CRISPR/Cas9 Landscape: Current State and Future Perspectives
Abstract
1. Introduction
2. Cas9 S. pyogenes—The Most Widely Used Cas9 Nuclease
3. Cas9 Variants with Improved Efficacy and Specificity
4. Cas9 S. pyogenes Nickases
5. Catalytically Inactive Cas9 S. pyogenes (dCas9) and Its Derivatives
5.1. dCas9 S. pyogenes
5.2. dCas9-Based Transcription Repressors
5.3. dCas9-Based Transcription Activators
5.4. dCas9-Mediated Epigenetic Editing
5.5. dCas9-Based Imaging Tools
5.6. dCas9-Based Methods for Isolation of Genomic Loci
6. Base Editors
7. Prime Editors
8. Cas-CLOVER
9. Cas-FokI
10. CRISPR/Cas9 Delivery Methods
11. CRISPR/Cas9-Based Diagnostics
12. CRISPR/Cas9-Based Therapeutics
13. Pre-Existing Immunity against CRISPR/Cas9 Proteins
14. CRISPR/Cas9 Activity Modulators
15. Cas9 Orthologs
16. Future Perspectives
17. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaj, T.; Sirk, S.J.; Shui, S.-L.; Liu, J. Genome-Editing Technologies: Principles and Applications. Cold Spring Harb. Perspect. Biol. 2016, 8, a023754. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of Genome Editing Technology in the Targeted Therapy of Human Diseases: Mechanisms, Advances and Prospects. Signal Transduct. Target. Ther. 2020, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, D.; Nomura, W. The History of Genome Editing: Advances from the Interface of Chemistry & Biology. Chem. Commun. 2023, 59, 7676–7684. [Google Scholar] [CrossRef]
- Bock, C.; Datlinger, P.; Chardon, F.; Coelho, M.A.; Dong, M.B.; Lawson, K.A.; Lu, T.; Maroc, L.; Norman, T.M.; Song, B.; et al. High-Content CRISPR Screening. Nat. Rev. Methods Primers 2022, 2, 8. [Google Scholar] [CrossRef]
- Grav, L.M.; la Cour Karottki, K.J.; Lee, J.S.; Kildegaard, H.F. Application of CRISPR/Cas9 Genome Editing to Improve Recombinant Protein Production in CHO Cells. In Methods in Molecular Biology; Springer: New York, NY, USA, 2017; pp. 101–118. ISBN 9781493969715. [Google Scholar]
- Chan, K.F.; Shahreel, W.; Wan, C.; Teo, G.; Hayati, N.; Tay, S.J.; Tong, W.H.; Yang, Y.; Rudd, P.M.; Zhang, P.; et al. Inactivation of GDP-fucose Transporter Gene (Slc35c1) in CHO Cells by ZFNs, TALENs and CRISPR-Cas9 for Production of Fucose-free Antibodies. Biotechnol. J. 2016, 11, 399–414. [Google Scholar] [CrossRef]
- Geurts, A.M.; Cost, G.J.; Freyvert, Y.; Zeitler, B.; Miller, J.C.; Choi, V.M.; Jenkins, S.S.; Wood, A.; Cui, X.; Meng, X.; et al. Knockout Rats via Embryo Microinjection of Zinc-Finger Nucleases. Science 2009, 325, 433. [Google Scholar] [CrossRef]
- Lee, H.; Yoon, D.E.; Kim, K. Genome Editing Methods in Animal Models. Anim. Cells Syst. 2020, 24, 8–16. [Google Scholar] [CrossRef]
- Lin, Y.; Li, J.; Li, C.; Tu, Z.; Li, S.; Li, X.-J.; Yan, S. Application of CRISPR/Cas9 System in Establishing Large Animal Models. Front. Cell Dev. Biol. 2022, 10, 919155. [Google Scholar] [CrossRef]
- Gan, W.C.; Ling, A.P.K. CRISPR/Cas9 in Plant Biotechnology: Applications and Challenges. BioTechnologia 2022, 103, 81–93. [Google Scholar] [CrossRef]
- Son, S.; Park, S.R. Challenges Facing CRISPR/Cas9-Based Genome Editing in Plants. Front. Plant Sci. 2022, 13, 902413. [Google Scholar] [CrossRef]
- Singh, P.; Ali, S.A. Impact of CRISPR-Cas9-Based Genome Engineering in Farm Animals. Vet. Sci. 2021, 8, 122. [Google Scholar] [CrossRef]
- Menchaca, A.; dos Santos-Neto, P.C.; Mulet, A.P.; Crispo, M. CRISPR in Livestock: From Editing to Printing. Theriogenology 2020, 150, 247–254. [Google Scholar] [CrossRef]
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell 2014, 156, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Li, H.; Chen, L.-L.; Xie, K. Recent Advances in Genome Editing Using CRISPR/Cas9. Front. Plant Sci. 2016, 7, 703. [Google Scholar] [CrossRef] [PubMed]
- Cradick, T.J.; Fine, E.J.; Antico, C.J.; Bao, G. CRISPR/Cas9 Systems Targeting β-Globin and CCR5 Genes Have Substantial off-Target Activity. Nucleic Acids Res. 2013, 41, 9584–9592. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-Frequency off-Target Mutagenesis Induced by CRISPR-Cas Nucleases in Human Cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA Targeting Specificity of RNA-Guided Cas9 Nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Mali, P.; Aach, J.; Stranges, P.B.; Esvelt, K.M.; Moosburner, M.; Kosuri, S.; Yang, L.; Church, G.M. CAS9 Transcriptional Activators for Target Specificity Screening and Paired Nickases for Cooperative Genome Engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef]
- Pattanayak, V.; Lin, S.; Guilinger, J.P.; Ma, E.; Doudna, J.A.; Liu, D.R. High-Throughput Profiling of off-Target DNA Cleavage Reveals RNA-Programmed Cas9 Nuclease Specificity. Nat. Biotechnol. 2013, 31, 839–843. [Google Scholar] [CrossRef]
- Tsai, S.Q.; Zheng, Z.; Nguyen, N.T.; Liebers, M.; Topkar, V.V.; Thapar, V.; Wyvekens, N.; Khayter, C.; Iafrate, A.J.; Le, L.P.; et al. GUIDE-Seq Enables Genome-Wide Profiling of off-Target Cleavage by CRISPR-Cas Nucleases. Nat. Biotechnol. 2015, 33, 187–197. [Google Scholar] [CrossRef]
- Kim, D.; Bae, S.; Park, J.; Kim, E.; Kim, S.; Yu, H.R.; Hwang, J.; Kim, J.-I.; Kim, J.-S. Digenome-Seq: Genome-Wide Profiling of CRISPR-Cas9 off-Target Effects in Human Cells. Nat. Methods 2015, 12, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Kuscu, C.; Arslan, S.; Singh, R.; Thorpe, J.; Adli, M. Genome-Wide Analysis Reveals Characteristics of off-Target Sites Bound by the Cas9 Endonuclease. Nat. Biotechnol. 2014, 32, 677–683. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-Fidelity CRISPR–Cas9 Nucleases with No Detectable Genome-Wide off-Target Effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.W.; Li, Z.; Peterson, R.T.; Yeh, J.-R.J.; et al. Engineered CRISPR-Cas9 Nucleases with Altered PAM Specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef]
- Davis, K.M.; Pattanayak, V.; Thompson, D.B.; Zuris, J.A.; Liu, D.R. Small Molecule–Triggered Cas9 Protein with Improved Genome-Editing Specificity. Nat. Chem. Biol. 2015, 11, 316–318. [Google Scholar] [CrossRef] [PubMed]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally Engineered Cas9 Nucleases with Improved Specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Ge, X.; Yang, F.; Wang, B.; Li, S.; Duan, J.; Lv, X.; Cheng, C.; Song, Z.; Liu, C.; et al. High-Fidelity SaCas9 Identified by Directional Screening in Human Cells. PLoS Biol. 2020, 18, e3000747. [Google Scholar] [CrossRef]
- Casini, A.; Olivieri, M.; Petris, G.; Montagna, C.; Reginato, G.; Maule, G.; Lorenzin, F.; Prandi, D.; Romanel, A.; Demichelis, F.; et al. A Highly Specific SpCas9 Variant Is Identified by in Vivo Screening in Yeast. Nat. Biotechnol. 2018, 36, 265–271. [Google Scholar] [CrossRef]
- Vakulskas, C.A.; Dever, D.P.; Rettig, G.R.; Turk, R.; Jacobi, A.M.; Collingwood, M.A.; Bode, N.M.; McNeill, M.S.; Yan, S.; Camarena, J.; et al. A High-Fidelity Cas9 Mutant Delivered as a Ribonucleoprotein Complex Enables Efficient Gene Editing in Human Hematopoietic Stem and Progenitor Cells. Nat. Med. 2018, 24, 1216–1224. [Google Scholar] [CrossRef]
- Chen, J.S.; Dagdas, Y.S.; Kleinstiver, B.P.; Welch, M.M.; Sousa, A.A.; Harrington, L.B.; Sternberg, S.H.; Joung, J.K.; Yildiz, A.; Doudna, J.A. Enhanced Proofreading Governs CRISPR–Cas9 Targeting Accuracy. Nature 2017, 550, 407–410. [Google Scholar] [CrossRef]
- Tan, Y.; Chu, A.H.Y.; Bao, S.; Hoang, D.A.; Kebede, F.T.; Xiong, W.; Ji, M.; Shi, J.; Zheng, Z. Rationally Engineered Staphylococcus Aureus Cas9 Nucleases with High Genome-Wide Specificity. Proc. Natl. Acad. Sci. USA 2019, 116, 20969–20976. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Jeong, E.; Lee, J.; Jung, M.; Shin, E.; Kim, Y.-H.; Lee, K.; Jung, I.; Kim, D.; Kim, S.; et al. Directed Evolution of CRISPR-Cas9 to Increase Its Specificity. Nat. Commun. 2018, 9, 3048. [Google Scholar] [CrossRef]
- Cerchione, D.; Loveluck, K.; Tillotson, E.L.; Harbinski, F.; DaSilva, J.; Kelley, C.P.; Keston-Smith, E.; Fernandez, C.A.; Myer, V.E.; Jayaram, H.; et al. SMOOT Libraries and Phage-Induced Directed Evolution of Cas9 to Engineer Reduced off-Target Activity. PLoS ONE 2020, 15, e0231716. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.-Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef]
- Bravo, J.P.K.; Liu, M.-S.; Hibshman, G.N.; Dangerfield, T.L.; Jung, K.; McCool, R.S.; Johnson, K.A.; Taylor, D.W. Structural Basis for Mismatch Surveillance by CRISPR–Cas9. Nature 2022, 603, 343–347. [Google Scholar] [CrossRef]
- Schmid-Burgk, J.L.; Gao, L.; Li, D.; Gardner, Z.; Strecker, J.; Lash, B.; Zhang, F. Highly Parallel Profiling of Cas9 Variant Specificity. Mol. Cell 2020, 78, 794–800.e8. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 Variants with Broad PAM Compatibility and High DNA Specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef]
- Yuen, C.T.L.; Thean, D.G.L.; Chan, B.K.C.; Zhou, P.; Kwok, C.C.S.; Chu, H.Y.; Cheung, M.S.H.; Wang, B.; Chan, Y.M.; Mak, S.Y.L.; et al. High-Fidelity KKH Variant of Staphylococcus aureus Cas9 Nucleases with Improved Base Mismatch Discrimination. Nucleic Acids Res. 2022, 50, 1650–1660. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Ruan, J.; Song, J.; Wen, L.; Yang, D.; Zhao, J.; Xia, X.; Chen, Y.E.; Zhang, J.; Xu, J. MiCas9 Increases Large Size Gene Knock-in Rates and Reduces Undesirable on-Target and off-Target Indel Edits. Nat. Commun. 2020, 11, 6082. [Google Scholar] [CrossRef]
- Tsai, S.Q.; Wyvekens, N.; Khayter, C.; Foden, J.A.; Thapar, V.; Reyon, D.; Goodwin, M.J.; Aryee, M.J.; Joung, J.K. Dimeric CRISPR RNA-Guided FokI Nucleases for Highly Specific Genome Editing. Nat. Biotechnol. 2014, 32, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Kurt, I.C.; Zhou, R.; Iyer, S.; Garcia, S.P.; Miller, B.R.; Langner, L.M.; Grünewald, J.; Joung, J.K. CRISPR C-to-G Base Editors for Inducing Targeted DNA Transversions in Human Cells. Nat. Biotechnol. 2021, 39, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable Editing of a Target Base in Genomic DNA without Double-Stranded DNA Cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable Base Editing of A•T to G•C in Genomic DNA without DNA Cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Molla, K.A.; Yang, Y. CRISPR/Cas-Mediated Base Editing: Technical Considerations and Practical Applications. Trends Biotechnol. 2019, 37, 1121–1142. [Google Scholar] [CrossRef]
- Yang, B.; Yang, L.; Chen, J. Development and Application of Base Editors. CRISPR J. 2019, 2, 91–104. [Google Scholar] [CrossRef] [PubMed]
- McGrath, E.; Shin, H.; Zhang, L.; Phue, J.-N.; Wu, W.W.; Shen, R.-F.; Jang, Y.-Y.; Revollo, J.; Ye, Z. Targeting Specificity of APOBEC-Based Cytosine Base Editor in Human IPSCs Determined by Whole Genome Sequencing. Nat. Commun. 2019, 10, 5353. [Google Scholar] [CrossRef]
- Xin, X.; Li, J.; Zhao, D.; Li, S.; Xie, Q.; Li, Z.; Fan, F.; Bi, C.; Zhang, X. Double-Check Base Editing for Efficient A to G Conversions. ACS Synth. Biol. 2019, 8, 2629–2634. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, X.; Wang, L.; Yin, S.; Zhu, B.; Xie, L.; Duan, Q.; Hu, H.; Zheng, R.; Wei, Y.; et al. Increasing Targeting Scope of Adenosine Base Editors in Mouse and Rat Embryos through Fusion of TadA Deaminase with Cas9 Variants. Protein Cell 2018, 9, 814–819. [Google Scholar] [CrossRef]
- Li, C.; Zong, Y.; Wang, Y.; Jin, S.; Zhang, D.; Song, Q.; Zhang, R.; Gao, C. Expanded Base Editing in Rice and Wheat Using a Cas9-Adenosine Deaminase Fusion. Genome Biol. 2018, 19, 59. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Lam, D.K.; Rees, H.A.; Solá-Esteves, N.M.; Barrera, L.A.; Born, D.A.; Edwards, A.; Gehrke, J.M.; Lee, S.-J.; Liquori, A.J.; et al. Directed Evolution of Adenine Base Editors with Increased Activity and Therapeutic Application. Nat. Biotechnol. 2020, 38, 892–900. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-Replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Nelson, J.W.; Randolph, P.B.; Shen, S.P.; Everette, K.A.; Chen, P.J.; Anzalone, A.V.; An, M.; Newby, G.A.; Chen, J.C.; Hsu, A.; et al. Engineered PegRNAs Improve Prime Editing Efficiency. Nat. Biotechnol. 2022, 40, 402–410. [Google Scholar] [CrossRef]
- Ledford, H. Super-Precise New CRISPR Tool Could Tackle a Plethora of Genetic Diseases. Nature 2019, 574, 464–465. [Google Scholar] [CrossRef]
- Madison, B.B.; Patil, D.; Richter, M.; Li, X.; Tong, M.; Cranert, S.; Wang, X.; Martin, R.; Xi, H.; Tan, Y.; et al. Cas-CLOVER Is a Novel High-Fidelity Nuclease for Safe and Robust Generation of TSCM-Enriched Allogeneic CAR-T Cells. Mol. Ther. Nucleic Acids 2022, 29, 979–995. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, L.; Ntui, V.O.; Tripathi, J.N.; Norman, D.; Crawford, J. A New and Novel High-fidelity Genome Editing Tool for Banana Using Cas-CLOVER. Plant Biotechnol. J. 2023, 21, 1731–1733. [Google Scholar] [CrossRef] [PubMed]
- GEN. Cas-CLOVER for Creating Improved Platform Cell Lines and Animal Models. Genet. Eng. Biotechnol. News 2022, 42, 47. [Google Scholar] [CrossRef]
- Wyvekens, N.; Topkar, V.V.; Khayter, C.; Joung, J.K.; Tsai, S.Q. Dimeric CRISPR RNA-Guided FokI-DCas9 Nucleases Directed by Truncated GRNAs for Highly Specific Genome Editing. Hum. Gene Ther. 2015, 26, 425–431. [Google Scholar] [CrossRef]
- Aouida, M.; Eid, A.; Ali, Z.; Cradick, T.; Lee, C.; Deshmukh, H.; Atef, A.; AbuSamra, D.; Gadhoum, S.Z.; Merzaban, J.; et al. Efficient FdCas9 Synthetic Endonuclease with Improved Specificity for Precise Genome Engineering. PLoS ONE 2015, 10, e0133373. [Google Scholar] [CrossRef] [PubMed]
- Guilinger, J.P.; Thompson, D.B.; Liu, D.R. Fusion of Catalytically Inactive Cas9 to FokI Nuclease Improves the Specificity of Genome Modification. Nat. Biotechnol. 2014, 32, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Saifaldeen, M.; Al-Ansari, D.E.; Ramotar, D.; Aouida, M. CRISPR FokI Dead Cas9 System: Principles and Applications in Genome Engineering. Cells 2020, 9, 2518. [Google Scholar] [CrossRef]
- Luo, Z.; Gao, M.; Huang, N.; Wang, X.; Yang, Z.; Yang, H.; Huang, Z.; Feng, W. Efficient Disruption of Bcr-Abl Gene by CRISPR RNA-Guided FokI Nucleases Depresses the Oncogenesis of Chronic Myeloid Leukemia Cells. J. Exp. Clin. Cancer Res. 2019, 38, 224. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Sander, J.D.; Reyon, D.; Cascio, V.M.; Joung, J.K. Improving CRISPR-Cas Nuclease Specificity Using Truncated Guide RNAs. Nat. Biotechnol. 2014, 32, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Mateos, M.A.; Vejnar, C.E.; Beaudoin, J.-D.; Fernandez, J.P.; Mis, E.K.; Khokha, M.K.; Giraldez, A.J. CRISPRscan: Designing Highly Efficient SgRNAs for CRISPR-Cas9 Targeting in Vivo. Nat. Methods 2015, 12, 982–988. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, C.; Wang, B.; Li, B.; Wang, Q.; Liu, D.; Wang, H.; Zhou, Y.; Shi, L.; Lan, F.; et al. Optimized CRISPR Guide RNA Design for Two High-Fidelity Cas9 Variants by Deep Learning. Nat. Commun. 2019, 10, 4284. [Google Scholar] [CrossRef]
- Kim, D.; Kim, S.; Kim, S.; Park, J.; Kim, J.-S. Genome-Wide Target Specificities of CRISPR-Cas9 Nucleases Revealed by Multiplex Digenome-Seq. Genome Res. 2016, 26, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Cromwell, C.R.; Sung, K.; Park, J.; Krysler, A.R.; Jovel, J.; Kim, S.K.; Hubbard, B.P. Incorporation of Bridged Nucleic Acids into CRISPR RNAs Improves Cas9 Endonuclease Specificity. Nat. Commun. 2018, 9, 1448. [Google Scholar] [CrossRef]
- Economos, N.G.; Oyaghire, S.; Quijano, E.; Ricciardi, A.S.; Saltzman, W.M.; Glazer, P.M. Peptide Nucleic Acids and Gene Editing: Perspectives on Structure and Repair. Molecules 2020, 25, 735. [Google Scholar] [CrossRef]
- Ho, P.Y.; Zhang, Z.; Hayes, M.E.; Curd, A.; Dib, C.; Rayburn, M.; Tam, S.N.; Srivastava, T.; Hriniak, B.; Li, X.-J.; et al. Peptide Nucleic Acid–Dependent Artifact Can Lead to False-Positive Triplex Gene Editing Signals. Proc. Natl. Acad. Sci. USA 2021, 118, e2109175118. [Google Scholar] [CrossRef]
- Economos, N.G.; Quijano, E.; Carufe, K.E.W.; Perera, J.D.R.; Glazer, P.M. Antispacer Peptide Nucleic Acids for Sequence-Specific CRISPR-Cas9 Modulation. Nucleic Acids Res. 2022, 50, e59. [Google Scholar] [CrossRef]
- ChRDNA Technology. Available online: https://www.cariboubio.com/technology/chrdna_technology/ (accessed on 12 October 2023).
