPT2020.pdf (accessed on 3 June 2023).
- Pisapia, P.; Malapelle, U.; Troncone, G. Liquid Biopsy and Lung Cancer. Acta. Cytol. 2019, 63, 489–496. [Google Scholar] [CrossRef]
- Malapelle, U.; Tiseo, M.; Vivancos, A.; Kapp, J.; Serrano, M.J.; Tiemann, M. Liquid Biopsy for Biomarker Testing in Non-Small Cell Lung Cancer: A European Perspective. J. Mol. Pathol. 2021, 2, 255–273. [Google Scholar] [CrossRef]
- Andree, K.C.; van Dalum, G.; Terstappen, L.W. Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol. 2016, 10, 395–407. [Google Scholar] [CrossRef]
- Micalizzi, D.S.; Maheswaran, S.; Haber, D.A. A conduit to metastasis: Circulating tumor cell biology. Genes Dev. 2017, 31, 1827–1840. [Google Scholar] [CrossRef]
- Aceto, N.; Toner, M.; Maheswaran, S.; Haber, D.A. En Route to Metastasis: Circulating Tumor Cell Clusters and Epithelial-To-Mesenchymal Transition. Trends Cancer 2015, 1, 44–52. [Google Scholar] [CrossRef]
- Aceto, N. Bring along your friends: Homotypic and heterotypic circulating tumor cell clustering to accelerate metastasis. Biomed. J. 2020, 43, 18–23. [Google Scholar] [CrossRef]
- Yu, M. Metastasis Stemming from Circulating Tumor Cell Clusters. Trends Cell Biol. 2019, 29, 275–276. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Pantel, K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016, 6, 479–491. [Google Scholar] [CrossRef]
- Szczerba, B.M.; Castro-Giner, F.; Vetter, M.; Vetter, M.; Krol, I.; Gkountela, S.; Landin, J.; Aceto, N. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 2019, 566, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Rozenberg, J.M.; Buzdin, A.A.; Mohammad, T.; Rakitina, O.A.; Didych, D.A.; Pleshkan, V.V.; Alekseenko, I.V. Molecules promoting circulating clusters of cancer cells suggest novel therapeutic targets for treatment of metastatic cancers. Front. Immunol. 2023, 14, 1099921. [Google Scholar] [CrossRef] [PubMed]
- Baccelli, I.; Schneeweiss, A.; Riethdorf, S.; Stenzinger, A.; Schillert, A.; Vogel, V.; Klein, C.; Saini, M.; Bäuerle, T.; Wallwiener, M.; et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 2013, 31, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Mu, Z.; Ye, Z.; Zhang, Z.; Abu-Khalaf, M.M.; Silver, D.P.; Palazzo, J.P.; Jagannathan, G.; Fellin, F.M.; Bhattacharya, S.; et al. Prognostic value of HER2 status on circulating tumor cells in advanced-stage breast cancer patients with HER2-negative tumors. Breast Cancer Res. Treat. 2020, 181, 679–689. [Google Scholar] [CrossRef]
- Paoletti, C.; Regan, M.M.; Niman, S.M.; Dolce, E.M.; Darga, E.P.; Liu, M.C.; Marcom, P.K.; Hart, L.L.; Smith, J.W., II; Tedesco, K.L.; et al. Circulating tumor cell number and endocrine therapy index in ER positive metastatic breast cancer patients. NPJ Breast Cancer 2021, 7, 77. [Google Scholar] [CrossRef]
- de Kruijff, I.E.; Sieuwerts, A.M.; Onstenk, W.; Kraan, J.; Smid, M.; Van, M.N.; van der Vlugt-Daane, M.; Hoop, E.O.; Mathijssen, R.H.J.; Lolkema, M.P.; et al. Circulating Tumor Cell Enumeration and Characterization in Metastatic Castration-Resistant Prostate Cancer Patients Treated with Cabazitaxel. Cancers 2019, 11, 1212. [Google Scholar] [CrossRef]
- Lindsay, C.R.; Faugeroux, V.; Michiels, S.; Pailler, E.; Facchinetti, F.; Ou, D.; Bluthgen, M.V.; Pannet, C.; Ngo-Camus, M.; Bescher, G.; et al. A prospective examination of circulating tumor cell profiles in non-small-cell lung cancer molecular subgroups. Ann. Oncol. 2017, 28, 1523–1531. [Google Scholar] [CrossRef]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [Google Scholar] [CrossRef] [PubMed]
- Gleghorn, J.P.; Pratt, E.D.; Denning, D.; Liu, H.; Bander, N.H.; Tagawa, S.T.; Nanus, D.M.; Giannakakou, P.A.; Kirby, B.J. Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody. Lab Chip 2010, 10, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Casavant, B.P.; Mosher, R.; Warrick, J.W.; Maccoux, L.J.; Berry, S.M.; Becker, J.T.; Chen, V.; Lang, J.M.; McNeel, D.G.; Beebe, D.J. A negative selection methodology using a microfluidic platform for the isolation and enumeration of circulating tumor cells. Methods 2013, 64, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro-Samy, S.; Oliveira, M.I.; Pereira-Veiga, T.; Muinelo-Romay, L.; Carvalho, S.; Gaspar, J.; Freitas, P.P.; López-López, R.; Costa, C.; Diéguez, L. Fast and efficient microfluidic cell filter for isolation of circulating tumor cells from unprocessed whole blood of colorectal cancer patients. Sci. Rep. 2019, 9, 8032. [Google Scholar] [CrossRef]