- Shin, J.; Jiang, F.; Liu, J.-J.; Bray, N.L.; Rauch, B.J.; Baik, S.H.; Nogales, E.; Bondy-Denomy, J.; Corn, J.E.; Doudna, J.A. Disabling Cas9 by an Anti-CRISPR DNA Mimic. Sci. Adv. 2017, 3, e1701620. [Google Scholar] [CrossRef]
- Eitzinger, S.; Asif, A.; Watters, K.E.; Iavarone, A.T.; Knott, G.J.; Doudna, J.A.; Minhas, F.U.A.A. Machine Learning Predicts New Anti-CRISPR Proteins. Nucleic Acids Res. 2020, 48, 4698–4708. [Google Scholar] [CrossRef]
- Pawluk, A.; Amrani, N.; Zhang, Y.; Garcia, B.; Hidalgo-Reyes, Y.; Lee, J.; Edraki, A.; Shah, M.; Sontheimer, E.J.; Maxwell, K.L.; et al. Naturally Occurring Off-Switches for CRISPR-Cas9. Cell 2016, 167, 1829–1838.e9. [Google Scholar] [CrossRef]
- Rauch, B.J.; Silvis, M.R.; Hultquist, J.F.; Waters, C.S.; McGregor, M.J.; Krogan, N.J.; Bondy-Denomy, J. Inhibition of CRISPR-Cas9 with Bacteriophage Proteins. Cell 2017, 168, 150–158.e10. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Tian, R.; Huang, Z.; Jin, Z.; Li, L.; Liu, J.; Huang, Z.; Xie, H.; Liu, D.; Mo, H.; et al. FrCas9 Is a CRISPR/Cas9 System with High Editing Efficiency and Fidelity. Nat. Commun. 2022, 13, 1425. [Google Scholar] [CrossRef]
- Amrani, N.; Gao, X.D.; Liu, P.; Edraki, A.; Mir, A.; Ibraheim, R.; Gupta, A.; Sasaki, K.E.; Wu, T.; Donohoue, P.D.; et al. NmeCas9 Is an Intrinsically High-Fidelity Genome-Editing Platform. Genome Biol. 2018, 19, 214. [Google Scholar] [CrossRef]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In Vivo Genome Editing Using Staphylococcus Aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, B.; Xie, H.; Ren, Q.; Liu, X.; Li, F.; Lv, X.; He, X.; Cheng, C.; Deng, R.; et al. Efficient Human Genome Editing Using SaCas9 Ribonucleoprotein Complexes. Biotechnol. J. 2019, 14, 1800689. [Google Scholar] [CrossRef]
- Xie, H.; Tang, L.; He, X.; Liu, X.; Zhou, C.; Liu, J.; Ge, X.; Li, J.; Liu, C.; Zhao, J.; et al. SaCas9 Requires 5′-NNGRRT-3′ PAM for Sufficient Cleavage and Possesses Higher Cleavage Activity than SpCas9 or FnCpf1 in Human Cells. Biotechnol. J. 2018, 13, e1700561. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Topkar, V.V.; Zheng, Z.; Joung, J.K. Broadening the Targeting Range of Staphylococcus Aureus CRISPR-Cas9 by Modifying PAM Recognition. Nat. Biotechnol. 2015, 33, 1293–1298. [Google Scholar] [CrossRef]
- Luan, B.; Xu, G.; Feng, M.; Cong, L.; Zhou, R. Combined Computational–Experimental Approach to Explore the Molecular Mechanism of SaCas9 with a Broadened DNA Targeting Range. J. Am. Chem. Soc. 2019, 141, 6545–6552. [Google Scholar] [CrossRef]
- Ma, D.; Xu, Z.; Zhang, Z.; Chen, X.; Zeng, X.; Zhang, Y.; Deng, T.; Ren, M.; Sun, Z.; Jiang, R.; et al. Engineer Chimeric Cas9 to Expand PAM Recognition Based on Evolutionary Information. Nat. Commun. 2019, 10, 560. [Google Scholar] [CrossRef] [PubMed]
- Kulcsár, P.I.; Tálas, A.; Ligeti, Z.; Krausz, S.L.; Welker, E. SuperFi-Cas9 Exhibits Remarkable Fidelity but Severely Reduced Activity yet Works Effectively with ABE8e. Nat. Commun. 2022, 13, 6858. [Google Scholar] [CrossRef]
- Bolukbasi, M.F.; Gupta, A.; Oikemus, S.; Derr, A.G.; Garber, M.; Brodsky, M.H.; Zhu, L.J.; Wolfe, S.A. DNA-Binding-Domain Fusions Enhance the Targeting Range and Precision of Cas9. Nat. Methods 2015, 12, 1150–1156. [Google Scholar] [CrossRef] [PubMed]
- Bolukbasi, M.F.; Liu, P.; Luk, K.; Kwok, S.F.; Gupta, A.; Amrani, N.; Sontheimer, E.J.; Zhu, L.J.; Wolfe, S.A. Orthogonal Cas9–Cas9 Chimeras Provide a Versatile Platform for Genome Editing. Nat. Commun. 2018, 9, 4856. [Google Scholar] [CrossRef] [PubMed]
- Trevino, A.E.; Zhang, F. Genome Editing Using Cas9 Nickases. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2014; pp. 161–174. [Google Scholar]
- Wang, Q.; Liu, J.; Janssen, J.M.; Gonçalves, M.A.F.V. Precise Homology-Directed Installation of Large Genomic Edits in Human Cells with Cleaving and Nicking High-Specificity Cas9 Variants. Nucleic Acids Res. 2023, 51, 3465–3484. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.; Janssen, J.M.; Le Bouteiller, M.; Frock, R.L.; Gonçalves, M.A.F.V. Precise and Broad Scope Genome Editing Based on High-Specificity Cas9 Nickases. Nucleic Acids Res. 2021, 49, 1173–1198. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Larson, M.H.; Gilbert, L.A.; Wang, X.; Lim, W.A.; Weissman, J.S.; Qi, L.S. CRISPR Interference (CRISPRi) for Sequence-Specific Control of Gene Expression. Nat. Protoc. 2013, 8, 2180–2196. [Google Scholar] [CrossRef]
- Jusiak, B.; Cleto, S.; Perez-Piñera, P.; Lu, T.K. Engineering Synthetic Gene Circuits in Living Cells with CRISPR Technology. Trends Biotechnol. 2016, 34, 535–547. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Hawkins, J.S.; Wong, S.; Peters, J.M.; Almeida, R.; Qi, L.S. Targeted Transcriptional Repression in Bacteria Using CRISPR Interference (CRISPRi). In Methods in Molecular Biology; Springer: New York, NY, USA, 2015; pp. 349–362. ISBN 9781493926862. [Google Scholar]
- Replogle, J.M.; Bonnar, J.L.; Pogson, A.N.; Liem, C.R.; Maier, N.K.; Ding, Y.; Russell, B.J.; Wang, X.; Leng, K.; Guna, A.; et al. Maximizing CRISPRi Efficacy and Accessibility with Dual-SgRNA Libraries and Optimal Effectors. eLife 2022, 11, e81856. [Google Scholar] [CrossRef]
- Byun, G.; Yang, J.; Seo, S.W. CRISPRi-Mediated Tunable Control of Gene Expression Level with Engineered Single-Guide RNA in Escherichia coli. Nucleic Acids Res. 2023, 51, 4650–4659. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef]
- Gao, Y.; Xiong, X.; Wong, S.; Charles, E.J.; Lim, W.A.; Qi, L.S. Complex Transcriptional Modulation with Orthogonal and Inducible DCas9 Regulators. Nat. Methods 2016, 13, 1043–1049. [Google Scholar] [CrossRef]
- Li, R.; Xia, X.; Wang, X.; Sun, X.; Dai, Z.; Huo, D.; Zheng, H.; Xiong, H.; He, A.; Wu, X. Generation and Validation of Versatile Inducible CRISPRi Embryonic Stem Cell and Mouse Model. PLoS Biol. 2020, 18, e3000749. [Google Scholar] [CrossRef]
- Mandegar, M.A.; Huebsch, N.; Frolov, E.B.; Shin, E.; Truong, A.; Olvera, M.P.; Chan, A.H.; Miyaoka, Y.; Holmes, K.; Spencer, C.I.; et al. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human IPSCs. Cell Stem Cell 2016, 18, 541–553. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.E.; Zhang, F. High-Throughput Functional Genomics Using CRISPR–Cas9. Nat. Rev. Genet. 2015, 16, 299–311. [Google Scholar] [CrossRef]
- Yeo, N.C.; Chavez, A.; Lance-Byrne, A.; Chan, Y.; Menn, D.; Milanova, D.; Kuo, C.-C.; Guo, X.; Sharma, S.; Tung, A.; et al. An Enhanced CRISPR Repressor for Targeted Mammalian Gene Regulation. Nat. Methods 2018, 15, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Alerasool, N.; Segal, D.; Lee, H.; Taipale, M. An Efficient KRAB Domain for CRISPRi Applications in Human Cells. Nat. Methods 2020, 17, 1093–1096. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.; Riching, A.; Keller, A.; Stombaugh, J.; Haupt, A.; Maksimova, E.; Dickerson, S.M.; Anderson, E.; Hemphill, K.; Ebmeier, C.; et al. A Novel CRISPR Interference Effector Enabling Functional Gene Characterization with Synthetic Guide RNAs. CRISPR J. 2022, 5, 769–786. [Google Scholar] [CrossRef] [PubMed]
- Thakore, P.I.; Kwon, J.B.; Nelson, C.E.; Rouse, D.C.; Gemberling, M.P.; Oliver, M.L.; Gersbach, C.A. RNA-Guided Transcriptional Silencing in Vivo with S. Aureus CRISPR-Cas9 Repressors. Nat. Commun. 2018, 9, 1674. [Google Scholar] [CrossRef] [PubMed]
- Lowder, L.G.; Paul, J.W., III; Qi, Y. Multiplexed Transcriptional Activation or Repression in Plants Using CRISPR-DCas9-Based Systems. In Methods in Molecular Biology; Springer: New York, NY, USA, 2017; pp. 167–184. ISBN 9781493971244. [Google Scholar]
- Moradpour, M.; Abdulah, S.N.A. CRISPR/DCas9 Platforms in Plants: Strategies and Applications beyond Genome Editing. Plant Biotechnol. J. 2020, 18, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Karlson, C.K.S.; Mohd-Noor, S.N.; Nolte, N.; Tan, B.C. CRISPR/DCas9-Based Systems: Mechanisms and Applications in Plant Sciences. Plants 2021, 10, 2055. [Google Scholar] [CrossRef] [PubMed]
- La Russa, M.F.; Qi, L.S. The New State of the Art: Cas9 for Gene Activation and Repression. Mol. Cell. Biol. 2015, 35, 3800–3809. [Google Scholar] [CrossRef]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-Scale Transcriptional Activation by an Engineered CRISPR-Cas9 Complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef]
- Morita, S.; Horii, T.; Hatada, I. Regulation of Gene Expression Using DCas9-SunTag Platforms. In Methods in Molecular Biology; Springer: New York, NY, USA, 2023; pp. 189–195. ISBN 9781071627235. [Google Scholar]
- Zhou, H.; Liu, J.; Zhou, C.; Gao, N.; Rao, Z.; Li, H.; Hu, X.; Li, C.; Yao, X.; Shen, X.; et al. In Vivo Simultaneous Transcriptional Activation of Multiple Genes in the Brain Using CRISPR–DCas9-Activator Transgenic Mice. Nat. Neurosci. 2018, 21, 440–446. [Google Scholar] [CrossRef]
- Chavez, A.; Scheiman, J.; Vora, S.; Pruitt, B.W.; Tuttle, M.; P R Iyer, E.; Lin, S.; Kiani, S.; Guzman, C.D.; Wiegand, D.J.; et al. Highly Efficient Cas9-Mediated Transcriptional Programming. Nat. Methods 2015, 12, 326–328. [Google Scholar] [CrossRef]
- Chavez, A.; Tuttle, M.; Pruitt, B.W.; Ewen-Campen, B.; Chari, R.; Ter-Ovanesyan, D.; Haque, S.J.; Cecchi, R.J.; Kowal, E.J.K.; Buchthal, J.; et al. Comparison of Cas9 Activators in Multiple Species. Nat. Methods 2016, 13, 563–567. [Google Scholar] [CrossRef]
- Zhang, Y.; Yin, C.; Zhang, T.; Li, F.; Yang, W.; Kaminski, R.; Fagan, P.R.; Putatunda, R.; Young, W.-B.; Khalili, K.; et al. CRISPR/GRNA-Directed Synergistic Activation Mediator (SAM) Induces Specific, Persistent and Robust Reactivation of the HIV-1 Latent Reservoirs. Sci. Rep. 2015, 5, 16277. [Google Scholar] [CrossRef]
- Bialek, J.K.; Dunay, G.A.; Voges, M.; Schäfer, C.; Spohn, M.; Stucka, R.; Hauber, J.; Lange, U.C. Targeted HIV-1 Latency Reversal Using CRISPR/Cas9-Derived Transcriptional Activator Systems. PLoS ONE 2016, 11, e0158294. [Google Scholar] [CrossRef] [PubMed]
- Riedmayr, L.M.; Hinrichsmeyer, K.S.; Karguth, N.; Böhm, S.; Splith, V.; Michalakis, S.; Becirovic, E. DCas9-VPR-Mediated Transcriptional Activation of Functionally Equivalent Genes for Gene Therapy. Nat. Protoc. 2022, 17, 781–818. [Google Scholar] [CrossRef] [PubMed]
- Casas-Mollano, J.A.; Zinselmeier, M.H.; Erickson, S.E.; Smanski, M.J. CRISPR-Cas Activators for Engineering Gene Expression in Higher Eukaryotes. CRISPR J. 2020, 3, 350–364. [Google Scholar] [CrossRef]
- Hilton, I.B.; D’Ippolito, A.M.; Vockley, C.M.; Thakore, P.I.; Crawford, G.E.; Reddy, T.E.; Gersbach, C.A. Epigenome Editing by a CRISPR-Cas9-Based Acetyltransferase Activates Genes from Promoters and Enhancers. Nat. Biotechnol. 2015, 33, 510–517. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, W.; Zhang, J.; Zhao, A.; Jiang, H. Epigenome Editing Based on CRISPR/DCas9p300 Facilitates Transdifferentiation of Human Fibroblasts into Leydig-like Cells. Exp. Cell Res. 2023, 425, 113551. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, J.P.; Zhang, H.; Wandling, G.M.; He, D.; Kyzar, E.J.; Lasek, A.W.; Pandey, S.C. Targeted Epigenomic Editing Ameliorates Adult Anxiety and Excessive Drinking after Adolescent Alcohol Exposure. Sci. Adv. 2022, 8, eabn2748. [Google Scholar] [CrossRef]
- Kearns, N.A.; Pham, H.; Tabak, B.; Genga, R.M.; Silverstein, N.J.; Garber, M.; Maehr, R. Functional Annotation of Native Enhancers with a Cas9–Histone Demethylase Fusion. Nat. Methods 2015, 12, 401–403. [Google Scholar] [CrossRef]
- Rudolph, T.; Beuch, S.; Reuter, G. Lysine-Specific Histone Demethylase LSD1 and the Dynamic Control of Chromatin. Biol. Chem. 2013, 394, 1019–1028. [Google Scholar] [CrossRef]
- Xu, X.; Tao, Y.; Gao, X.; Zhang, L.; Li, X.; Zou, W.; Ruan, K.; Wang, F.; Xu, G.-L.; Hu, R. A CRISPR-Based Approach for Targeted DNA Demethylation. Cell Discov. 2016, 2, 16009. [Google Scholar] [CrossRef]
- Choudhury, S.R.; Cui, Y.; Lubecka, K.; Stefanska, B.; Irudayaraj, J. CRISPR-DCas9 Mediated TET1 Targeting for Selective DNA Demethylation at BRCA1 Promoter. Oncotarget 2016, 7, 46545–46556. [Google Scholar] [CrossRef]
- Wang, M.; He, L.; Chen, B.; Wang, Y.; Wang, L.; Zhou, W.; Zhang, T.; Cao, L.; Zhang, P.; Xie, L.; et al. Transgenerationally Transmitted DNA Demethylation of a Spontaneous Epialleles Using CRISPR/DCas9-TET1cd Targeted Epigenetic Editing in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 10492. [Google Scholar] [CrossRef]
- Vojta, A.; Dobrinić, P.; Tadić, V.; Bočkor, L.; Korać, P.; Julg, B.; Klasić, M.; Zoldoš, V. Repurposing the CRISPR-Cas9 System for Targeted DNA Methylation. Nucleic Acids Res. 2016, 44, 5615–5628. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Shin, J.; Kim, Y.; Saito, T.; Saido, T.C.; Kim, J. CRISPR/DCas9-Dnmt3a-Mediated Targeted DNA Methylation of APP Rescues Brain Pathology in a Mouse Model of Alzheimer’s Disease. Transl. Neurodegener. 2022, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Pflueger, C.; Tan, D.; Swain, T.; Nguyen, T.; Pflueger, J.; Nefzger, C.; Polo, J.M.; Ford, E.; Lister, R. A Modular DCas9-SunTag DNMT3A Epigenome Editing System Overcomes Pervasive off-Target Activity of Direct Fusion DCas9-DNMT3A Constructs. Genome Res. 2018, 28, 1193–1206. [Google Scholar] [CrossRef]
- Nuñez, J.K.; Chen, J.; Pommier, G.C.; Cogan, J.Z.; Replogle, J.M.; Adriaens, C.; Ramadoss, G.N.; Shi, Q.; Hung, K.L.; Samelson, A.J.; et al. Genome-Wide Programmable Transcriptional Memory by CRISPR-Based Epigenome Editing. Cell 2021, 184, 2503–2519.e17. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Wu, J.; Zhang, X.; Luo, J.; Wang, H.; Ming, D. The Advance of CRISPR-Cas9-Based and NIR/CRISPR-Cas9-Based Imaging System. Front. Chem. 2021, 9, 786354. [Google Scholar] [CrossRef]
- Ma, H.; Tu, L.-C.; Naseri, A.; Huisman, M.; Zhang, S.; Grunwald, D.; Pederson, T. Multiplexed Labeling of Genomic Loci with DCas9 and Engineered SgRNAs Using CRISPRainbow. Nat. Biotechnol. 2016, 34, 528–530. [Google Scholar] [CrossRef]
- Chen, B.; Gilbert, L.A.; Cimini, B.A.; Schnitzbauer, J.; Zhang, W.; Li, G.-W.; Park, J.; Blackburn, E.H.; Weissman, J.S.; Qi, L.S.; et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 2013, 155, 1479–1491. [Google Scholar] [CrossRef]
- Clow, P.A.; Du, M.; Jillette, N.; Taghbalout, A.; Zhu, J.J.; Cheng, A.W. CRISPR-Mediated Multiplexed Live Cell Imaging of Nonrepetitive Genomic Loci with One Guide RNA per Locus. Nat. Commun. 2022, 13, 1871. [Google Scholar] [CrossRef]
- Mao, S.; Ying, Y.; Wu, X.; Krueger, C.J.; Chen, A.K. CRISPR/Dual-FRET Molecular Beacon for Sensitive Live-Cell Imaging of Non-Repetitive Genomic Loci. Nucleic Acids Res. 2019, 47, e131. [Google Scholar] [CrossRef]
- Ma, H.; Naseri, A.; Reyes-Gutierrez, P.; Wolfe, S.A.; Zhang, S.; Pederson, T. Multicolor CRISPR Labeling of Chromosomal Loci in Human Cells. Proc. Natl. Acad. Sci. USA 2015, 112, 3002–3007. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Tu, L.-C.; Naseri, A.; Chung, Y.-C.; Grunwald, D.; Zhang, S.; Pederson, T. CRISPR-Sirius: RNA Scaffolds for Signal Amplification in Genome Imaging. Nat. Methods 2018, 15, 928–931. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.W.; Jillette, N.; Lee, P.; Plaskon, D.; Fujiwara, Y.; Wang, W.; Taghbalout, A.; Wang, H. Casilio: A Versatile CRISPR-Cas9-Pumilio Hybrid for Gene Regulation and Genomic Labeling. Cell Res. 2016, 26, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Yuno, M.; Fujii, H. EnChIP Systems Using Different CRISPR Orthologues and Epitope Tags. BMC Res. Notes 2018, 11, 154. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Yuno, M.; Fujii, H. An EnChIP System for the Analysis of Bacterial Genome Functions. BMC Res. Notes 2018, 11, 387. [Google Scholar] [CrossRef]
- Fujita, T.; Fujii, H. Purification of Specific DNA Species Using the CRISPR System. Biol. Methods Protoc. 2019, 4, bpz008. [Google Scholar] [CrossRef]
- Fujita, T.; Kitaura, F.; Oji, A.; Tanigawa, N.; Yuno, M.; Ikawa, M.; Taniuchi, I.; Fujii, H. Transgenic Mouse Lines Expressing the 3xFLAG-dCas9 Protein for EnChIP Analysis. Genes Cells 2018, 23, 318–325. [Google Scholar] [CrossRef]
- Fujita, T.; Fujii, H. Efficient Isolation of Specific Genomic Regions and Identification of Associated Proteins by Engineered DNA-Binding Molecule-Mediated Chromatin Immunoprecipitation (EnChIP) Using CRISPR. Biochem. Biophys. Res. Commun. 2013, 439, 132–136. [Google Scholar] [CrossRef]
- Fujita, T.; Fujii, H. Isolation of Specific Genomic Regions and Identification of Associated Molecules by Engineered DNA-Binding Molecule-Mediated Chromatin Immunoprecipitation (EnChIP) Using CRISPR. In Methods in Molecular Biology; Springer: New York, NY, USA, 2015; pp. 43–52. ISBN 9781493924738. [Google Scholar]
- Schmidtmann, E.; Anton, T.; Rombaut, P.; Herzog, F.; Leonhardt, H. Determination of Local Chromatin Composition by CasID. Nucleus 2016, 7, 476–484. [Google Scholar] [CrossRef]
- Tsui, C.; Inouye, C.; Levy, M.; Lu, A.; Florens, L.; Washburn, M.P.; Tjian, R. DCas9-Targeted Locus-Specific Protein Isolation Method Identifies Histone Gene Regulators. Proc. Natl. Acad. Sci. USA 2018, 115, E2734–E2741. [Google Scholar] [CrossRef]
- Gao, X.D.; Tu, L.-C.; Mir, A.; Rodriguez, T.; Ding, Y.; Leszyk, J.; Dekker, J.; Shaffer, S.A.; Zhu, L.J.; Wolfe, S.A.; et al. C-BERST: Defining Subnuclear Proteomic Landscapes at Genomic Elements with DCas9–APEX2. Nat. Methods 2018, 15, 433–436. [Google Scholar] [CrossRef]
- Huang, T.P.; Newby, G.A.; Liu, D.R. Precision Genome Editing Using Cytosine and Adenine Base Editors in Mammalian Cells. Nat. Protoc. 2021, 16, 1089–1128. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.K.; Song, B.; Bae, S. Current Status and Challenges of DNA Base Editing Tools. Mol. Ther. 2020, 28, 1938–1952. [Google Scholar] [CrossRef] [PubMed]
- Porto, E.M.; Komor, A.C. In the Business of Base Editors: Evolution from Bench to Bedside. PLoS Biol. 2023, 21, e3002071. [Google Scholar] [CrossRef] [PubMed]
- Kantor, A.; McClements, M.; MacLaren, R. CRISPR-Cas9 DNA Base-Editing and Prime-Editing. Int. J. Mol. Sci. 2020, 21, 6240. [Google Scholar] [CrossRef] [PubMed]
- Neugebauer, M.E.; Hsu, A.; Arbab, M.; Krasnow, N.A.; McElroy, A.N.; Pandey, S.; Doman, J.L.; Huang, T.P.; Raguram, A.; Banskota, S.; et al. Evolution of an Adenine Base Editor into a Small, Efficient Cytosine Base Editor with Low off-Target Activity. Nat. Biotechnol. 2023, 41, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Lam, D.K.; Feliciano, P.R.; Arif, A.; Bohnuud, T.; Fernandez, T.P.; Gehrke, J.M.; Grayson, P.; Lee, K.D.; Ortega, M.A.; Sawyer, C.; et al. Improved Cytosine Base Editors Generated from TadA Variants. Nat. Biotechnol. 2023, 41, 686–697. [Google Scholar] [CrossRef]
- Chen, L.; Zhu, B.; Ru, G.; Meng, H.; Yan, Y.; Hong, M.; Zhang, D.; Luan, C.; Zhang, S.; Wu, H.; et al. Re-Engineering the Adenine Deaminase TadA-8e for Efficient and Specific CRISPR-Based Cytosine Base Editing. Nat. Biotechnol. 2023, 41, 663–672. [Google Scholar] [CrossRef]
- Richter, M.F.; Zhao, K.T.; Eton, E.; Lapinaite, A.; Newby, G.A.; Thuronyi, B.W.; Wilson, C.; Koblan, L.W.; Zeng, J.; Bauer, D.E.; et al. Phage-Assisted Evolution of an Adenine Base Editor with Improved Cas Domain Compatibility and Activity. Nat. Biotechnol. 2020, 38, 883–891. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, W.; Wang, F.; Zhao, S.; Feng, F.; Song, J.; Zhang, C.; Yang, J. Increasing Cytosine Base Editing Scope and Efficiency with Engineered Cas9-PmCDA1 Fusions and the Modified SgRNA in Rice. Front. Genet. 2019, 10, 379. [Google Scholar] [CrossRef]
- Komor, A.C.; Zhao, K.T.; Packer, M.S.; Gaudelli, N.M.; Waterbury, A.L.; Koblan, L.W.; Kim, Y.B.; Badran, A.H.; Liu, D.R. Improved Base Excision Repair Inhibition and Bacteriophage Mu Gam Protein Yields C:G-to-T:A Base Editors with Higher Efficiency and Product Purity. Sci. Adv. 2017, 3, eaao4774. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.D.; Huang, M.; Dai, P.; Liu, T.; Fan, S.; Cheng, X.; Zhao, Y.; Yeap, L.-S.; Meng, F.-L. Intrinsic Nucleotide Preference of Diversifying Base Editors Guides Antibody Ex Vivo Affinity Maturation. Cell Rep. 2018, 25, 884–892.e3. [Google Scholar] [CrossRef] [PubMed]
- Knipping, F.; Newby, G.A.; Eide, C.R.; McElroy, A.N.; Nielsen, S.C.; Smith, K.; Fang, Y.; Cornu, T.I.; Costa, C.; Gutierrez-Guerrero, A.; et al. Disruption of HIV-1 Co-Receptors CCR5 and CXCR4 in Primary Human T Cells and Hematopoietic Stem and Progenitor Cells Using Base Editing. Mol. Ther. 2022, 30, 130–144. [Google Scholar] [CrossRef]
- Alves, C.R.R.; Ha, L.L.; Yaworski, R.; Lazzarotto, C.R.; Christie, K.A.; Reilly, A.; Beauvais, A.; Doll, R.M.; de la Cruz, D.; Maguire, C.A.; et al. Base Editing as a Genetic Treatment for Spinal Muscular Atrophy. bioRxiv 2023. [Google Scholar] [CrossRef]
- Arbab, M.; Matuszek, Z.; Kray, K.M.; Du, A.; Newby, G.A.; Blatnik, A.J.; Raguram, A.; Richter, M.F.; Zhao, K.T.; Levy, J.M.; et al. Base Editing Rescue of Spinal Muscular Atrophy in Cells and in Mice. Science 2023, 380, eadg6518. [Google Scholar] [CrossRef] [PubMed]
- Musunuru, K.; Chadwick, A.C.; Mizoguchi, T.; Garcia, S.P.; DeNizio, J.E.; Reiss, C.W.; Wang, K.; Iyer, S.; Dutta, C.; Clendaniel, V.; et al. In Vivo CRISPR Base Editing of PCSK9 Durably Lowers Cholesterol in Primates. Nature 2021, 593, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Li, J.; Li, S.; Xin, X.; Hu, M.; Price, M.A.; Rosser, S.J.; Bi, C.; Zhang, X. Glycosylase Base Editors Enable C-to-A and C-to-G Base Changes. Nat. Biotechnol. 2021, 39, 35–40. [Google Scholar] [CrossRef]
- Technologies ASH, Head of Gene Editing Platform Applied Base Editing to Treat Beta-Hemoglobinopathies. Available online: https://beamtx.com/media/kmkew24k/202111-ash-applied-base-editing-to-treat-beta-hemoglobinopathies-vfinal.pdf (accessed on 12 October 2023).
- Haihua Chu, S.; Lam, D.; Packer, M.S.; Olins, J.; Liquori, A.; Rehberger, K.; Rinaldi, C.; Marshall, J.; Lee, C.; Yan, B.; et al. Adenine Base Editing of the Sickle Allele in CD34 Hematopoietic Stem and Progenitor Cells Eliminates Hemoglobin S. Available online: https://beamtx.com/media/m2qaoudq/202012-ash-beam102-makassar-at-ash-v5.pdf (accessed on 12 October 2023).
- Verve Therapeutics Announces VERVE-101 Awarded Innovation Passport by the UK MHRA for the Treatment of Heterozygous Familial Hypercholesterolemia. Available online: https://ir.vervetx.com/news-releases/news-release-details/verve-therapeutics-announces-verve-101-awarded-innovation/ (accessed on 12 October 2023).