- Warkiani, M.E.; Khoo, B.L.; Tan, D.S.; Bhagat, A.A.; Lim, W.T.; Yap, Y.S.; Lee, S.C.; Soo, R.A.; Han, J.; Lim, C.T. An ultra-high-throughput spiral microfluidic biochip for the enrichment of circulating tumor cells. Analyst 2014, 139, 3245–3255. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Hou, J.M.; Sloane, R.; Lancashire, L.; Priest, L.; Nonaka, D.; Ward, T.H.; Backen, A.; Clack, G.; Hughes, A.; et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J. Thorac. Oncol. 2012, 7, 306–315. [Google Scholar] [CrossRef]
- Krebs, M.G.; Sloane, R.; Priest, L.; Lancashire, L.; Hou, J.M.; Greystoke, A.; Ward, T.H.; Ferraldeschi, R.; Hughes, A.; Clack, G.; et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 2011, 29, 1556–1563. [Google Scholar] [CrossRef]
- He, Y.; Shi, J.; Shi, G.; Xu, X.; Liu, Q.; Liu, C.; Gao, Z.; Bai, J.; Shan, B. Using the New CellCollector to Capture Circulating Tumor Cells from Blood in Different Groups of Pulmonary Disease: A Cohort Study. Sci. Rep. 2017, 7, 9542. [Google Scholar] [CrossRef]
- Qian, H.; Zhang, Y.; Xu, J.; He, J.; Gao, W. Progress and application of circulating tumor cells in non-small cell lung cancer. Mol. Ther. Oncolytics 2021, 22, 72–84. [Google Scholar] [CrossRef]
- Xu, L.; Mao, X.; Imrali, A.; Syed, F.; Mutsvangwa, K.; Berney, D.; Cathcart, P.; Hines, J.; Shamash, J.; Lu, Y.J. Optimization and Evaluation of a Novel Size Based Circulating Tumor Cell Isolation System. PLoS ONE 2015, 10, e0138032. [Google Scholar] [CrossRef]
- Wu, S.; Liu, S.; Liu, Z.; Huang, J.; Pu, X.; Li, J.; Yang, D.; Deng, H.; Yang, N.; Xu, J. Classification of circulating tumor cells by epithelial-mesenchymal transition markers. PLoS ONE 2015, 10, e0123976. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Wang, S.; Fang, Y.; Zheng, L.; Zhi, X.; Cheng, B.; Chen, Y.; Zhang, C.; Shi, D.; Song, H.; et al. Feasibility of a novel one-stop ISET device to capture CTCs and its clinical application. Oncotarget 2017, 8, 3029–3041. [Google Scholar] [CrossRef] [PubMed]
- Gires, O.; Pan, M.; Schinke, H.; Canis, M.; Baeuerle, P.A. Expression and function of epithelial cell adhesion molecule EpCAM: Where are we after 40 years? Cancer Metastasis Rev. 2020, 39, 969–987. [Google Scholar] [CrossRef]
- Cohen, S.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 3213–3221, Erratum in: J. Clin. Oncol. 2009, 27, 1923. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Hayes, D.F.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Reuben, J.M.; Doyle, G.V.; Matera, J.; Allard, W.J.; Miller, M.C.; et al. Circulating tumor cells: A novel prognostic factor for newly diagnosed metastatic breast cancer. J. Clin. Oncol. 2005, 23, 1420–1430, Erratum in: J. Clin. Oncol. 2005, 23, 4808. [Google Scholar] [CrossRef] [PubMed]
- de Bono, J.S.; Attard, G.; Adjei, A.; Pollak, M.N.; Fong, P.C.; Haluska, P.; Roberts, L.; Melvin, C.; Repollet, M.; Chianese, D.; et al. Potential applications for circulating tumor cells expressing the insulin-like growth factor-I receptor. Clin. Cancer Res. 2007, 13, 3611–3616. [Google Scholar] [CrossRef]
- Hayes, D.F.; Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Miller, M.C.; Matera, J.; Allard, W.J.; Doyle, G.V.; Terstappen, L.W. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 2006, 12, 4218–4224. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.; Uhr, J.W.; Terstappen, L.W. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Shao, W.; Li, Z.; Zhao, R.; Ye, Z. Strategies for enrichment of circulating tumor cells. Transl. Cancer Res. 2020, 9, 2012–2025. [Google Scholar] [CrossRef]