- Scholefield, J.; Harrison, P.T. Prime Editing—An Update on the Field. Gene Ther. 2021, 28, 396–401. [Google Scholar] [CrossRef]
- Huang, Z.; Liu, G. Current Advancement in the Application of Prime Editing. Front. Bioeng. Biotechnol. 2023, 11, 1039315. [Google Scholar] [CrossRef]
- Lu, C.; Kuang, J.; Shao, T.; Xie, S.; Li, M.; Zhu, L.; Zhu, L. Prime Editing: An All-Rounder for Genome Editing. Int. J. Mol. Sci. 2022, 23, 9862. [Google Scholar] [CrossRef]
- Tao, R.; Wang, Y.; Jiao, Y.; Hu, Y.; Li, L.; Jiang, L.; Zhou, L.; Qu, J.; Chen, Q.; Yao, S. Bi-PE: Bi-Directional Priming Improves CRISPR/Cas9 Prime Editing in Mammalian Cells. Nucleic Acids Res. 2022, 50, 6423–6434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Petri, K.; Ma, J.; Lee, H.; Tsai, C.-L.; Joung, J.K.; Yeh, J.-R.J. Enhancing CRISPR Prime Editing by Reducing Misfolded PegRNA Interactions. bioRxiv 2023. [Google Scholar] [CrossRef]
- Zhao, Z.; Shang, P.; Mohanraju, P.; Geijsen, N. Prime Editing: Advances and Therapeutic Applications. Trends Biotechnol. 2023, 41, 1000–1012. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Lee, W.-J.; Hur, J.K.; Song, W.J.; Lee, Y.; Kim, H.; Gwon, L.W.; Kim, Y.-H.; Park, Y.-H.; Kim, C.H.; et al. Expansion of the Prime Editing Modality with Cas9 from Francisella Novicida. Genome Biol. 2022, 23, 92. [Google Scholar] [CrossRef]
- Henry, J.; Oh, D.; Eskew, J.; Baranda, J.; Rodriguez Rivera, I.I.; Dumbrava, E.; Cohen, E.; Belani, R.; McCaigue, J.; Shedlock, D.; et al. 728 Phase 1 Study of P-MUC1C-ALLO1 Allogeneic CAR-T Cells in Patients with Epithelial-Derived Cancers. In Proceedings of the Regular and Young Investigator Award Abstracts; BMJ Publishing Group Ltd.: London, UK, 2022. [Google Scholar]
- Kocoglu, M.H.; Asch, A.; Ramakrishnan, A.; Bachier, C.; Martin, T.; Rodriguez, T.; McArthur, K.; Martin, C.; Namini, H.; Ostertag, E.; et al. 47P Phase I Study to Assess the Safety and Efficacy of P-BCMA-ALLO1: A Fully Allogeneic CAR-T Therapy, in Patients with Relapsed/Refractory Multiple Myeloma (RRMM). Immuno-Oncol. Technol. 2022, 16, 100152. [Google Scholar] [CrossRef]
- Pan, Y.; Shen, N.; Jung-Klawitter, S.; Betzen, C.; Hoffmann, G.F.; Hoheisel, J.D.; Blau, N. CRISPR RNA-Guided FokI Nucleases Repair a PAH Variant in a Phenylketonuria Model. Sci. Rep. 2016, 6, 35794. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-X.; Li, M.; Lee, C.M.; Chakraborty, S.; Kim, H.-W.; Bao, G.; Leong, K.W. CRISPR/Cas9-Based Genome Editing for Disease Modeling and Therapy: Challenges and Opportunities for Nonviral Delivery. Chem. Rev. 2017, 117, 9874–9906. [Google Scholar] [CrossRef]
- Kotterman, M.A.; Chalberg, T.W.; Schaffer, D.V. Viral Vectors for Gene Therapy: Translational and Clinical Outlook. Annu. Rev. Biomed. Eng. 2015, 17, 63–89. [Google Scholar] [CrossRef]
- Xu, C.L.; Ruan, M.Z.C.; Mahajan, V.B.; Tsang, S.H. Viral Delivery Systems for CRISPR. Viruses 2019, 11, 28. [Google Scholar] [CrossRef]
- Wan, T.; Niu, D.; Wu, C.; Xu, F.-J.; Church, G.; Ping, Y. Material Solutions for Delivery of CRISPR/Cas-Based Genome Editing Tools: Current Status and Future Outlook. Mater. Today 2019, 26, 40–66. [Google Scholar] [CrossRef]
- Navarro-Serna, S.; Vilarino, M.; Park, I.; Gadea, J.; Ross, P.J. Livestock Gene Editing by One-Step Embryo Manipulation. J. Equine Vet. Sci. 2020, 89, 103025. [Google Scholar] [CrossRef] [PubMed]
- Bhandawat, A.; Sharma, V.; Rishi, V.; Roy, J.K. Biolistic Delivery of Programmable Nuclease (CRISPR/Cas9) in Bread Wheat. In Methods in Molecular Biology; Springer: New York, NY, USA, 2020; pp. 309–329. ISBN 9781071603550. [Google Scholar]
- Liang, Z.; Chen, K.; Gao, C. Biolistic Delivery of CRISPR/Cas9 with Ribonucleoprotein Complex in Wheat. In Methods in Molecular Biology; Springer: New York, NY, USA, 2019; pp. 327–335. ISBN 9781493989904. [Google Scholar]
- Seki, A.; Rutz, S. Optimized RNP Transfection for Highly Efficient CRISPR/Cas9-Mediated Gene Knockout in Primary T Cells. J. Exp. Med. 2018, 215, 985–997. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.-S.; Shih, H.-A.; Lai, M.-C.; Chang, Y.-J.; Lin, S. Enhanced NK-92 Cytotoxicity by CRISPR Genome Engineering Using Cas9 Ribonucleoproteins. Front. Immunol. 2020, 11, 1008. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.-S. Highly Efficient RNA-Guided Genome Editing in Human Cells via Delivery of Purified Cas9 Ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Modarai, S.R.; Man, D.; Bialk, P.; Rivera-Torres, N.; Bloh, K.; Kmiec, E.B. Efficient Delivery and Nuclear Uptake Is Not Sufficient to Detect Gene Editing in CD34+ Cells Directed by a Ribonucleoprotein Complex. Mol. Ther. Nucleic Acids 2018, 11, 116–129. [Google Scholar] [CrossRef]
- Hiatt, J.; Cavero, D.A.; McGregor, M.J.; Zheng, W.; Budzik, J.M.; Roth, T.L.; Haas, K.M.; Wu, D.; Rathore, U.; Meyer-Franke, A.; et al. Efficient Generation of Isogenic Primary Human Myeloid Cells Using CRISPR-Cas9 Ribonucleoproteins. Cell Rep. 2021, 35, 109105. [Google Scholar] [CrossRef]
- Han, X.; Liu, Z.; Ma, Y.; Zhang, K.; Qin, L. Cas9 Ribonucleoprotein Delivery via Microfluidic Cell-deformation Chip for Human T-cell Genome Editing and Immunotherapy. Adv. Biosyst. 2017, 1, e1600007. [Google Scholar] [CrossRef]
- Jarrell, J.A.; Sytsma, B.J.; Wilson, L.H.; Pan, F.L.; Lau, K.H.W.J.; Kirby, G.T.S.; Lievano, A.A.; Pawell, R.S. Genome Editing Human Primary T Cells with Microfluidic Vortex Shedding & CRISPR Cas9. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yen, J.; Fiorino, M.; Liu, Y.; Paula, S.; Clarkson, S.; Quinn, L.; Tschantz, W.R.; Klock, H.; Guo, N.; Russ, C.; et al. TRIAMF: A New Method for Delivery of Cas9 Ribonucleoprotein Complex to Human Hematopoietic Stem Cells. Sci. Rep. 2018, 8, 16304. [Google Scholar] [CrossRef]
- Chen, Y.; Aslanoglou, S.; Murayama, T.; Gervinskas, G.; Fitzgerald, L.I.; Sriram, S.; Tian, J.; Johnston, A.P.R.; Morikawa, Y.; Suu, K.; et al. Silicon-nanotube-mediated Intracellular Delivery Enables Ex Vivo Gene Editing. Adv. Mater. 2020, 32, e2000036. [Google Scholar] [CrossRef]
- Kholosy, W.M.; Visscher, M.; Ogink, K.; Buttstedt, H.; Griffin, K.; Beier, A.; Gerlach, J.P.; Molenaar, J.J.; Geijsen, N.; de Boer, M.; et al. Simple, Fast and Efficient ITOP-Mediated Delivery of CRISPR/Cas9 RNP in Difficult-to-Transduce Human Cells Including Primary T Cells. J. Biotechnol. 2021, 338, 71–80. [Google Scholar] [CrossRef]
- Wei, T.; Cheng, Q.; Min, Y.-L.; Olson, E.N.; Siegwart, D.J. Systemic Nanoparticle Delivery of CRISPR-Cas9 Ribonucleoproteins for Effective Tissue Specific Genome Editing. Nat. Commun. 2020, 11, 3232. [Google Scholar] [CrossRef]
- Gustafsson, O.; Rädler, J.; Roudi, S.; Lehto, T.; Hällbrink, M.; Lehto, T.; Gupta, D.; Andaloussi, S.E.L.; Nordin, J.Z. Efficient Peptide-Mediated in Vitro Delivery of Cas9 RNP. Pharmaceutics 2021, 13, 878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Shen, J.; Li, D.; Cheng, Y. Strategies in the Delivery of Cas9 Ribonucleoprotein for CRISPR/Cas9 Genome Editing. Theranostics 2021, 11, 614–648. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wan, T.; Wang, H.; Zhang, S.; Ping, Y.; Cheng, Y. A Boronic Acid–Rich Dendrimer with Robust and Unprecedented Efficiency for Cytosolic Protein Delivery and CRISPR-Cas9 Gene Editing. Sci. Adv. 2019, 5, eaaw8922. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Sun, W.; Lin, S.; Jin, R.; Ma, L.; Liu, Y. Cytosolic Delivery of CRISPR/Cas9 Ribonucleoproteins for Genome Editing Using Chitosan-Coated Red Fluorescent Protein. Chem. Commun. 2019, 55, 4707–4710. [Google Scholar] [CrossRef]
- Ding, F.; Huang, X.; Gao, X.; Xie, M.; Pan, G.; Li, Q.; Song, J.; Zhu, X.; Zhang, C. A Non-Cationic Nucleic Acid Nanogel for the Delivery of the CRISPR/Cas9 Gene Editing Tool. Nanoscale 2019, 11, 17211–17215. [Google Scholar] [CrossRef]
- Shahbazi, R.; Sghia-Hughes, G.; Reid, J.L.; Kubek, S.; Haworth, K.G.; Humbert, O.; Kiem, H.-P.; Adair, J.E. Targeted Homology-Directed Repair in Blood Stem and Progenitor Cells with CRISPR Nanoformulations. Nat. Mater. 2019, 18, 1124–1132. [Google Scholar] [CrossRef]
- Alyami, M.Z.; Alsaiari, S.K.; Li, Y.; Qutub, S.S.; Aleisa, F.A.; Sougrat, R.; Merzaban, J.S.; Khashab, N.M. Cell-Type-Specific CRISPR/Cas9 Delivery by Biomimetic Metal Organic Frameworks. J. Am. Chem. Soc. 2020, 142, 1715–1720. [Google Scholar] [CrossRef]
- Yue, H.; Zhou, X.; Cheng, M.; Xing, D. Graphene Oxide-Mediated Cas9/SgRNA Delivery for Efficient Genome Editing. Nanoscale 2018, 10, 1063–1071. [Google Scholar] [CrossRef]
- Zhou, W.; Cui, H.; Ying, L.; Yu, X.-F. Enhanced Cytosolic Delivery and Release of CRISPR/Cas9 by Black Phosphorus Nanosheets for Genome Editing. Angew. Chem. Int. Ed. Engl. 2018, 57, 10268–10272. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Song, Z.; Liu, C.; Chen, X.-L.; Han, H. Biomimetic Mineralization-Based CRISPR/Cas9 Ribonucleoprotein Nanoparticles for Gene Editing. ACS Appl. Mater. Interfaces 2019, 11, 47762–47770. [Google Scholar] [CrossRef] [PubMed]
- Tyumentseva, M.A.; Tyumentsev, A.I.; Akimkin, V.G. Protocol for Assessment of the Efficiency of CRISPR/Cas RNP Delivery to Different Types of Target Cells. PLoS ONE 2021, 16, e0259812. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic Acid Detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a Target Binding Unleashes Indiscriminate Single-Stranded DNase Activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and Portable Nucleic Acid Detection Platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-Deployable Viral Diagnostics Using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef]
- Müller, V.; Rajer, F.; Frykholm, K.; Nyberg, L.K.; Quaderi, S.; Fritzsche, J.; Kristiansson, E.; Ambjörnsson, T.; Sandegren, L.; Westerlund, F. Direct Identification of Antibiotic Resistance Genes on Single Plasmid Molecules Using CRISPR/Cas9 in Combination with Optical DNA Mapping. Sci. Rep. 2016, 6, 37938. [Google Scholar] [CrossRef]
- Zhou, W.; Hu, L.; Ying, L.; Zhao, Z.; Chu, P.K.; Yu, X.-F. A CRISPR–Cas9-Triggered Strand Displacement Amplification Method for Ultrasensitive DNA Detection. Nat. Commun. 2018, 9, 5012. [Google Scholar] [CrossRef]
- Huang, M.; Zhou, X.; Wang, H.; Xing, D. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Triggered Isothermal Amplification for Site-Specific Nucleic Acid Detection. Anal. Chem. 2018, 90, 2193–2200. [Google Scholar] [CrossRef]
- Hajian, R.; Balderston, S.; Tran, T.; deBoer, T.; Etienne, J.; Sandhu, M.; Wauford, N.A.; Chung, J.-Y.; Nokes, J.; Athaiya, M.; et al. Detection of Unamplified Target Genes via CRISPR–Cas9 Immobilized on a Graphene Field-Effect Transistor. Nat. Biomed. Eng. 2019, 3, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Langelier, C.; Kuchta, A.; Batson, J.; Teyssier, N.; Lyden, A.; Caldera, S.; McGeever, A.; Dimitrov, B.; King, R.; et al. FLASH: A next-Generation CRISPR Diagnostic for Multiplexed Detection of Antimicrobial Resistance Sequences. Nucleic Acids Res. 2019, 47, e83. [Google Scholar] [CrossRef] [PubMed]
- Azhar, M.; Phutela, R.; Kumar, M.; Ansari, A.H.; Rauthan, R.; Gulati, S.; Sharma, N.; Sinha, D.; Sharma, S.; Singh, S.; et al. Rapid and Accurate Nucleobase Detection Using FnCas9 and Its Application in COVID-19 Diagnosis. Biosens. Bioelectron. 2021, 183, 113207. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xiong, E.; Tian, T.; Cheng, M.; Lin, W.; Sun, J.; Zhou, X. CASLFA: CRISPR/Cas9-Mediated Lateral Flow Nucleic Acid Assay. bioRxiv 2019, 702209. [Google Scholar] [CrossRef]
- Marsic, T.; Ali, Z.; Tehseen, M.; Mahas, A.; Hamdan, S.; Mahfouz, M. Vigilant: An Engineered VirD2-Cas9 Complex for Lateral Flow Assay-Based Detection of SARS-CoV2. Nano Lett. 2021, 21, 3596–3603. [Google Scholar] [CrossRef]
- Osborn, M.J.; Bhardwaj, A.; Bingea, S.P.; Knipping, F.; Feser, C.J.; Lees, C.J.; Collins, D.P.; Steer, C.J.; Blazar, B.R.; Tolar, J. CRISPR/Cas9-Based Lateral Flow and Fluorescence Diagnostics. Bioengineering 2021, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.; Sharma, S.; Dugar, G.; Peeck, N.L.; Bischler, T.; Wimmer, F.; Yu, Y.; Barquist, L.; Schoen, C.; Kurzai, O.; et al. Noncanonical CrRNAs Derived from Host Transcripts Enable Multiplexable RNA Detection by Cas9. Science 2021, 372, 941–948. [Google Scholar] [CrossRef]
- Ali, Z.; Sánchez, E.; Tehseen, M.; Mahas, A.; Marsic, T.; Aman, R.; Sivakrishna Rao, G.; Alhamlan, F.S.; Alsanea, M.S.; Al-Qahtani, A.A.; et al. Bio-SCAN: A CRISPR/DCas9-Based Lateral Flow Assay for Rapid, Specific, and Sensitive Detection of SARS-CoV-2. ACS Synth. Biol. 2022, 11, 406–419. [Google Scholar] [CrossRef]
- Wu, S.-S.; Li, Q.-C.; Yin, C.-Q.; Xue, W.; Song, C.-Q. Advances in CRISPR/Cas-Based Gene Therapy in Human Genetic Diseases. Theranostics 2020, 10, 4374–4382. [Google Scholar] [CrossRef]
- Papasavva, P.; Kleanthous, M.; Lederer, C.W. Rare Opportunities: CRISPR/Cas-Based Therapy Development for Rare Genetic Diseases. Mol. Diagn. Ther. 2019, 23, 201–222. [Google Scholar] [CrossRef]
- Kennedy, E.M.; Cullen, B.R. Gene Editing: A New Tool for Viral Disease. Annu. Rev. Med. 2017, 68, 401–411. [Google Scholar] [CrossRef]
- Xiong, X.; Chen, M.; Lim, W.A.; Zhao, D.; Qi, L.S. CRISPR/Cas9 for Human Genome Engineering and Disease Research. Annu. Rev. Genom. Hum. Genet. 2016, 17, 131–154. [Google Scholar] [CrossRef] [PubMed]
- Ferdosi, S.R.; Ewaisha, R.; Moghadam, F.; Krishna, S.; Park, J.G.; Ebrahimkhani, M.R.; Kiani, S.; Anderson, K.S. Multifunctional CRISPR-Cas9 with Engineered Immunosilenced Human T Cell Epitopes. Nat. Commun. 2019, 10, 1842. [Google Scholar] [CrossRef] [PubMed]
- Doms, R.W. Chemokine Receptors and HIV Entry. AIDS 2001, 15, S34–S35. [Google Scholar] [CrossRef]
- Koujah, L.; Shukla, D.; Naqvi, A.R. CRISPR-Cas Based Targeting of Host and Viral Genes as an Antiviral Strategy. Semin. Cell Dev. Biol. 2019, 96, 53–64. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, S.; Jin, X.; Wang, Q.; Yang, K.; Li, C.; Xiao, Q.; Hou, P.; Liu, S.; Wu, S.; et al. Genome Editing of the HIV Co-Receptors CCR5 and CXCR4 by CRISPR-Cas9 Protects CD4+ T Cells from HIV-1 Infection. Cell Biosci. 2017, 7, 47. [Google Scholar] [CrossRef]
- Xu, L.; Yang, H.; Gao, Y.; Chen, Z.; Xie, L.; Liu, Y.; Liu, Y.; Wang, X.; Li, H.; Lai, W.; et al. CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance in Vivo. Mol. Ther. 2017, 25, 1782–1789. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Guan, X.; Du, T.; Jin, W.; Wu, B.; Liu, Y.; Wang, P.; Hu, B.; Griffin, G.E.; Shattock, R.J.; et al. Inhibition of HIV-1 Infection of Primary CD4+ T-Cells by Gene Editing of CCR5 Using Adenovirus-Delivered CRISPR/Cas9. J. Gen. Virol. 2015, 96, 2381–2393. [Google Scholar] [CrossRef]
- Cho, S.W.; Kim, S.; Kim, Y.; Kweon, J.; Kim, H.S.; Bae, S.; Kim, J.-S. Analysis of Off-Target Effects of CRISPR/Cas-Derived RNA-Guided Endonucleases and Nickases. Genome Res. 2014, 24, 132–141. [Google Scholar] [CrossRef]
- Hou, P.; Chen, S.; Wang, S.; Yu, X.; Chen, Y.; Jiang, M.; Zhuang, K.; Ho, W.; Hou, W.; Huang, J.; et al. Genome Editing of CXCR4 by CRISPR/Cas9 Confers Cells Resistant to HIV-1 Infection. Sci. Rep. 2015, 5, 15577. [Google Scholar] [CrossRef] [PubMed]
- Schumann, K.; Lin, S.; Boyer, E.; Simeonov, D.R.; Subramaniam, M.; Gate, R.E.; Haliburton, G.E.; Ye, C.J.; Bluestone, J.A.; Doudna, J.A.; et al. Generation of Knock-in Primary Human T Cells Using Cas9 Ribonucleoproteins. Proc. Natl. Acad. Sci. USA 2015, 112, 10437–10442. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, Q.; Yu, X.; Li, Y.; Guo, Y.; Liu, Z.; Sun, F.; Hou, W.; Li, C.; Wu, L.; et al. HIV-1 Inhibition in Cells with CXCR4 Mutant Genome Created by CRISPR-Cas9 and PiggyBac Recombinant Technologies. Sci. Rep. 2018, 8, 8573. [Google Scholar] [CrossRef]
- Strich, J.R.; Chertow, D.S. CRISPR-Cas Biology and Its Application to Infectious Diseases. J. Clin. Microbiol. 2019, 57, e01307-18. [Google Scholar] [CrossRef]
- Hu, W.; Kaminski, R.; Yang, F.; Zhang, Y.; Cosentino, L.; Li, F.; Luo, B.; Alvarez-Carbonell, D.; Garcia-Mesa, Y.; Karn, J.; et al. RNA-Directed Gene Editing Specifically Eradicates Latent and Prevents New HIV-1 Infection. Proc. Natl. Acad. Sci. USA 2014, 111, 11461–11466. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhao, N.; Berkhout, B.; Das, A.T. CRISPR-Cas Based Antiviral Strategies against HIV-1. Virus Res. 2018, 244, 321–332. [Google Scholar] [CrossRef]
- Yin, C.; Zhang, T.; Qu, X.; Zhang, Y.; Putatunda, R.; Xiao, X.; Li, F.; Xiao, W.; Zhao, H.; Dai, S.; et al. In Vivo Excision of HIV-1 Provirus by SaCas9 and Multiplex Single-Guide RNAs in Animal Models. Mol. Ther. 2017, 25, 1168–1186. [Google Scholar] [CrossRef]
- Bella, R.; Kaminski, R.; Mancuso, P.; Young, W.-B.; Chen, C.; Sariyer, R.; Fischer, T.; Amini, S.; Ferrante, P.; Jacobson, J.M.; et al. Removal of HIV DNA by CRISPR from Patient Blood Engrafts in Humanized Mice. Mol. Ther. Nucleic Acids 2018, 12, 275–282. [Google Scholar] [CrossRef]
- Li, H.; Sheng, C.; Wang, S.; Yang, L.; Liang, Y.; Huang, Y.; Liu, H.; Li, P.; Yang, C.; Yang, X.; et al. Removal of Integrated Hepatitis B Virus DNA Using CRISPR-Cas9. Front. Cell. Infect. Microbiol. 2017, 7, 91. [Google Scholar] [CrossRef]
- Scott, T.; Moyo, B.; Nicholson, S.; Maepa, M.B.; Watashi, K.; Ely, A.; Weinberg, M.S.; Arbuthnot, P. SsAAVs Containing Cassettes Encoding SaCas9 and Guides Targeting Hepatitis B Virus Inactivate Replication of the Virus in Cultured Cells. Sci. Rep. 2017, 7, 7401. [Google Scholar] [CrossRef]
- van Diemen, F.R.; Kruse, E.M.; Hooykaas, M.J.G.; Bruggeling, C.E.; Schürch, A.C.; van Ham, P.M.; Imhof, S.M.; Nijhuis, M.; Wiertz, E.J.H.J.; Lebbink, R.J. CRISPR/Cas9-Mediated Genome Editing of Herpesviruses Limits Productive and Latent Infections. PLoS Pathog. 2016, 12, e1005701. [Google Scholar] [CrossRef] [PubMed]
- Roehm, P.C.; Shekarabi, M.; Wollebo, H.S.; Bellizzi, A.; He, L.; Salkind, J.; Khalili, K. Inhibition of HSV-1 Replication by Gene Editing Strategy. Sci. Rep. 2016, 6, 23146. [Google Scholar] [CrossRef]
- Wollebo, H.S.; Bellizzi, A.; Kaminski, R.; Hu, W.; White, M.K.; Khalili, K. CRISPR/Cas9 System as an Agent for Eliminating Polyomavirus JC Infection. PLoS ONE 2015, 10, e0136046. [Google Scholar] [CrossRef]
- Kennedy, E.M.; Kornepati, A.V.R.; Goldstein, M.; Bogerd, H.P.; Poling, B.C.; Whisnant, A.W.; Kastan, M.B.; Cullen, B.R. Inactivation of the Human Papillomavirus E6 or E7 Gene in Cervical Carcinoma Cells by Using a Bacterial CRISPR/Cas RNA-Guided Endonuclease. J. Virol. 2014, 88, 11965–11972. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, S.; Salahudeen, A.A.; Sellers, Z.M.; Bravo, D.T.; Choi, S.S.; Batish, A.; Le, W.; Baik, R.; de la O, S.; Kaushik, M.P.; et al. High-Efficiency, Selection-Free Gene Repair in Airway Stem Cells from Cystic Fibrosis Patients Rescues CFTR Function in Differentiated Epithelia. Cell Stem Cell 2020, 26, 161–171.e4. [Google Scholar] [CrossRef] [PubMed]
- Schwank, G.; Koo, B.-K.; Sasselli, V.; Dekkers, J.F.; Heo, I.; Demircan, T.; Sasaki, N.; Boymans, S.; Cuppen, E.; van der Ent, C.K.; et al. Functional Repair of CFTR by CRISPR/Cas9 in Intestinal Stem Cell Organoids of Cystic Fibrosis Patients. Cell Stem Cell 2013, 13, 653–658. [Google Scholar] [CrossRef]
- Firth, A.L.; Menon, T.; Parker, G.S.; Qualls, S.J.; Lewis, B.M.; Ke, E.; Dargitz, C.T.; Wright, R.; Khanna, A.; Gage, F.H.; et al. Functional Gene Correction for Cystic Fibrosis in Lung Epithelial Cells Generated from Patient IPSCs. Cell Rep. 2015, 12, 1385–1390. [Google Scholar] [CrossRef]
- Wang, G. Genome Editing for Cystic Fibrosis. Cells 2023, 12, 1555. [Google Scholar] [CrossRef]
- Lomova, A.; Clark, D.N.; Campo-Fernandez, B.; Flores-Bjurström, C.; Kaufman, M.L.; Fitz-Gibbon, S.; Wang, X.; Miyahira, E.Y.; Brown, D.; DeWitt, M.A.; et al. Improving Gene Editing Outcomes in Human Hematopoietic Stem and Progenitor Cells by Temporal Control of DNA Repair. Stem Cells 2019, 37, 284–294. [Google Scholar] [CrossRef]
- Martin, R.M.; Ikeda, K.; Cromer, M.K.; Uchida, N.; Nishimura, T.; Romano, R.; Tong, A.J.; Lemgart, V.T.; Camarena, J.; Pavel-Dinu, M.; et al. Highly Efficient and Marker-Free Genome Editing of Human Pluripotent Stem Cells by CRISPR-Cas9 RNP and AAV6 Donor-Mediated Homologous Recombination. Cell Stem Cell 2019, 24, 821–828.e5. [Google Scholar] [CrossRef]
- Park, S.H.; Bao, G. CRISPR/Cas9 Gene Editing for Curing Sickle Cell Disease. Transfus. Apher. Sci. 2021, 60, 103060. [Google Scholar] [CrossRef] [PubMed]
- Wattanapanitch, M.; Damkham, N.; Potirat, P.; Trakarnsanga, K.; Janan, M.; U-pratya, Y.; Kheolamai, P.; Klincumhom, N.; Issaragrisil, S. One-Step Genetic Correction of Hemoglobin E/Beta-Thalassemia Patient-Derived IPSCs by the CRISPR/Cas9 System. Stem Cell Res. Ther. 2018, 9, 46. [Google Scholar] [CrossRef]
- Patsali, P.; Turchiano, G.; Papasavva, P.; Romito, M.; Loucari, C.C.; Stephanou, C.; Christou, S.; Sitarou, M.; Mussolino, C.; Cornu, T.I.; et al. Correction of IVS I-110(G>A) β-Thalassemia by CRISPR/Cas-and TALEN-Mediated Disruption of Aberrant Regulatory Elements in Human Hematopoietic Stem and Progenitor Cells. Haematologica 2019, 104, e497–e501. [Google Scholar] [CrossRef]
- Niu, X.; He, W.; Song, B.; Ou, Z.; Fan, D.; Chen, Y.; Fan, Y.; Sun, X. Combining Single Strand Oligodeoxynucleotides and CRISPR/Cas9 to Correct Gene Mutations in β-Thalassemia-Induced Pluripotent Stem Cells. J. Biol. Chem. 2016, 291, 16576–16585. [Google Scholar] [CrossRef] [PubMed]
- Gamage, U.; Warnakulasuriya, K.; Hansika, S.; Silva, G.N. CRISPR Gene Therapy: A Promising One-Time Therapeutic Approach for Transfusion-Dependent β-Thalassemia—CRISPR-Cas9 Gene Editing for β-Thalassemia. Thalass. Rep. 2023, 13, 51–69. [Google Scholar] [CrossRef]
- Monteys, A.M.; Ebanks, S.A.; Keiser, M.S.; Davidson, B.L. CRISPR/Cas9 Editing of the Mutant Huntingtin Allele in Vitro and in Vivo. Mol. Ther. 2017, 25, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Chang, R.; Yang, H.; Zhao, T.; Hong, Y.; Kong, H.E.; Sun, X.; Qin, Z.; Jin, P.; Li, S.; et al. CRISPR/Cas9-Mediated Gene Editing Ameliorates Neurotoxicity in Mouse Model of Huntington’s Disease. J. Clin. Investig. 2017, 127, 2719–2724. [Google Scholar] [CrossRef]
- Merienne, N.; Vachey, G.; de Longprez, L.; Meunier, C.; Zimmer, V.; Perriard, G.; Canales, M.; Mathias, A.; Herrgott, L.; Beltraminelli, T.; et al. The Self-Inactivating KamiCas9 System for the Editing of CNS Disease Genes. Cell Rep. 2017, 20, 2980–2991. [Google Scholar] [CrossRef]
- Ekman, F.K.; Ojala, D.S.; Adil, M.M.; Lopez, P.A.; Schaffer, D.V.; Gaj, T. CRISPR-Cas9-Mediated Genome Editing Increases Lifespan and Improves Motor Deficits in a Huntington’s Disease Mouse Model. Mol. Ther. Nucleic Acids 2019, 17, 829–839. [Google Scholar] [CrossRef]
- Alkanli, S.S.; Alkanli, N.; Ay, A.; Albeniz, I. CRISPR/Cas9 Mediated Therapeutic Approach in Huntington’s Disease. Mol. Neurobiol. 2023, 60, 1486–1498. [Google Scholar] [CrossRef]
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Rivera, R.M.C.; Madhavan, S.; Pan, X.; Ran, F.A.; Yan, W.X.; et al. In Vivo Genome Editing Improves Muscle Function in a Mouse Model of Duchenne Muscular Dystrophy. Science 2016, 351, 403–407. [Google Scholar] [CrossRef]
- Nelson, C.E.; Wu, Y.; Gemberling, M.P.; Oliver, M.L.; Waller, M.A.; Bohning, J.D.; Robinson-Hamm, J.N.; Bulaklak, K.; Castellanos Rivera, R.M.; Collier, J.H.; et al. Long-Term Evaluation of AAV-CRISPR Genome Editing for Duchenne Muscular Dystrophy. Nat. Med. 2019, 25, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Min, Y.-L.; Li, H.; Rodriguez-Caycedo, C.; Mireault, A.A.; Huang, J.; Shelton, J.M.; McAnally, J.R.; Amoasii, L.; Mammen, P.P.A.; Bassel-Duby, R.; et al. CRISPR-Cas9 Corrects Duchenne Muscular Dystrophy Exon 44 Deletion Mutations in Mice and Human Cells. Sci. Adv. 2019, 5, eaav4324. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, N.E.; Hall, J.K.; Odom, G.L.; Phelps, M.P.; Andrus, C.R.; Hawkins, R.D.; Hauschka, S.D.; Chamberlain, J.R.; Chamberlain, J.S. Muscle-Specific CRISPR/Cas9 Dystrophin Gene Editing Ameliorates Pathophysiology in a Mouse Model for Duchenne Muscular Dystrophy. Nat. Commun. 2017, 8, 14454. [Google Scholar] [CrossRef] [PubMed]
- Amoasii, L.; Hildyard, J.C.W.; Li, H.; Sanchez-Ortiz, E.; Mireault, A.; Caballero, D.; Harron, R.; Stathopoulou, T.-R.; Massey, C.; Shelton, J.M.; et al. Gene Editing Restores Dystrophin Expression in a Canine Model of Duchenne Muscular Dystrophy. Science 2018, 362, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Long, C.; Li, H.; McAnally, J.R.; Baskin, K.K.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. CRISPR-Cpf1 Correction of Muscular Dystrophy Mutations in Human Cardiomyocytes and Mice. Sci. Adv. 2017, 3, e1602814. [Google Scholar] [CrossRef]
- Agrawal, P.; Harish, V.; Mohd, S.; Singh, S.K.; Tewari, D.; Tatiparthi, R.; Harshita; Vishwas, S.; Sutrapu, S.; Dua, K.; et al. Role of CRISPR/Cas9 in the Treatment of Duchenne Muscular Dystrophy and Its Delivery Strategies. Life Sci. 2023, 330, 122003. [Google Scholar] [CrossRef]
- Stephens, C.J.; Lauron, E.J.; Kashentseva, E.; Lu, Z.H.; Yokoyama, W.M.; Curiel, D.T. Long-Term Correction of Hemophilia B Using Adenoviral Delivery of CRISPR/Cas9. J. Control. Release 2019, 298, 128–141. [Google Scholar] [CrossRef]
- Hu, Z.; Zhou, M.; Wu, Y.; Li, Z.; Liu, X.; Wu, L.; Liang, D. SsODN-Mediated in-Frame Deletion with CRISPR/Cas9 Restores FVIII Function in Hemophilia A-Patient-Derived IPSCs and ECs. Mol. Ther. Nucleic Acids 2019, 17, 198–209. [Google Scholar] [CrossRef]
- Lyu, C.; Shen, J.; Wang, R.; Gu, H.; Zhang, J.; Xue, F.; Liu, X.; Liu, W.; Fu, R.; Zhang, L.; et al. Targeted Genome Engineering in Human Induced Pluripotent Stem Cells from Patients with Hemophilia B Using the CRISPR-Cas9 System. Stem Cell Res. Ther. 2018, 9, 92. [Google Scholar] [CrossRef]
- Chen, H.; Shi, M.; Gilam, A.; Zheng, Q.; Zhang, Y.; Afrikanova, I.; Li, J.; Gluzman, Z.; Jiang, R.; Kong, L.-J.; et al. Hemophilia A Ameliorated in Mice by CRISPR-Based in Vivo Genome Editing of Human Factor VIII. Sci. Rep. 2019, 9, 16838. [Google Scholar] [CrossRef] [PubMed]