- Wang, J.; Lu, W.; Tang, C.; Liu, Y.; Sun, J.; Mu, X.; Zhang, L.; Dai, B.; Li, X.; Zhuo, H.; et al. Label-Free Isolation and mRNA Detection of Circulating Tumor Cells from Patients with Metastatic Lung Cancer for Disease Diagnosis and Monitoring Therapeutic Efficacy. Anal. Chem. 2015, 87, 11893–11900. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Pantazaka, E.; Vardas, V.; Roumeliotou, A.; Kakavogiannis, S.; Kallergi, G. Clinical Relevance of Mesenchymal- and Stem-Associated Phenotypes in Circulating Tumor Cells Isolated from Lung Cancer Patients. Cancers 2021, 13, 2158. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Men, Y.; Wang, J.; Xing, P.; Zhao, J.; Li, J.; Xu, D.; Hui, Z.; Cui, W. Epithelial circulating tumor cells with a heterogeneous phenotype are associated with metastasis in NSCLC. J. Cancer Res. Clin. Oncol. 2022, 148, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wei, L.; Li, J.; Zheng, J.; Zhang, S.; Zhou, J. Epithelial-mesenchymal transition phenotype of circulating tumor cells is associated with distant metastasis in patients with NSCLC. Mol. Med. Rep. 2019, 19, 601–608. [Google Scholar] [CrossRef]
- Kallergi, G.; Aggouraki, D.; Zacharopoulou, N.; Stournaras, C.; Georgoulias, V.; Martin, S.S. Evaluation of α-tubulin, detyrosinated α-tubulin, and vimentin in CTCs: Identification of the interaction between CTCs and blood cells through cytoskeletal elements. Breast Cancer Res. 2018, 20, 67. [Google Scholar] [CrossRef]
- Kallergi, G.; Papadaki, M.A.; Politaki, E.; Mavroudis, D.; Georgoulias, V.; Agelaki, S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011, 13, R59. [Google Scholar] [CrossRef]
- Eslami-S, Z.; Cortés-Hernández, L.E.; Alix-Panabières, C. Epithelial Cell Adhesion Molecule: An Anchor to Isolate Clinically Relevant Circulating Tumor Cells. Cells 2020, 9, 1836. [Google Scholar] [CrossRef]
- Farace, F.; Massard, C.; Vimond, N.; Drusch, F.; Jacques, N.; Billiot, F.; Laplanche, A.; Chauchereau, A.; Lacroix, L.; Planchard, D.; et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br. J. Cancer 2011, 105, 847–853. [Google Scholar] [CrossRef]
- Miller, M.C.; Robinson, P.S.; Wagner, C.; O’Shannessy, D.J. The Parsortix™ Cell Separation System-A versatile liquid biopsy platform. Cytometry A 2018, 93, 1234–1239. [Google Scholar] [CrossRef]
- Papadaki, M.A.; Sotiriou, A.I.; Vasilopoulou, C.; Filika, M.; Aggouraki, D.; Tsoulfas, P.G.; Apostolopoulou, C.A.; Rounis, K.; Mavroudis, D.; Agelaki, S. Optimization of the Enrichment of Circulating Tumor Cells for Downstream Phenotypic Analysis in Patients with Non-Small Cell Lung Cancer Treated with Anti-PD-1 Immunotherapy. Cancers 2020, 12, 1556. [Google Scholar] [CrossRef]
- Pandey, C.M.; Augustine, S.; Kumar, S.; Kumar, S.; Nara, S.; Srivastava, S.; Malhotra, B.D. Microfluidics Based Point-Of-Care Diagnostics. Biotechnol. J. 2018, 13, 1700047. [Google Scholar] [CrossRef]
- Kamińska, A.; Szymborski, T.; Witkowska, E.; Kijeńska-Gawrońska, E.; Świeszkowski, W.; Niciński, K.; Trzcińska-Danielewicz, J.; Girstun, A. Detection of Circulating Tumor Cells Using Membrane-Based SERS Platform: A New Diagnostic Approach for ‘Liquid Biopsy’. Nanomaterials 2019, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Fu, B.; Liu, W.; Huang, W.; Li, M.; Liu, Y.; Kang, Y.; Wang, J.; Bai, S.; Lu, C.; et al. Efficient detection of single circulating tumor cell in blood using Raman mapping based on Aptamer-SERS bio-probe coupled with micropore membrane filtration. Talanta 2023, 267, 125220. [Google Scholar] [CrossRef] [PubMed]
- Premachandran, S.; Dhinakaran, A.K.; Das, S.; Venkatakrishnan, K.; Tan, B.; Sharma, M. Detection of lung cancer metastasis from blood using L-MISC nanosensor: Targeting circulating metastatic cues for improved diagnosis. Biosens. Bioelectron. 2023, 243, 115782. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, D.; Peng, L.; Huang, T.; He, X.; Wang, J.; Ou, C. Single-cell sequencing: A promising approach for uncovering the mechanisms of tumor metastasis. J. Hematol. Oncol. 2022, 15, 59. [Google Scholar] [CrossRef] [PubMed]
- Owen, S.; Lo, T.W.; Fouladdel, S.; Zeinali, M.; Keller, E.; Azizi, E.; Ramnath, N.; Nagrath, S. Simultaneous Single Cell Gene Expression and EGFR Mutation Analysis of Circulating Tumor Cells Reveals Distinct Phenotypes in NSCLC. Adv. Biosyst. 2020, 4, e2000110. [Google Scholar] [CrossRef]
- Rowlands, V.; Rutkowski, A.J.; Meuser, E.; Carr, T.H.; Harrington, E.A.; Barrett, J.C. Optimisation of robust singleplex and multiplex droplet digital PCR assays for high confidence mutation detection in circulating tumour DNA. Sci. Rep. 2019, 9, 12620. [Google Scholar] [CrossRef]
- Denis, J.A.; Patroni, A.; Guillerm, E.; Pépin, D.; Benali-Furet, N.; Wechsler, J.; Manceau, G.; Bernard, M.; Coulet, F.; Larsen, A.K.; et al. Droplet digital PCR of circulating tumor cells from colorectal cancer patients can predict KRAS mutations before surgery. Mol. Oncol. 2016, 10, 1221–1231. [Google Scholar] [CrossRef]
- Zhang, Q.; Nong, J.; Wang, J.; Yan, Z.; Yi, L.; Gao, X.; Liu, Z.; Zhang, H.; Zhang, S. Isolation of circulating tumor cells and detection of EGFR mutations in patients with non-small-cell lung cancer. Oncol. Lett. 2019, 17, 3799–3807. [Google Scholar] [CrossRef]
- Pailler, E.; Faugeroux, V.; Oulhen, M.; Mezquita, L.; Laporte, M.; Honoré, A.; Lecluse, Y.; Queffelec, P.; NgoCamus, M.; Nicotra, C.; et al. Acquired Resistance Mutations to ALK Inhibitors Identified by Single Circulating Tumor Cell Sequencing in ALK-Rearranged Non-Small-Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 6671–6682. [Google Scholar] [CrossRef]
- Chang, Y.; Wang, Y.; Li, B.; Lu, X.; Wang, R.; Li, H.; Yan, B.; Gu, A.; Wang, W.; Huang, A.; et al. Whole-Exome Sequencing on Circulating Tumor Cells Explores Platinum-Drug Resistance Mutations in Advanced Non-small Cell Lung Cancer. Front. Genet. 2021, 12, 722078. [Google Scholar] [CrossRef] [PubMed]
- Negishi, R.; Yamakawa, H.; Kobayashi, T.; Horikawa, M.; Shimoyama, T.; Koizumi, F.; Sawada, T.; Oboki, K.; Omuro, Y.; Funasaka, C.; et al. Transcriptomic profiling of single circulating tumor cells provides insight into human metastatic gastric cancer. Commun. Biol. 2022, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, Y.; Zhang, Q.; Ren, X.; Zhang, Z. Direct Comparative Analyses of 10X Genomics Chromium and Smart-seq2. Genom. Proteom. Bioinform. 2021, 19, 253–266. [Google Scholar] [CrossRef]
- Rieckmann, L.M.; Spohn, M.; Selbuz, E.; Schubert, C.; Agorku, D.; Becker, L.; Borchers, A.; Krause, J.; Ruff, L.; Heinemann, S.; et al. Abstract 3374: Large-scale single-cell whole transcriptomic analyses reveal distinct malignant phenotypes of CTCs from NSCLC patients. Cancer Res. 2022, 82 (Suppl. 12), 3374. [Google Scholar] [CrossRef]
- Zhou, Q.; Liu, X.; Li, J.; Tong, B.; Xu, Y.; Chen, M.; Liu, X.; Gao, X.; Shi, Y.; Zhao, J.; et al. Circulating tumor cells PD-L1 expression detection and correlation of therapeutic efficacy of immune checkpoint inhibition in advanced non-small-cell lung cancer. Thorac. Cancer 2023, 14, 470–478. [Google Scholar] [CrossRef]
- Guibert, N.; Delaunay, M.; Lusque, A.; Boubekeur, N.; Rouquette, I.; Clermont, E.; Mourlanette, J.; Gouin, S.; Dormoy, I.; Favre, G.; et al. PD-L1 expression in circulating tumor cells of advanced non-small cell lung cancer patients treated with nivolumab. Lung Cancer 2018, 120, 108–112. [Google Scholar] [CrossRef]
- Dall’Olio, F.G.; Gelsomino, F.; Conci, N.; Marcolin, L.; De Giglio, A.; Grilli, G.; Sperandi, F.; Fontana, F.; Terracciano, M.; Fragomeno, B.; et al. PD-L1 Expression in Circulating Tumor Cells as a Promising Prognostic Biomarker in Advanced Non-small-cell Lung Cancer Treated with Immune Checkpoint Inhibitors. Clin. Lung Cancer 2021, 22, 423–431. [Google Scholar] [CrossRef]
- Bergmann, S.; Coym, A.; Ott, L.; Soave, A.; Rink, M.; Janning, M.; Stoupiec, M.; Coith, C.; Peine, S.; von Amsberg, G.; et al. Evaluation of PD-L1 expression on circulating tumor cells (CTCs) in patients with advanced urothelial carcinoma (UC). Oncoimmunology 2020, 9, 1738798. [Google Scholar] [CrossRef]