- Hiramoto, T.; Kashiwakura, Y.; Hayakawa, M.; Baatartsogt, N.; Kamoshita, N.; Abe, T.; Inaba, H.; Nishimasu, H.; Uosaki, H.; Hanazono, Y.; et al. PAM-Flexible Cas9-Mediated Base Editing of a Hemophilia B Mutation in Induced Pluripotent Stem Cells. Commun. Med. 2023, 3, 56. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.Y.; Ain, Q.U.; Song, Y.; Yong, S.-B.; Kim, Y.-H. Targeted Delivery of CRISPR Interference System against Fabp4 to White Adipocytes Ameliorates Obesity, Inflammation, Hepatic Steatosis, and Insulin Resistance. Genome Res. 2019, 29, 1442–1452. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, K.G.; Augsornworawat, P.; Velazco-Cruz, L.; Kim, M.H.; Asada, R.; Hogrebe, N.J.; Morikawa, S.; Urano, F.; Millman, J.R. Gene-Edited Human Stem Cell–Derived β Cells from a Patient with Monogenic Diabetes Reverse Preexisting Diabetes in Mice. Sci. Transl. Med. 2020, 12, eaax9106. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.Y.; Ryu, J.-Y.; Lee, H.A.R.; Hong, S.H.; Park, H.S.; Hong, K.S.; Park, S.-G.; Kim, H.P.; Yoon, T.-J. Lecithin Nano-Liposomal Particle as a CRISPR/Cas9 Complex Delivery System for Treating Type 2 Diabetes. J. Nanobiotechnol. 2019, 17, 19. [Google Scholar] [CrossRef]
- Grotz, A.K.; Abaitua, F.; Navarro-Guerrero, E.; Hastoy, B.; Ebner, D.; Gloyn, A.L. A CRISPR/Cas9 Genome Editing Pipeline in the EndoC-ΒH1 Cell Line to Study Genes Implicated in Beta Cell Function. Wellcome Open Res. 2020, 4, 150. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, H.; Li, M. The Promise of CRISPR/Cas9 Technology in Diabetes Mellitus Therapy: How Gene Editing Is Revolutionizing Diabetes Research and Treatment. J. Diabetes Complicat. 2023, 37, 108524. [Google Scholar] [CrossRef]
- Olivaes, J.; Bonamino, M.H.; Markoski, M.M. CRISPR/Cas 9 System for the Treatment of Dilated Cardiomyopathy: A Hypothesis Related to Function of a MAP Kinase. Med. Hypotheses 2019, 128, 91–93. [Google Scholar] [CrossRef]
- Zhao, H.; Li, Y.; He, L.; Pu, W.; Yu, W.; Li, Y.; Wu, Y.-T.; Xu, C.; Wei, Y.; Ding, Q.; et al. In Vivo AAV-CRISPR/Cas9–Mediated Gene Editing Ameliorates Atherosclerosis in Familial Hypercholesterolemia. Circulation 2020, 141, 67–79. [Google Scholar] [CrossRef]
- Caron, J.; Pène, V.; Tolosa, L.; Villaret, M.; Luce, E.; Fourrier, A.; Heslan, J.-M.; Saheb, S.; Bruckert, E.; Gómez-Lechón, M.J.; et al. Low-Density Lipoprotein Receptor-Deficient Hepatocytes Differentiated from Induced Pluripotent Stem Cells Allow Familial Hypercholesterolemia Modeling, CRISPR/Cas-Mediated Genetic Correction, and Productive Hepatitis C Virus Infection. Stem Cell Res. Ther. 2019, 10, 221. [Google Scholar] [CrossRef]
- Wang, X.; Raghavan, A.; Chen, T.; Qiao, L.; Zhang, Y.; Ding, Q.; Musunuru, K. CRISPR-Cas9 Targeting of PCSK9 in Human Hepatocytes in Vivo—Brief Report. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 783–786. [Google Scholar] [CrossRef] [PubMed]
- Musunuru, K. CRISPR and Cardiovascular Diseases. Cardiovasc. Res. 2023, 119, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Y.; Cheng, C.; Cheng, A.W.; Zhang, X.; Li, N.; Xia, C.; Wei, X.; Liu, X.; Wang, H. CRISPR-Cas9-Mediated Multiplex Gene Editing in CAR-T Cells. Cell Res. 2017, 27, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.D.; Yu, X.; Castano, A.P.; Darr, H.; Henderson, D.B.; Bouffard, A.A.; Larson, R.C.; Scarfò, I.; Bailey, S.R.; Gerhard, G.M.; et al. CRISPR-Cas9 Disruption of PD-1 Enhances Activity of Universal EGFRvIII CAR T Cells in a Preclinical Model of Human Glioblastoma. J. Immunother. Cancer 2019, 7, 304. [Google Scholar] [CrossRef]
- McGowan, E.; Lin, Q.; Ma, G.; Yin, H.; Chen, S.; Lin, Y. PD-1 Disrupted CAR-T Cells in the Treatment of Solid Tumors: Promises and Challenges. Biomed. Pharmacother. 2020, 121, 109625. [Google Scholar] [CrossRef]
- Guo, X.; Jiang, H.; Shi, B.; Zhou, M.; Zhang, H.; Shi, Z.; Du, G.; Luo, H.; Wu, X.; Wang, Y.; et al. Disruption of PD-1 Enhanced the Anti-Tumor Activity of Chimeric Antigen Receptor T Cells against Hepatocellular Carcinoma. Front. Pharmacol. 2018, 9, 1118. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, G.; Zhang, L.; Zhao, Q. Building Potent Chimeric Antigen Receptor T Cells with CRISPR Genome Editing. Front. Immunol. 2019, 10, 456. [Google Scholar] [CrossRef]
- Nakazawa, T.; Natsume, A.; Nishimura, F.; Morimoto, T.; Matsuda, R.; Nakamura, M.; Yamada, S.; Nakagawa, I.; Motoyama, Y.; Park, Y.-S.; et al. Effect of CRISPR/Cas9-Mediated PD-1-Disrupted Primary Human Third-Generation CAR-T Cells Targeting EGFRvIII on in Vitro Human Glioblastoma Cell Growth. Cells 2020, 9, 998. [Google Scholar] [CrossRef]
- Ren, J.; Zhang, X.; Liu, X.; Fang, C.; Jiang, S.; June, C.H.; Zhao, Y. A Versatile System for Rapid Multiplex Genome-Edited CAR T Cell Generation. Oncotarget 2017, 8, 17002–17011. [Google Scholar] [CrossRef]
- Ren, J.; Liu, X.; Fang, C.; Jiang, S.; June, C.H.; Zhao, Y. Multiplex Genome Editing to Generate Universal CAR T Cells Resistant to PD1 Inhibition. Clin. Cancer Res. 2017, 23, 2255–2266. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Cheng, C.; Mu, W.; Liu, X.; Li, N.; Wei, X.; Liu, X.; Xia, C.; Wang, H. CRISPR-Cas9 Mediated LAG-3 Disruption in CAR-T Cells. Front. Med. 2017, 11, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Blaeschke, F.; Willier, S.; Stenger, D.; Lepenies, M.; Horstmann, M.A.; Escherich, G.; Zimmermann, M.; Rojas Ringeling, F.; Canzar, S.; Kaeuferle, T.; et al. Leukemia-Induced Dysfunctional TIM-3+CD4+ Bone Marrow T Cells Increase Risk of Relapse in Pediatric B-Precursor ALL Patients. Leukemia 2020, 34, 2607–2620. [Google Scholar] [CrossRef] [PubMed]
- Stadtmauer, E.A.; Fraietta, J.A.; Davis, M.M.; Cohen, A.D.; Weber, K.L.; Lancaster, E.; Mangan, P.A.; Kulikovskaya, I.; Gupta, M.; Chen, F.; et al. CRISPR-Engineered T Cells in Patients with Refractory Cancer. Science 2020, 367, eaba7365. [Google Scholar] [CrossRef]
- Wei, W.; Chen, Z.-N.; Wang, K. CRISPR/Cas9: A Powerful Strategy to Improve CAR-T Cell Persistence. Int. J. Mol. Sci. 2023, 24, 12317. [Google Scholar] [CrossRef] [PubMed]
- Dimitri, A.; Herbst, F.; Fraietta, J.A. Engineering the Next-Generation of CAR T-Cells with CRISPR-Cas9 Gene Editing. Mol. Cancer 2022, 21, 78. [Google Scholar] [CrossRef]
- Eyquem, J.; Mansilla-Soto, J.; Giavridis, T.; van der Stegen, S.J.C.; Hamieh, M.; Cunanan, K.M.; Odak, A.; Gönen, M.; Sadelain, M. Targeting a CAR to the TRAC Locus with CRISPR/Cas9 Enhances Tumour Rejection. Nature 2017, 543, 113–117. [Google Scholar] [CrossRef]
- MacLeod, D.T.; Antony, J.; Martin, A.J.; Moser, R.J.; Hekele, A.; Wetzel, K.J.; Brown, A.E.; Triggiano, M.A.; Hux, J.A.; Pham, C.D.; et al. Integration of a CD19 CAR into the TCR Alpha Chain Locus Streamlines Production of Allogeneic Gene-Edited CAR T Cells. Mol. Ther. 2017, 25, 949–961. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, Y. CRISPR/Cas9 Genome Editing: Fueling the Revolution in Cancer Immunotherapy. Curr. Res. Transl. Med. 2018, 66, 39–42. [Google Scholar] [CrossRef]
- Roth, T.L.; Puig-Saus, C.; Yu, R.; Shifrut, E.; Carnevale, J.; Li, P.J.; Hiatt, J.; Saco, J.; Krystofinski, P.; Li, H.; et al. Reprogramming Human T Cell Function and Specificity with Non-Viral Genome Targeting. Nature 2018, 559, 405–409. [Google Scholar] [CrossRef]
- Mehta, A.; Merkel, O.M. Immunogenicity of Cas9 Protein. J. Pharm. Sci. 2020, 109, 62–67. [Google Scholar] [CrossRef]
- Simhadri, V.L.; McGill, J.; McMahon, S.; Wang, J.; Jiang, H.; Sauna, Z.E. Prevalence of Pre-Existing Antibodies to CRISPR-Associated Nuclease Cas9 in the USA Population. Mol. Ther. Methods Clin. Dev. 2018, 10, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, C.T.; Deshpande, P.S.; Dever, D.P.; Camarena, J.; Lemgart, V.T.; Cromer, M.K.; Vakulskas, C.A.; Collingwood, M.A.; Zhang, L.; Bode, N.M.; et al. Identification of Preexisting Adaptive Immunity to Cas9 Proteins in Humans. Nat. Med. 2019, 25, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.-Z.E.; Tan, S.X.; Hoon, S.; Yeo, G.W. Pre-Existing Adaptive Immunity to the RNA-Editing Enzyme Cas13d in Humans. Nat. Med. 2022, 28, 1372–1376. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Lin, Q.; Liang, Z.; Wang, J.; Yang, X.; Liang, Y.; Liang, H.; Pan, H.; Yang, J.; Zhu, Y.; et al. Reduction of Pre-Existing Adaptive Immune Responses against SaCas9 in Humans Using Epitope Mapping and Identification. CRISPR J. 2022, 5, 445–456. [Google Scholar] [CrossRef]
- Toral, M.A.; Charlesworth, C.T.; Ng, B.; Chemudupati, T.; Homma, S.; Nakauchi, H.; Bassuk, A.G.; Porteus, M.H.; Mahajan, V.B. Investigation of Cas9 Antibodies in the Human Eye. Nat. Commun. 2022, 13, 1053. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.L.; Amini, L.; Wendering, D.J.; Burkhardt, L.-M.; Akyüz, L.; Reinke, P.; Volk, H.-D.; Schmueck-Henneresse, M. High Prevalence of Streptococcus Pyogenes Cas9-Reactive T Cells within the Adult Human Population. Nat. Med. 2019, 25, 242–248. [Google Scholar] [CrossRef]
- Liu, J.-J.; Orlova, N.; Oakes, B.L.; Ma, E.; Spinner, H.B.; Baney, K.L.M.; Chuck, J.; Tan, D.; Knott, G.J.; Harrington, L.B.; et al. CasX Enzymes Comprise a Distinct Family of RNA-Guided Genome Editors. Nature 2019, 566, 218–223. [Google Scholar] [CrossRef]
- Verdera, H.C.; Kuranda, K.; Mingozzi, F. AAV Vector Immunogenicity in Humans: A Long Journey to Successful Gene Transfer. Mol. Ther. 2020, 28, 723–746. [Google Scholar] [CrossRef]
- Mingozzi, F.; High, K.A. Overcoming the Host Immune Response to Adeno-Associated Virus Gene Delivery Vectors: The Race between Clearance, Tolerance, Neutralization, and Escape. Annu. Rev. Virol. 2017, 4, 511–534. [Google Scholar] [CrossRef]
- Mingozzi, F.; High, K.A. Immune Responses to AAV Vectors: Overcoming Barriers to Successful Gene Therapy. Blood 2013, 122, 23–36. [Google Scholar] [CrossRef]
- Samelson-Jones, B.J.; Finn, J.D.; Favaro, P.; Wright, J.F.; Arruda, V.R. Timing of Intensive Immunosuppression Impacts Risk of Transgene Antibodies after AAV Gene Therapy in Nonhuman Primates. Mol. Ther. Methods Clin. Dev. 2020, 17, 1129–1138. [Google Scholar] [CrossRef]
- Velazquez, V.M.; Meadows, A.S.; Pineda, R.J.; Camboni, M.; McCarty, D.M.; Fu, H. Effective Depletion of Pre-Existing Anti-AAV Antibodies Requires Broad Immune Targeting. Mol. Ther. Methods Clin. Dev. 2017, 4, 159–168. [Google Scholar] [CrossRef]
- Essawi, K.; Hakami, W.; Naeem Khan, M.B.; Martin, R.; Zeng, J.; Chu, R.; Uchida, N.; Bonifacino, A.C.; Krouse, A.E.; Linde, N.S.; et al. Pre-Existing Immunity Does Not Impair the Engraftment of CRISPR-Cas9-Edited Cells in Rhesus Macaques Conditioned with Busulfan or Radiation. Mol. Ther. Methods Clin. Dev. 2023, 29, 483–493. [Google Scholar] [CrossRef]
- Dong, C.; Hao, G.-F.; Hua, H.-L.; Liu, S.; Labena, A.A.; Chai, G.; Huang, J.; Rao, N.; Guo, F.-B. Anti-CRISPRdb: A Comprehensive Online Resource for Anti-CRISPR Proteins. Nucleic Acids Res. 2018, 46, D393–D398. [Google Scholar] [CrossRef]
- Dong, C.; Wang, X.; Ma, C.; Zeng, Z.; Pu, D.-K.; Liu, S.; Wu, C.-S.; Chen, S.; Deng, Z.; Guo, F.-B. Anti-CRISPRdb v2.2: An Online Repository of Anti-CRISPR Proteins Including Information on Inhibitory Mechanisms, Activities and Neighbors of Curated Anti-CRISPR Proteins. Database 2022, 2022, baac010. [Google Scholar] [CrossRef] [PubMed]
- Hynes, A.P.; Rousseau, G.M.; Lemay, M.-L.; Horvath, P.; Romero, D.A.; Fremaux, C.; Moineau, S. An Anti-CRISPR from a Virulent Streptococcal Phage Inhibits Streptococcus Pyogenes Cas9. Nat. Microbiol. 2017, 2, 1374–1380. [Google Scholar] [CrossRef]
- Lee, J.; Mir, A.; Edraki, A.; Garcia, B.; Amrani, N.; Lou, H.E.; Gainetdinov, I.; Pawluk, A.; Ibraheim, R.; Gao, X.D.; et al. Potent Cas9 Inhibition in Bacterial and Human Cells by AcrIIC4 and AcrIIC5 Anti-CRISPR Proteins. mBio 2018, 9, e02321-18. [Google Scholar] [CrossRef] [PubMed]
- Uribe, R.V.; van der Helm, E.; Misiakou, M.-A.; Lee, S.-W.; Kol, S.; Sommer, M.O.A. Discovery and Characterization of Cas9 Inhibitors Disseminated across Seven Bacterial Phyla. Cell Host Microbe 2019, 25, 233–241.e5. [Google Scholar] [CrossRef]
- Hynes, A.P.; Rousseau, G.M.; Agudelo, D.; Goulet, A.; Amigues, B.; Loehr, J.; Romero, D.A.; Fremaux, C.; Horvath, P.; Doyon, Y.; et al. Widespread Anti-CRISPR Proteins in Virulent Bacteriophages Inhibit a Range of Cas9 Proteins. Nat. Commun. 2018, 9, 2919. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, K.J.; Bhatt, I.V.; Schmidtke, D.T.; Javanmardi, K.; Dillard, K.E.; Stoddard, B.L.; Finkelstein, I.J.; Kaiser, B.K.; Malik, H.S. Functional Metagenomics-Guided Discovery of Potent Cas9 Inhibitors in the Human Microbiome. eLife 2019, 8, e46540. [Google Scholar] [CrossRef] [PubMed]
- Watters, K.E.; Shivram, H.; Fellmann, C.; Lew, R.J.; McMahon, B.; Doudna, J.A. Potent CRISPR-Cas9 Inhibitors from Staphylococcus Genomes. Proc. Natl. Acad. Sci. USA 2020, 117, 6531–6539. [Google Scholar] [CrossRef] [PubMed]
- Mahendra, C.; Christie, K.A.; Osuna, B.A.; Pinilla-Redondo, R.; Kleinstiver, B.P.; Bondy-Denomy, J. Broad-Spectrum Anti-CRISPR Proteins Facilitate Horizontal Gene Transfer. Nat. Microbiol. 2020, 5, 620–629. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, K.J.; Schmidtke, D.T.; Werther, R.; Uribe, R.V.; Hausman, D.; Sommer, M.O.A.; Stoddard, B.L.; Kaiser, B.K.; Malik, H.S. The Novel Anti-CRISPR AcrIIA22 Relieves DNA Torsion in Target Plasmids and Impairs SpyCas9 Activity. PLoS Biol. 2021, 19, e3001428. [Google Scholar] [CrossRef]