- Mazel, M.; Jacot, W.; Pantel, K.; Bartkowiak, K.; Topart, D.; Cayrefourcq, L.; Rossille, D.; Maudelonde, T.; Fest, T.; Alix-Panabières, C. Frequent expression of PD-L1 on circulating breast cancer cells. Mol. Oncol. 2015, 9, 1773–1782. [Google Scholar] [CrossRef]
- Di Trapani, M.; Manaresi, N.; Medoro, G. DEPArray™ system: An automatic image-based sorter for isolation of pure circulating tumor cells. Cytometry A 2018, 93, 1260–1266. [Google Scholar] [CrossRef]
- Cappelletti, V.; Verzoni, E.; Ratta, R.; Vismara, M.; Silvestri, M.; Montone, R.; Miodini, P.; Reduzzi, C.; Claps, M.; Sepe, P.; et al. Analysis of Single Circulating Tumor Cells in Renal Cell Carcinoma Reveals Phenotypic Heterogeneity and Genomic Alterations Related to Progression. Int. J. Mol. Sci. 2020, 21, 1475. [Google Scholar] [CrossRef] [PubMed]
- Nieva, J.; Wendel, M.; Luttgen, M.S.; Marrinucci, D.; Bazhenova, L.; Kolatkar, A.; Santala, R.; Whittenberger, B.; Burke, J.; Torrey, M.; et al. High-definition imaging of circulating tumor cells and associated cellular events in non-small cell lung cancer patients: A longitudinal analysis. Phys. Biol. 2012, 9, 016004. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Muinelo-Romay, L.; Vieito, M.; Abalo, A.; Nocelo, M.A.; Barón, F.; Anido, U.; Brozos, E.; Vázquez, F.; Aguín, S.; Abal, M.; et al. Evaluation of Circulating Tumor Cells and Related Events as Prognostic Factors and Surrogate Biomarkers in Advanced NSCLC Patients Receiving First-Line Systemic Treatment. Cancers 2014, 6, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, C.R.; Blackhall, F.H.; Carmel, A.; Fernandez-Gutierrez, F.; Gazzaniga, P.; Groen, H.J.M.; Hiltermann, T.J.; Krebs, M.G.; Loges, S.; López-López, R.; et al. EPAC-lung: Pooled analysis of circulating tumour cells in advanced non-small cell lung cancer. Eur. J. Cancer 2019, 117, 60–68. [Google Scholar] [CrossRef]
- Hirose, T.; Murata, Y.; Oki, Y.; Sugiyama, T.; Kusumoto, S.; Ishida, H.; Shirai, T.; Nakashima, M.; Yamaoka, T.; Okuda, K.; et al. Relationship of circulating tumor cells to the effectiveness of cytotoxic chemotherapy in patients with metastatic non-small-cell lung cancer. Oncol. Res. 2012, 20, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Juan, O.; Vidal, J.; Gisbert, R.; Muñoz, J.; Maciá, S.; Gómez-Codina, J. Prognostic significance of circulating tumor cells in advanced non-small cell lung cancer patients treated with docetaxel and gemcitabine. Clin. Transl. Oncol. 2014, 16, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xiao, Y.; Zhao, J.; Chen, M.; Xu, Y.; Zhong, W.; Xing, J.; Wang, M. Relationship between circulating tumour cell count and prognosis following chemotherapy in patients with advanced non-small-cell lung cancer. Respirology 2016, 21, 519–525. [Google Scholar] [CrossRef]
- Pawlikowska, P.; Faugeroux, V.; Oulhen, M.; Aberlenc, A.; Tayoun, T.; Pailler, E.; Farace, F. Circulating tumor cells (CTCs) for the noninvasive monitoring and personalization of non-small cell lung cancer (NSCLC) therapies. J. Thorac. Dis. 2019, 11, S45–S56. [Google Scholar] [CrossRef]
- Tong, B.; Xu, Y.; Zhao, J.; Chen, M.; Zhong, W.; Xing, J.; Wang, M. Prognostic role of circulating tumor cells in patients with EGFR-mutated or ALK-rearranged non-small cell lung cancer. Thorac. Cancer 2018, 9, 640–645. [Google Scholar] [CrossRef]
- Yang, B.; Zheng, D.; Zeng, Υ.; Qin, A.; Gao, J.; Yu, G. Circulating tumor cells predict prognosis following secondline AZD 9291 treatment in EGFR-T790M mutant non-small cell lung cancer patients. J. BUON 2018, 23, 1077–1081. [Google Scholar]
- Isobe, K.; Hata, Y.; Kobayashi, K.; Hirota, N.; Sato, K.; Sano, G.; Sugino, K.; Sakamoto, S.; Takai, Y.; Shibuya, K.; et al. Clinical significance of circulating tumor cells and free DNA in non-small cell lung cancer. Anticancer Res. 2012, 32, 3339–3344. [Google Scholar] [PubMed]
- Li, J. Significance of Circulating Tumor Cells in Nonsmall-Cell Lung Cancer Patients: Prognosis, Chemotherapy Efficacy, and Survival. J. Healthc. Eng. 2021, 2021, 2680526. [Google Scholar] [CrossRef] [PubMed]
- Alama, A.; Coco, S.; Genova, C.; Rossi, G.; Fontana, V.; Tagliamento, M.; Giovanna Dal Bello, M.; Rosa, A.; Boccardo, S.; Rijavec, E.; et al. Prognostic Relevance of Circulating Tumor Cells and Circulating Cell-Free DNA Association in Metastatic Non-Small Cell Lung Cancer Treated with Nivolumab. J. Clin. Med. 2019, 8, 1011. [Google Scholar] [CrossRef]
- Zhou, J.; Dong, F.; Cui, F.; Xu, R.; Tang, X. The role of circulating tumor cells in evaluation of prognosis and treatment response in advanced non-small-cell lung cancer. Cancer Chemother. Pharmacol. 2017, 79, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Ilié, M.; Szafer-Glusman, E.; Hofman, V.; Chamorey, E.; Lalvée, S.; Selva, E.; Leroy, S.; Marquette, C.H.; Kowanetz, M.; Hedge, P.; et al. Detection of PD-L1 in circulating tumor cells and white blood cells from patients with advanced non-small-cell lung cancer. Ann. Oncol. 2018, 29, 193–199. [Google Scholar] [CrossRef]
- Kallergi, G.; Vetsika, E.K.; Aggouraki, D.; Lagoudaki, E.; Koutsopoulos, A.; Koinis, F.; Katsarlinos, P.; Trypaki, M.; Messaritakis, I.; Stournaras, C.; et al. Evaluation of PD-L1/PD-1 on circulating tumor cells in patients with advanced non-small cell lung cancer. Ther. Adv. Med. Oncol. 2018, 10, 1758834017750121. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, T.; Lv, X.; Li, R.; Yuan, L.; Shen, J.; Li, Y.; Yan, T.; Liu, B.; Wang, L. Detection of PD-L1 Expression and Its Clinical Significance in Circulating Tumor Cells from Patients with Non-Small-Cell Lung Cancer. Cancer Manag. Res. 2020, 12, 2069–2078. [Google Scholar] [CrossRef]
- Sinoquet, L.; Jacot, W.; Gauthier, L.; Pouderoux, S.; Viala, M.; Cayrefourcq, L.; Quantin, X.; Alix-Panabières, C. Programmed Cell Death Ligand 1-Expressing Circulating Tumor Cells: A New Prognostic Biomarker in Non-Small Cell Lung Cancer. Clin. Chem. 2021, 67, 1503–1512. [Google Scholar] [CrossRef]
- Rossi, E.; Aieta, M.; Tartarone, A.; Pezzuto, A.; Facchinetti, A.; Santini, D.; Ulivi, P.; Ludovini, V.; Possidente, L.; Fiduccia, P.; et al. A fully automated assay to detect the expression of pan-cytokeratins and of EML4-ALK fusion protein in circulating tumour cells (CTCs) predicts outcome of non-small cell lung cancer (NSCLC) patients. Transl. Lung Cancer Res. 2021, 10, 80–92. [Google Scholar] [CrossRef]
- Kulasinghe, A.; Kapeleris, J.; Kimberley, R.; Mattarollo, S.R.; Thompson, E.W.; Thiery, J.P.; Kenny, L.; O’Byrne, K.; Punyadeera, C. The prognostic significance of circulating tumor cells in head and neck and non-small-cell lung cancer. Cancer Med. 2018, 7, 5910–5919. [Google Scholar] [CrossRef]
- Tamminga, M.; de Wit, S.; Schuuring, E.; Timens, W.; Terstappen, L.W.M.M.; Hiltermann, T.J.N.; Groen, H.J.M. Circulating tumor cells in lung cancer are prognostic and predictive for worse tumor response in both targeted- and chemotherapy. Transl. Lung Cancer Res. 2019, 8, 854–861. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.H.; Zhou, J.; Pan, X.F. Detecting circulating tumor cells in patients with advanced non-small cell lung cancer. Genet. Mol. Res. 2015, 14, 10352–10358. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, X.C.; Feng, W.N.; Zhang, L.; Liu, X.Q.; Guo, W.B.; Deng, Y.M.; Zou, Q.F.; Yang, J.J.; Zhou, Q.; et al. Circulating tumor cells dynamics during chemotherapy predict survival and response in advanced non-small-cell lung cancer patients. Ther. Adv. Med. Oncol. 2023, 15, 17588359231167818. [Google Scholar] [CrossRef] [PubMed]
- Gorges, T.M.; Penkalla, N.; Schalk, T.; Joosse, S.A.; Riethdorf, S.; Tucholski, J.; Lücke, K.; Wikman, H.; Jackson, S.; Brychta, N.; et al. Enumeration and Molecular Characterization of Tumor Cells in Lung Cancer Patients Using a Novel In Vivo Device for Capturing Circulating Tumor Cells. Clin. Cancer Res. 2016, 22, 2197–2206. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.J.; Li, J.; Zhu, W.F.; Wu, Y.; Tang, X.P.; Wang, Y.; Hu, Y.M. Survivin mRNA-circulating tumor cells predict treatment efficacy of chemotherapy and survival for advanced non-small cell lung cancer patients. Tumor Biol. 2014, 35, 4499–4507. [Google Scholar] [CrossRef]