- Song, G.; Zhang, F.; Tian, C.; Gao, X.; Zhu, X.; Fan, D.; Tian, Y. Discovery of Potent and Versatile CRISPR–Cas9 Inhibitors Engineered for Chemically Controllable Genome Editing. Nucleic Acids Res. 2022, 50, 2836–2853. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Mou, H.; Ibraheim, R.; Liang, S.-Q.; Liu, P.; Xue, W.; Sontheimer, E.J. Tissue-Restricted Genome Editing in Vivo Specified by MicroRNA-Repressible Anti-CRISPR Proteins. RNA 2019, 25, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Sui, T.; Liu, Z.; Chen, M.; Liu, H.; Shan, H.; Lai, L.; Li, Z. AcrIIA5 Suppresses Base Editors and Reduces Their Off-Target Effects. Cells 2020, 9, 1786. [Google Scholar] [CrossRef]
- Jia, N.; Patel, D.J. Structure-Based Functional Mechanisms and Biotechnology Applications of Anti-CRISPR Proteins. Nat. Rev. Mol. Cell Biol. 2021, 22, 563–579. [Google Scholar] [CrossRef]
- Aschenbrenner, S.; Kallenberger, S.M.; Hoffmann, M.D.; Huck, A.; Eils, R.; Niopek, D. Coupling Cas9 to Artificial Inhibitory Domains Enhances CRISPR-Cas9 Target Specificity. Sci. Adv. 2020, 6, eaay0187. [Google Scholar] [CrossRef]
- Chen, S.; Chen, D.; Liu, B.; Haisma, H.J. Modulating CRISPR/Cas9 Genome-Editing Activity by Small Molecules. Drug Discov. Today 2022, 27, 951–966. [Google Scholar] [CrossRef]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.-Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In Vivo Genome Editing with a Small Cas9 Orthologue Derived from Campylobacter Jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef]
- Hou, Z.; Zhang, Y.; Propson, N.E.; Howden, S.E.; Chu, L.-F.; Sontheimer, E.J.; Thomson, J.A. Efficient Genome Engineering in Human Pluripotent Stem Cells Using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 2013, 110, 15644–15649. [Google Scholar] [CrossRef] [PubMed]
- Hirano, H.; Gootenberg, J.S.; Horii, T.; Abudayyeh, O.O.; Kimura, M.; Hsu, P.D.; Nakane, T.; Ishitani, R.; Hatada, I.; Zhang, F.; et al. Structure and Engineering of Francisella Novicida Cas9. Cell 2016, 164, 950–961. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.; Lee, J.; Nip, L.; Koseki, S.R.T.; Tysinger, E.; Sontheimer, E.J.; Jacobson, J.M.; Jakimo, N. A Cas9 with PAM Recognition for Adenine Dinucleotides. Nat. Commun. 2020, 11, 2474. [Google Scholar] [CrossRef]
- Hu, Z.; Wang, S.; Zhang, C.; Gao, N.; Li, M.; Wang, D.; Wang, D.; Liu, D.; Liu, H.; Ong, S.-G.; et al. A Compact Cas9 Ortholog from Staphylococcus Auricularis (SauriCas9) Expands the DNA Targeting Scope. PLoS Biol. 2020, 18, e3000686. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Lee, C.M.; Gasiunas, G.; Davis, T.H.; Cradick, T.J.; Siksnys, V.; Bao, G.; Cathomen, T.; Mussolino, C. Streptococcus Thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol. Ther. 2016, 24, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Glemzaite, M.; Balciunaite, E.; Karvelis, T.; Gasiunas, G.; Grusyte, M.M.; Alzbutas, G.; Jurcyte, A.; Anderson, E.M.; Maksimova, E.; Smith, A.J.; et al. Targeted Gene Editing by Transfection of in vitro Reconstituted Streptococcus thermophilus Cas9 Nuclease Complex. RNA Biol. 2015, 12, 1–4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, L.; Huang, Y.; Xu, D.; Yang, L.; Qian, K.; Chang, G.; Gong, Y.; Zhou, X.; Ma, K. Biochemical Characterization of a Thermostable HNH Endonuclease from Deep-Sea Thermophilic Bacteriophage GVE2. Appl. Microbiol. Biotechnol. 2016, 100, 8003–8012. [Google Scholar] [CrossRef]
- Harrington, L.B.; Doxzen, K.W.; Ma, E.; Liu, J.-J.; Knott, G.J.; Edraki, A.; Garcia, B.; Amrani, N.; Chen, J.S.; Cofsky, J.C.; et al. A Broad-Spectrum Inhibitor of CRISPR-Cas9. Cell 2017, 170, 1224–1233.e15. [Google Scholar] [CrossRef]
- Bai, Y.; Müller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional Overlap of the Arabidopsis Leaf and Root Microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef]
- Sun, Z.; Harris, H.M.B.; McCann, A.; Guo, C.; Argimón, S.; Zhang, W.; Yang, X.; Jeffery, I.B.; Cooney, J.C.; Kagawa, T.F.; et al. Expanding the Biotechnology Potential of Lactobacilli through Comparative Genomics of 213 Strains and Associated Genera. Nat. Commun. 2015, 6, 8322. [Google Scholar] [CrossRef]
- Green, N.M.; Zhang, S.; Porcella, S.F.; Nagiec, M.J.; Barbian, K.D.; Beres, S.B.; LeFebvre, R.B.; Musser, J.M. Genome Sequence of a Serotype M28 Strain of Group A Streptococcus: Potential New Insights into Puerperal Sepsis and Bacterial Disease Specificity. J. Infect. Dis. 2005, 192, 760–770. [Google Scholar] [CrossRef] [PubMed]
- Probst, A.J.; Castelle, C.J.; Singh, A.; Brown, C.T.; Anantharaman, K.; Sharon, I.; Hug, L.A.; Burstein, D.; Emerson, J.B.; Thomas, B.C.; et al. Genomic Resolution of a Cold Subsurface Aquifer Community Provides Metabolic Insights for Novel Microbes Adapted to High CO2 Concentrations. Environ. Microbiol. 2017, 19, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Bonasio, R.; Zhang, G.; Ye, C.; Mutti, N.S.; Fang, X.; Qin, N.; Donahue, G.; Yang, P.; Li, Q.; Li, C.; et al. Genomic Comparison of the Ants Camponotus floridanus and Harpegnathos saltator. Science 2010, 329, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Singleton, D.R.; Dickey, A.N.; Scholl, E.H.; Wright, F.A.; Aitken, M.D. Complete Genome Sequence of a Novel Bacterium within the Family Rhodocyclaceae That Degrades Polycyclic Aromatic Hydrocarbons. Genome Announc. 2015, 3, e00251-15. [Google Scholar] [CrossRef] [PubMed]
- Pridgeon, J.W.; Zhang, D.; Zhang, L. Complete Genome Sequence of the Attenuated Novobiocin-Resistant Streptococcus Iniae Vaccine Strain ISNO. Genome Announc. 2014, 2, e00510-14. [Google Scholar] [CrossRef] [PubMed]
- Pridgeon, J.W.; Zhang, D.; Zhang, L. Complete Genome Sequence of a Virulent Strain, Streptococcus Iniae ISET0901, Isolated from Diseased Tilapia. Genome Announc. 2014, 2, e00553-14. [Google Scholar] [CrossRef]
- Tan, B.; Foght, J. Draft Genome Sequences of Campylobacterales (Epsilonproteobacteria) Obtained from Methanogenic Oil Sands Tailings Pond Metagenomes. Genome Announc. 2014, 2, e01034-14. [Google Scholar] [CrossRef]
- Gundogdu, O.; Bentley, S.D.; Holden, M.T.; Parkhill, J.; Dorrell, N.; Wren, B.W. Re-Annotation and Re-Analysis of the Campylobacter Jejuni NCTC11168 Genome Sequence. BMC Genom. 2007, 8, 162. [Google Scholar] [CrossRef]
- Haft, D.H.; Selengut, J.; Mongodin, E.F.; Nelson, K.E. A Guild of 45 CRISPR-Associated (Cas) Protein Families and Multiple CRISPR/Cas Subtypes Exist in Prokaryotic Genomes. PLoS Comput. Biol. 2005, 1, e60. [Google Scholar] [CrossRef]
- Zhang, B.-C.; Zhang, J.; Sun, L. Streptococcus Iniae SF1: Complete Genome Sequence, Proteomic Profile, and Immunoprotective Antigens. PLoS ONE 2014, 9, e91324. [Google Scholar] [CrossRef]
- Rosini, R.; Campisi, E.; De Chiara, M.; Tettelin, H.; Rinaudo, D.; Toniolo, C.; Metruccio, M.; Guidotti, S.; Sørensen, U.B.S.; Kilian, M.; et al. Genomic Analysis Reveals the Molecular Basis for Capsule Loss in the Group B Streptococcus Population. PLoS ONE 2015, 10, e0125985. [Google Scholar] [CrossRef] [PubMed]
- Gasiunas, G.; Young, J.K.; Karvelis, T.; Kazlauskas, D.; Urbaitis, T.; Jasnauskaite, M.; Grusyte, M.; Paulraj, S.; Wang, P.-H.; Hou, Z.; et al. Biochemically Diverse CRISPR-Cas9 Orthologs. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wang, S.; Tao, C.; Mao, H.; Hou, L.; Wang, Y.; Qi, T.; Yang, Y.; Ong, S.-G.; Hu, S.; Chai, R.; et al. Identification of SaCas9 Orthologs Containing a Conserved Serine Residue That Determines Simple NNGG PAM Recognition. PLoS Biol. 2022, 20, e3001897. [Google Scholar] [CrossRef]
- Wei, J.; Hou, L.; Liu, J.; Wang, Z.; Gao, S.; Qi, T.; Gao, S.; Sun, S.; Wang, Y. Closely Related Type II-C Cas9 Orthologs Recognize Diverse PAMs. eLife 2022, 11, e77825. [Google Scholar] [CrossRef]
- Chen, F. CRISPR/Cas Fusion Proteins and Systems. U.S. Patent 10947517B2, 16 March 2021. [Google Scholar]
- Chatterjee, P.; Jakimo, N.; Lee, J.; Amrani, N.; Rodríguez, T.; Koseki, S.R.T.; Tysinger, E.; Qing, R.; Hao, S.; Sontheimer, E.J.; et al. An Engineered ScCas9 with Broad PAM Range and High Specificity and Activity. Nat. Biotechnol. 2020, 38, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and Evolution of Class 2 CRISPR–Cas Systems. Nat. Rev. Microbiol. 2017, 15, 169–182. [Google Scholar] [CrossRef]
- Yamano, T.; Nishimasu, H.; Zetsche, B.; Hirano, H.; Slaymaker, I.M.; Li, Y.; Fedorova, I.; Nakane, T.; Makarova, K.S.; Koonin, E.V.; et al. Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA. Cell 2016, 165, 949–962. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Kim, D.; Kim, J.; Hur, J.K.; Been, K.W.; Yoon, S.-H.; Kim, J.-S. Genome-Wide Analysis Reveals Specificities of Cpf1 Endonucleases in Human Cells. Nat. Biotechnol. 2016, 34, 863–868. [Google Scholar] [CrossRef]
- Ma, S.; Liu, Y.; Liu, Y.; Chang, J.; Zhang, T.; Wang, X.; Shi, R.; Lu, W.; Xia, X.; Zhao, P.; et al. An Integrated CRISPR Bombyx Mori Genome Editing System with Improved Efficiency and Expanded Target Sites. Insect Biochem. Mol. Biol. 2017, 83, 13–20. [Google Scholar] [CrossRef]
- Endo, A.; Masafumi, M.; Kaya, H.; Toki, S. Efficient Targeted Mutagenesis of Rice and Tobacco Genomes Using Cpf1 from Francisella Novicida. Sci. Rep. 2016, 6, 38169. [Google Scholar] [CrossRef]
- Zetsche, B.; Abudayyeh, O.O.; Gootenberg, J.S.; Scott, D.A.; Zhang, F. A Survey of Genome Editing Activity for 16 Cas12a Orthologs. Keio J. Med. 2020, 69, 59–65. [Google Scholar] [CrossRef]
- Teng, F.; Li, J.; Cui, T.; Xu, K.; Guo, L.; Gao, Q.; Feng, G.; Chen, C.; Han, D.; Zhou, Q.; et al. Enhanced Mammalian Genome Editing by New Cas12a Orthologs with Optimized CrRNA Scaffolds. Genome Biol. 2019, 20, 15. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, T.; Ttofali, F.; Liao, C.; Manchalu, S.; Gray, B.N.; Beisel, C.L. Characterization of Cas12a Nucleases Reveals Diverse PAM Profiles between Closely-Related Orthologs. Nucleic Acids Res. 2020, 48, 5624–5638. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zuris, J.A.; Viswanathan, R.; Edelstein, J.N.; Turk, R.; Thommandru, B.; Rube, H.T.; Glenn, S.E.; Collingwood, M.A.; Bode, N.M.; et al. AsCas12a Ultra Nuclease Facilitates the Rapid Generation of Therapeutic Cell Medicines. Nat. Commun. 2021, 12, 3908. [Google Scholar] [CrossRef]
- Jiang, W.; Samai, P.; Marraffini, L.A. Degradation of Phage Transcripts by CRISPR-Associated RNases Enables Type III CRISPR-Cas Immunity. Cell 2016, 164, 710–721. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.T.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 Is a Single-Component Programmable RNA-Guided RNA-Targeting CRISPR Effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA Editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Tambe, A.; East-Seletsky, A.; Knott, G.J.; Doudna, J.A.; O’Connell, M.R. RNA Binding and HEPN-Nuclease Activation Are Decoupled in CRISPR-Cas13a. Cell Rep. 2018, 24, 1025–1036. [Google Scholar] [CrossRef]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173, 665–676.e14. [Google Scholar] [CrossRef]
- Yan, W.X.; Chong, S.; Zhang, H.; Makarova, K.S.; Koonin, E.V.; Cheng, D.R.; Scott, D.A. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol. Cell 2018, 70, 327–339.e5. [Google Scholar] [CrossRef]
- Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA Destruction by Miniature CRISPR-Cas14 Enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef] [PubMed]
- Karvelis, T.; Bigelyte, G.; Young, J.K.; Hou, Z.; Zedaveinyte, R.; Budre, K.; Paulraj, S.; Djukanovic, V.; Gasior, S.; Silanskas, A.; et al. PAM Recognition by Miniature CRISPR–Cas12f Nucleases Triggers Programmable Double-Stranded DNA Target Cleavage. Nucleic Acids Res. 2020, 48, 5016–5023. [Google Scholar] [CrossRef] [PubMed]
- Aquino-Jarquin, G. CRISPR-Cas14 Is Now Part of the Artillery for Gene Editing and Molecular Diagnostic. Nanomedicine 2019, 18, 428–431. [Google Scholar] [CrossRef] [PubMed]
- Altae-Tran, H.; Kannan, S.; Demircioglu, F.E.; Oshiro, R.; Nety, S.P.; McKay, L.J.; Dlakić, M.; Inskeep, W.P.; Makarova, K.S.; Macrae, R.K.; et al. The Widespread IS200/IS605 Transposon Family Encodes Diverse Programmable RNA-Guided Endonucleases. Science 2021, 374, 57–65. [Google Scholar] [CrossRef]
- Karvelis, T.; Druteika, G.; Bigelyte, G.; Budre, K.; Zedaveinyte, R.; Silanskas, A.; Kazlauskas, D.; Venclovas, Č.; Siksnys, V. Transposon-Associated TnpB Is a Programmable RNA-Guided DNA Endonuclease. Nature 2021, 599, 692–696. [Google Scholar] [CrossRef]
- Saito, M.; Xu, P.; Faure, G.; Maguire, S.; Kannan, S.; Altae-Tran, H.; Vo, S.; Desimone, A.; Macrae, R.K.; Zhang, F. Fanzor Is a Eukaryotic Programmable RNA-Guided Endonuclease. Nature 2023, 620, 660–668. [Google Scholar] [CrossRef]
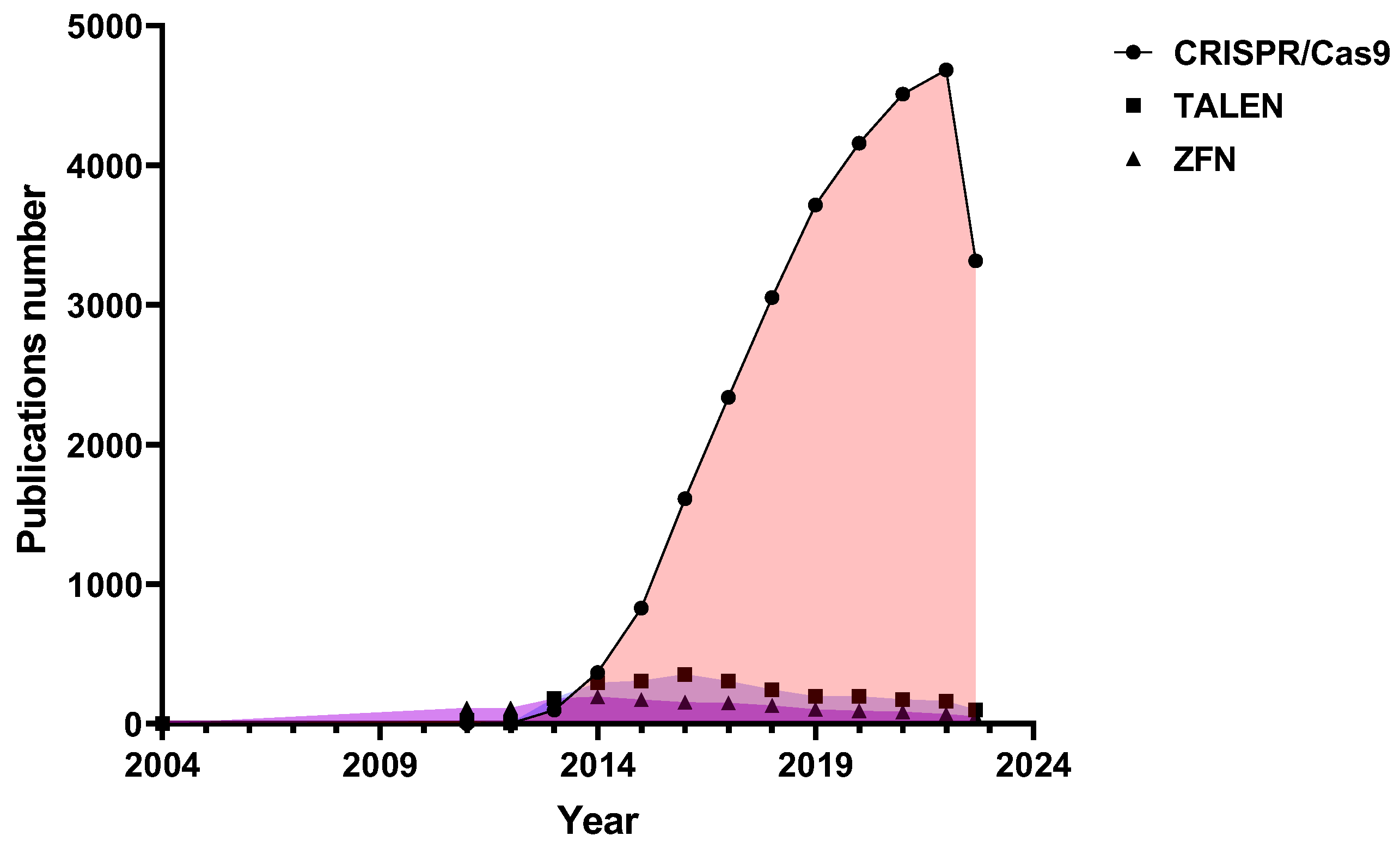
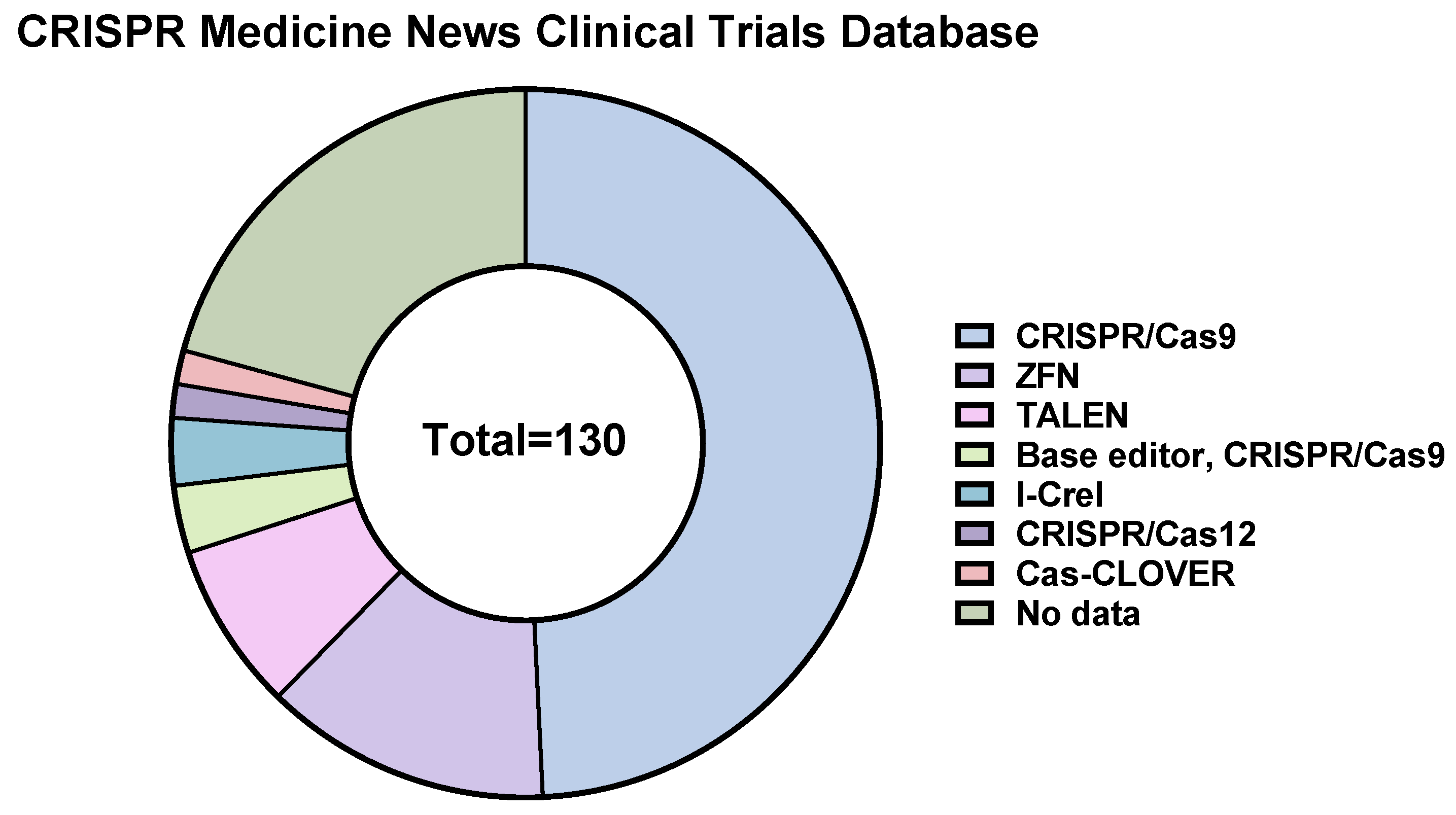
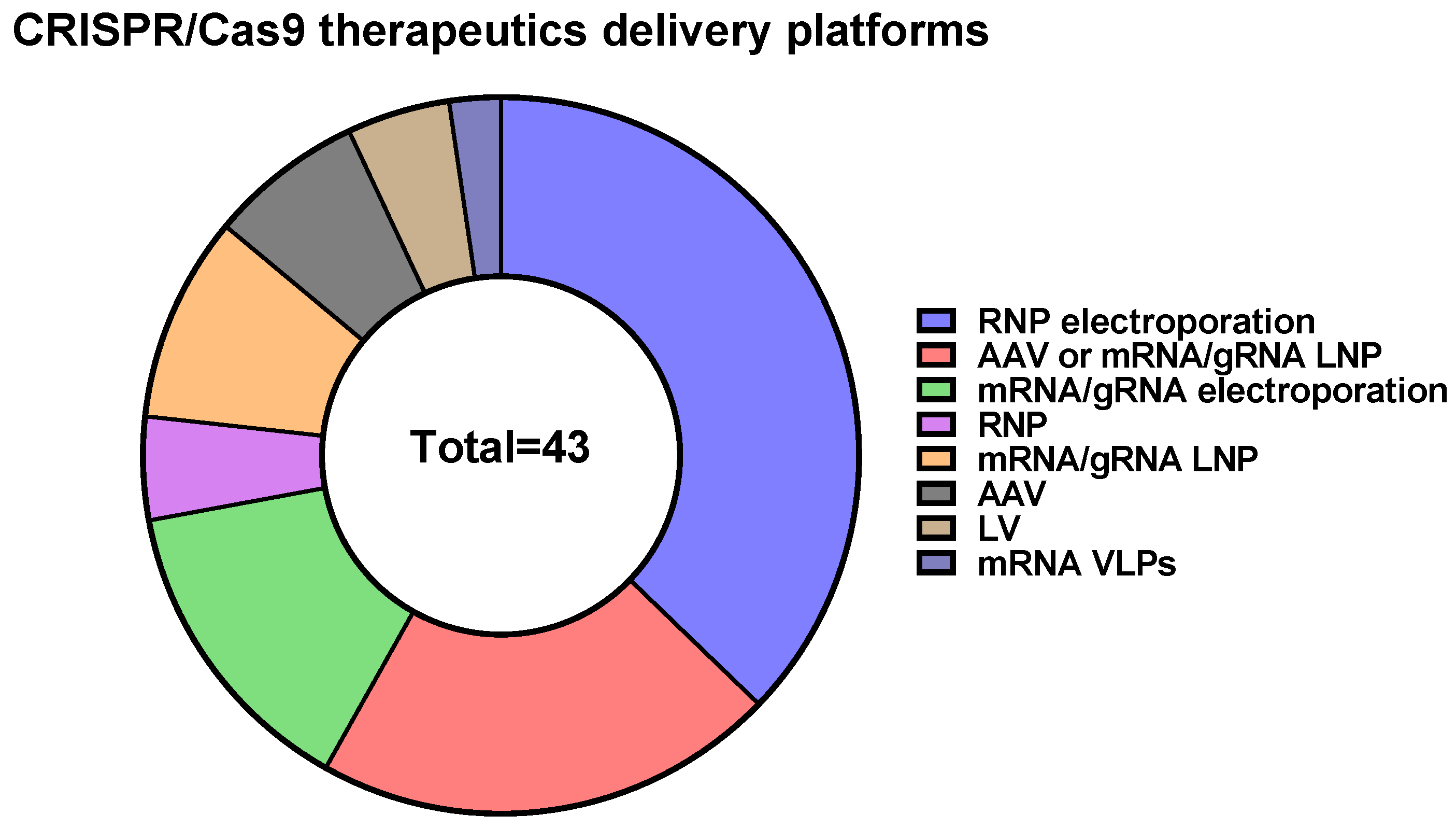

| Name | Key Features | Reference |
|---|---|---|
| Cas9 variants engineered using random mutagenesis combined with high-throughput screening | ||