- Punnoose, E.A.; Atwal, S.; Liu, W.; Raja, R.; Fine, B.M.; Hughes, B.G.; Hicks, R.J.; Hampton, G.M.; Amler, L.C.; Pirzkall, A.; et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: Association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin. Cancer Res. 2012, 18, 2391–2401. [Google Scholar] [CrossRef]
- He, W.; Li, W.; Jiang, B.; Chang, L.; Jin, C.; Tu, C.; Li, Y. Correlation between epidermal growth factor receptor tyrosine kinase inhibitor efficacy and circulating tumor cell levels in patients with advanced non-small cell lung cancer. OncoTargets Ther. 2016, 9, 7515–7520. [Google Scholar] [CrossRef]
- Breitenbuecher, F.; Hoffarth, S.; Worm, K.; Cortes-Incio, D.; Gauler, T.C.; Köhler, J.; Herold, T.; Schmid, K.W.; Freitag, L.; Kasper, S.; et al. Development of a highly sensitive and specific method for detection of circulating tumor cells harboring somatic mutations in non-small-cell lung cancer patients. PLoS ONE 2014, 9, e85350. [Google Scholar] [CrossRef]
- Pailler, E.; Adam, J.; Barthélémy, A.; Oulhen, M.; Auger, N.; Valent, A.; Borget, I.; Planchard, D.; Taylor, M.; André, F.; et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J. Clin. Oncol. 2013, 31, 2273–2281. [Google Scholar] [CrossRef]
- Ilie, M.; Hofman, V.; Leroy, S.; Cohen, C.; Heeke, S.; Cattet, F.; Bence, C.; Lalvée, S.; Mouroux, J.; Marquette, C.H.; et al. STALKLUNG01 and AIR Study Consortium Investigators. Use of circulating tumor cells in prospective clinical trials for NSCLC patients—Standardization of the pre-analytical conditions. Clin. Chem. Lab Med. 2018, 56, 980–989. [Google Scholar] [CrossRef]
- Provencio, M.; Pérez-Callejo, D.; Torrente, M.; Martin, P.; Calvo, V.; Gutiérrez, L.; Franco, F.; Coronado, M.J.; Cruz-Bermúdez, J.L.; Ruiz-Valdepeñas, A.M.; et al. Concordance between circulating tumor cells and clinical status during follow-up in anaplastic lymphoma kinase (ALK) non-small-cell lung cancer patients. Oncotarget 2017, 8, 59408–59416. [Google Scholar] [CrossRef] [PubMed]
- Pailler, E.; Oulhen, M.; Borget, I.; Remon, J.; Ross, K.; Auger, N.; Billiot, F.; Ngo Camus, M.; Commo, F.; Lindsay, C.R.; et al. Circulating Tumor Cells with Aberrant ALK Copy Number Predict Progression-Free Survival during Crizotinib Treatment in ALK-Rearranged Non-Small Cell Lung Cancer Patients. Cancer Res. 2017, 77, 2222–2230. [Google Scholar] [CrossRef] [PubMed]
- Ntzifa, A.; Strati, A.; Kallergi, G.; Kotsakis, A.; Georgoulias, V.; Lianidou, E. Gene expression in circulating tumor cells reveals a dynamic role of EMT and PD-L1 during osimertinib treatment in NSCLC patients. Sci. Rep. 2021, 11, 2313. [Google Scholar] [CrossRef]
- Kallergi, G.; Kontopodis, E.; Ntzifa, A.; Jordana-Ariza, N.; Karachaliou, N.; Pantazaka, E.; Charalambous, H.A.; Psyrri, A.; Tsaroucha, E.; Boukovinas, I.; et al. Effect of Osimertinib on CTCs and ctDNA in EGFR Mutant Non-Small Cell Lung Cancer Patients: The Prognostic Relevance of Liquid Biopsy. Cancers 2022, 14, 1574. [Google Scholar] [CrossRef] [PubMed]
- Pantazaka, E.; Ntzifa, A.; Roumeliotou, A.; Lianidou, E.; Georgoulias, V.; Kotsakis, A.; Kallergi, G. PD-L1/pS6 in Circulating Tumor Cells (CTCs) during Osimertinib Treatment in Patients with Non-Small Cell Lung Cancer (NSCLC). Biomedicines 2022, 10, 1893. [Google Scholar] [CrossRef] [PubMed]
- Isobe, K.; Yoshizawa, T.; Sekiya, M.; Miyoshi, S.; Nakamura, Y.; Urabe, N.; Isshiki, T.; Sakamoto, S.; Takai, Y.; Tomida, T.; et al. Quantification of BIM mRNA in circulating tumor cells of osimertinib-treated patients with EGFR mutation-positive lung cancer. Respir. Investig. 2021, 59, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Zhao, J.; Zhao, C.; Li, X.; Shen, J.; Zhou, J.; Ren, S.; Su, C.; Zhou, C.; O’Brien, M. Dynamic Monitoring and Predictive Value of Circulating Tumor Cells in EGFR-Mutated Advanced Non-Small-Cell Lung Cancer Patients Treated With First-Line EGFR Tyrosine Kinase Inhibitors. Clin. Lung Cancer 2019, 20, 124–133.e2. [Google Scholar] [CrossRef]
- Maheswaran, S.; Sequist, L.V.; Nagrath, S.; Ulkus, L.; Brannigan, B.; Collura, C.V.; Inserra, E.; Diederichs, S.; Iafrate, A.J.; Bell, D.W.; et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 2008, 359, 366–377. [Google Scholar] [CrossRef]
- Chen, X.; Luo, L.; Lu, Y.; Qiu, Y.; Liang, J.; Chen, Y.; Ning, Y.; Li, B. Construction of IMMS Containing Multi-site Liposomes for Dynamic Monitoring of Blood CTC in Patients with Osimertinib-resistant Non-small-cell Lung Cancer and its Mechanism. Anti-Cancer Agents Med. Chem. 2023, 23, 676–686. [Google Scholar] [CrossRef]
- Oulhen, M.; Pawlikowska, P.; Tayoun, T.; Garonzi, M.; Buson, G.; Forcato, C.; Manaresi, N.; Aberlenc, A.; Mezquita, L.; Lecluse, Y.; et al. Circulating tumor cell copy-number heterogeneity in ALK-rearranged non-small-cell lung cancer resistant to ALK inhibitors. NPJ Precis. Oncol. 2021, 5, 67. [Google Scholar] [CrossRef]
- Jia, Q.; Wang, A.; Yuan, Y.; Zhu, B.; Long, H. Heterogeneity of the tumour immune microenvironment and its clinical relevance. Exp. Hematol. Oncol. 2022, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Kloten, V.; Lampignano, R.; Krahn, T.; Schlange, T. Circulating Tumor Cell PDL1 Expression as Biomarker for Therapeutic Efficacy of Immune Checkpoint Inhibition in NSCLC. Cells 2019, 8, 809. [Google Scholar] [CrossRef] [PubMed]
- Nicolazzo, C.; Raimondi, C.; Mancini, M.; Caponnetto, S.; Gradilone, A.; Gandini, O.; Mastromartino, M.; Del Bene, G.; Prete, A.; Longo, F.; et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor Nivolumab. Sci. Rep. 2016, 6, 31726. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, C.; Carpino, G.; Nicolazzo, C.; Gradilone, A.; Gianni, W.; Gelibter, A.; Gaudio, E.; Cortesi, E.; Gazzaniga, P. PD-L1 and epithelial-mesenchymal transition in circulating tumor cells from non-small cell lung cancer patients: A molecular shield to evade immune system? Oncoimmunology 2017, 6, e1315488. [Google Scholar] [CrossRef]
- Spiliotaki, M.; Neophytou, C.M.; Vogazianos, P.; Stylianou, I.; Gregoriou, G.; Constantinou, A.I.; Deltas, C.; Charalambous, H. Dynamic monitoring of PD-L1 and Ki67 in circulating tumor cells of metastatic non-small cell lung cancer patients treated with pembrolizumab. Mol. Oncol. 2023, 17, 792–809. [Google Scholar] [CrossRef]
- Castello, A.; Carbone, F.G.; Rossi, S.; Monterisi, S.; Federico, D.; Toschi, L.; Lopci, E. Circulating Tumor Cells and Metabolic Parameters in NSCLC Patients Treated with Checkpoint Inhibitors. Cancers 2020, 12, 487. [Google Scholar] [CrossRef]
- Ikeda, M.; Koh, Y.; Teraoka, S.; Sato, K.; Oyanagi, J.; Hayata, A.; Tokudome, N.; Akamatsu, H.; Ozawa, Y.; Endo, K.; et al. Longitudinal Evaluation of PD-L1 Expression on Circulating Tumor Cells in Non-Small Cell Lung Cancer Patients Treated with Nivolumab. Cancers 2021, 13, 2290. [Google Scholar] [CrossRef]
- Janning, M.; Kobus, F.; Babayan, A.; Wikman, H.; Velthaus, J.L.; Bergmann, S.; Schatz, S.; Falk, M.; Berger, L.A.; Böttcher, L.M.; et al. Determination of PD-L1 Expression in Circulating Tumor Cells of NSCLC Patients and Correlation with Response to PD-1/PD-L1 Inhibitors. Cancers 2019, 11, 835. [Google Scholar] [CrossRef]
- Normanno, N.; Cervantes, A.; Ciardiello, F.; De Luca, A.; Pinto, C. The liquid biopsy in the management of colorectal cancer patients: Current applications and future scenarios. Cancer Treat. Rev. 2018, 70, 1–8. [Google Scholar] [CrossRef]
- Andor, N.; Graham, T.A.; Jansen, M.; Xia, L.C.; Aktipis, C.A.; Petritsch, C.; Ji, H.P.; Maley, C.C. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 2016, 22, 105–113. [Google Scholar] [CrossRef]
- Calabuig-Fariñas, S.; Jantus-Lewintre, E.; Herreros-Pomares, A.; Camps, C. Circulating tumor cells versus circulating tumor DNA in lung cancer-which one will win? Transl. Lung Cancer Res. 2016, 5, 466–482. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).