| Sniper-Cas9 | Sniper-Cas9 showed much higher on-target activities and comparable specificity at most off-target loci when compared with other engineered high-fidelity SpCas9 variants such as SpCas9-HF1, HypaCas9, evoCas9, and eSpCas9(1.1). However, Sniper-Cas9 showed stronger tolerance to single mismatches in the PAM-distal region (e.g., 16th, 18th, and 19th). Sniper-Cas9 showed on-target activities with extended or truncated sgRNAs and worked well in a preassembled ribonucleoprotein (RNP) format to allow DNA-free genome editing | [33] |
| HiFi Cas9 | HiFi Cas9 (R691A variant) demonstrated the highest on-target activity (82%) in the RNP format compared to other improved Cas9 variants | [30] |
| xCas9 | Cas9 variant with expanded PAM compatibility (xCas9 can recognize a broad range of PAM sequences, including NG, GAA, and GAT). xCas9 demonstrated lower genome-wide off-target activity at all NGG target sites, as well as minimal off-target activity when targeting genomic sites with non-NGG PAMs | [38] |
| SpartaCas | SpartaCas (D23A, T67L, Y128V, and D1251G variants) had reduced off-target events while maintaining high on-target editing in T-cells. The editing efficiency of SpartaCas was slightly reduced when compared to wild-type Cas9, but it demonstrated dramatically higher editing efficiency when compared to SpCas9-HF4 and eCas9 | [34] |
| efSaCas9 | efSaCas9 (N260D variant) is a high-fidelity, high-activity variant that could be harnessed for safe and reliable genome editing. efSaCas9 demonstrated dramatically reduced off-target effects (approximately 2- to 93-fold improvements) compared to wild-type SaCas9 | [28] |
| Cas9 variants engineered using a structure- and/or function-guided protein engineering approach | ||
| SpCas9-D1135E | SpCas9-D1135E possessed genome-wide specificity and demonstrated reduced off-target effects. The D1135E mutant was able to better discriminate between NGG and NGA PAMs compared with wild-type SpCas9. Moreover, it had decreased activity against non-canonical NAG, NGA, and NNGG PAMs relative to wild-type SpCas9 | [25] |
| eSpCas9 | eSpCas9 variants, also known as “enhanced specificity”, SpCas9 showed reduced off-target effects and maintained robust on-target cleavage. | [27] |
| SpCas9-HF1 (and -HF2, -HF3, -HF4) | SpCas9-HF are high-fidelity SpCas9 variants harboring alterations designed to reduce non-specific DNA contacts. SpCas9-HF retained on-target activities comparable to wild-type. SpCas9-HF rendered all or nearly all off-target events undetectable, even for atypical, repetitive target sites | [24] |
| HypaCas9 | HypaCas9 (N692A, M694A, Q695A, and H698A variants) exhibited dramatically improved genome-wide specificity compared to wild-type SpCas9 and showed equivalent or better genome-wide specificity relative to both SpCas9-HF1 and eSpCas9 variants | [31] |
| SuperFi-Cas9 | SuperFi-Cas9 was able to discriminate between on- and off-target substrates without compromising DNA cleavage efficiency, but a recent study showed that it had significantly reduced on-target activity in mammalian cells | [36,84] |
| LZ3 Cas9 | The LZ3 Cas9 variant is known to exhibit high activity, increased specificity, and a differential +1 insertion profile as compared to wild-type SpCas9. Further rational engineering of LZ3 Cas9 might provide novel opportunities for non-templated correction of disease-causing frameshift mutations in the human population | [37] |
| eSaCas9 | eSaCas9 is a high-fidelity version of SaCas9 obtained by weakening interactions between Cas9 and the target DNA. eSaCas9 demonstrated high on-target activities and comparable specificity at most off-target loci | [27] |
| SaCas9-HF | SaCas9-HF showed high genome-wide targeting accuracy without compromising on-target efficiency, as validated by rigorous evaluation of its on- and off-target activities across multiple endogenous sites | [32] |
| KKH-SaCas9-SAV1 (and -SAV2) | KKH-SaCas9-SAV1 and SAV2 are SaCas9 variants that exhibited low off-target and high on-target activities and revealed a pivotal role of the previously unreported Y239H substitution in determining target accuracy while maintaining the activity of KKH-SaCas9 | [39] |
| Cas9 variants engineered using directed evolution combined with structure-guided modeling | ||
| evoCas9 | EvoCas9 is a variant that has fidelity exceeding both wild-type (79-fold improvement) and rationally designed Cas9 variants (4-fold average improvement), while maintaining near-wild-type on-target editing efficiency (90% median residual activity) | [29] |
| SaCas9-Q414A | The Q414A variant of SaCas9 exhibited even higher fidelity than the N260D variant of SaCas9 while retaining the most on-target activity | [28] |
| Cas9-based fusion proteins with improved properties | ||
| miCas9 | Cas9 variant with improved homology-directed repairing capacity (2.5-fold higher). To improve Cas9’s homology-directed repair capacity, SpCas9 was fused to a minimal motif consisting of thirty-six amino acids (Brex27 motif) | [40] |
| Cas9-pDBD | Fusing a programmable DNA-binding domain (pDBD) to Cas9 combined with the attenuation of Cas9’s inherent DNA binding affinity produced a Cas9-pDBD chimera with dramatically improved precision and increased targeting range | [85] |
| Cas-Cas9 chimeras | RNA-programmable Cas9-Cas9 chimeras, in single- and dual-nuclease formats, were designed as versatile genome engineering systems. In both formats, Cas9-Cas9 fusions displayed an expanded targeting repertoire and achieved highly specific genome editing. Dual-nuclease Cas9-Cas9 chimeras had higher target site activity and generated predictable, precise deletion products between target sites | [86] |
| Action | Name | Function | Reference |
|---|---|---|---|
| Transcription repression | dCas9 (CRISPRi) | Steric hindrance | [92,93,94,95,96] |
| dCas9-KRAB, dSaCas9-KRAB | Histone methylation and deacetylation | [90,93,97,98,99,100,101,105] | |
| dCas9-KRAB-MeCP2 | Represses transcription from methylated promoters via binding | [102] | |
| ZIM3-dCas9 | Histone methylation and deacetylation | [95,103] | |
| dCas9-SALL1-SDS3 | Histone deacetylation | [104] | |
| dCas9-SRDX | Transcriptional repressor domain (in plants) | [106,107,108] | |
| Transcription activation | dCas9-VP64 | Transcriptional activator | [109] |
| dCas9-SAM | Multiple copies of transcriptional activators | [109] | |
| dCas9-SunTag | Multiple copies of transcriptional activators | [111,112] | |
| dCas9-SPH | Multiple copies of transcriptional activators | [113] | |
| dCas9-VPR | Combination of three transcriptional activators | [114,115] | |
| Epigenetic repression | dCas9-LSD1 | Histone demethylation | [123,124] |
| dCas9-DNMT3A | DNA methylation | [128] | |
| dCas9-SunTag-DNMT3A | DNA methylation (multiple copies) | [130] | |
| CRISPRoff | DNA methylation, histone methylation, and deacetylation (contains ZNF10 KRAB, DNMT3A, and DNMT3L protein domains) | [131] | |
| Epigenetic activation | dCas9-p300 | Histone acetylation | [120] |
| dCas9-TET1CD | DNA demethylation | [125] |
| CRISPR/Cas9 | Base Editors | Prime Editors | Cas-CLOVER | Cas-FokI | |
|---|---|---|---|---|---|
| DNA catalytic domain | RuvC and HNH (nuclease) | Cytidine deaminase or adenine deaminase | Cas9 H840A nickase, reverse transcriptase, and cellular endonuclease | Nuclease domain from Clostridium Clo051 | FokI |
| DNA recognition | sgRNA (crRNA in complex with tracrRNA) | sgRNA (crRNA in complex with tracrRNA) | pegRNA (containing a primer binding site and a donor template) | Left and right sgRNA | Left and right sgRNA |
| Mechanism | Double-strand breaks in edited DNA (repaired by non-homologous end joining, NHEJ, or homology-directed repair, HDR) | Single-base changes (C to T and A to G) in edited DNA without introducing double-strand breaks | Accurately introduces small targeted insertions into the edited DNA sequence and removes and replaces bases | High-fidelity site-specific nuclease that introduces double-strand breaks in edited DNA with undetectable off-target activity | High-fidelity site-specific nuclease, that introduces double-strand break in edited DNA with undetectable off-target activity |
| Specificity | Tolerates mismatches | Tolerates mismatches | Tolerates mismatches | Enhanced specificity | Enhanced specificity |
| Ease of delivery | Easily delivered using multiple techniques | Harder to deliver due to the increased size | Harder to deliver due to the increased size | Harder to deliver due to the increased size | Harder to deliver due to the increased size |
| Limitations | Possible off-target effects (especially for SpCas9) | Narrow range of editing in the immediate vicinity of the PAM, potential off-target effects | Overall complexity of the system | Difficult to deliver (size) | Difficult to deliver (size) |
| Multiplexing | Easy | Easy | Easy | Easy | Easy |
| Active clinical trials | Yes (64/130) | Yes (4/130) | No | Yes (2/130) | No |
| Use in academic laboratories | 26,717 results in PubMed® (https://pubmed.ncbi.nlm.nih.gov/, accessed on 2 November 2023 with search term “CRISPR Cas9” | 12,373 results in PubMed® (https://pubmed.ncbi.nlm.nih.gov/, accessed on 2 November 2023 with search term “CRISPR Cas9 base editing” | 306 results in PubMed® (https://pubmed.ncbi.nlm.nih.gov/, accessed on 2 November 2023 with search term “CRISPR Cas9 prime editing” | 3 results in PubMed® (https://pubmed.ncbi.nlm.nih.gov/, accessed on 2 November 2023 with search term “Cas-CLOVER” | 14 results in PubMed® (https://pubmed.ncbi.nlm.nih.gov/, accessed on 2 November 2023 with search term “FokI-dCas9” |
| First time mentioned | 2011 | 2016 | 2019 | 2020 | 2014 |
| Delivery Approach | Mode of Cas9 and Guide RNA Delivery | ||
|---|---|---|---|
| DNA | mRNA | RNP | |
| Electroporation | + | + | + |
| Viral vectors | + | + | − |
| Lipofection | + | + | + |
| Lipid nanoparticles | − | + | + |
| Polymer nanoparticles | − | − | + |
| Hydrogel nanoparticles | − | − | + |
| Gold nanoparticles | − | − | + |
| Graphene oxide | − | − | + |
| Metal−organic frameworks | − | − | + |
| Black phosphorus nanosheets | − | − | + |
| Cell-penetrating peptides | − | − | + |
| DNA nanostructures | − | − | + |
| CRISPR/Cas9 | CRISPR/Cas12 | CRISPR/Cas13 | CRISPR/Cas14 | Fanzor Proteins | |
|---|---|---|---|---|---|
| DNA catalytic domain | RuvC, HNH | RuvC-like nuclease domain, Nuc-domain | HEPN domains | RuvC | RuvC-like nuclease domain |
| Target | Double-stranded DNA | Double-stranded DNA | Single-stranded RNA | Single-stranded DNA | Double-stranded DNA |
| Collateral activity | No | Yes | Yes | Yes | No |
| DNA recognition | sgRNA (crRNA in complex with tracrRNA) | crRNA | crRNA | crRNA and tracrRNA | fRNA or ωRNA |
| PAM requirements | NGG, NAG for SpCas9, and other PAM variants for Cas9 orthologs (for details, see Table S4) | TTTN, TTTV (V = G, C, or A) For AsCpf1 from Acidaminococcus or LbCpf1 from Lachnospiraceae, and other PAM variants for Cas12 orthologs [362,363,364,365,366,367,368] | Requires protospacer flanking sequence–A, U, or C | None | Target adjacent motif preference is diverse, with a GC preference observed for the viral Fanzor proteins and AT preferences for the eukaryotic Fanzor proteins |
| Specificity | Regular SpCas9 tolerates mismatches, but high-fidelity variants exist | Cas12a has been successfully used for gene editing in vivo without any deleterious off-target effects | RNA-editor, no damage to DNA may occur | Cleaves ssDNA with high fidelity-sensitive to even a single mismatch in the target sequence | Needs further investigations |
| Ease of delivery | Easily delivered using multiple techniques | Easily delivered using multiple techniques | Easily delivered using multiple techniques | Easily delivered using multiple techniques | Needs further investigations |
| Limitations | GC-rich DNA targets Possible off-target effects (especially for SpCas9) | AT-rich DNA targets | Needs to be constitutively expressed to maintain the editing effect | Targets ssDNA | Unknown |
| Multiplexing | Easy | Easy | Easy | Easy | Possible |
| Active clinical trials | Yes (64/130) | Yes (2/130) | No | No | No |
| Use in academic laboratories | 26,717 results in PubMed® (https://pubmed.ncbi.nlm.nih.gov/, accessed on 2 November 2023 with search term “CRISPR Cas9” | 1684 results in PubMed® (https://pubmed.ncbi.nlm.nih.gov/, accessed on 2 November 2023 with search term “CRISPR Cas12a or Cpf1” | 373 results in PubMed® (https://pubmed.ncbi.nlm.nih.gov/, accessed on 2 November 2023 with search term “CRISPR Cas13” | 28 results in PubMed® (https://pubmed.ncbi.nlm.nih.gov/, accessed on 2 November 2023 with search term “CRISPR Cas14” | 4 results in PubMed® (https://pubmed.ncbi.nlm.nih.gov/, accessed on 2-November 2023 with search term “Fanzor” |
| First time mentioned | 2011 | 2017 | 2017 | 2018 | 2013, 2023 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tyumentseva, M.; Tyumentsev, A.; Akimkin, V. CRISPR/Cas9 Landscape: Current State and Future Perspectives. Int. J. Mol. Sci. 2023, 24, 16077. https://doi.org/10.3390/ijms242216077
Tyumentseva M, Tyumentsev A, Akimkin V. CRISPR/Cas9 Landscape: Current State and Future Perspectives. International Journal of Molecular Sciences. 2023; 24(22):16077. https://doi.org/10.3390/ijms242216077
Chicago/Turabian StyleTyumentseva, Marina, Aleksandr Tyumentsev, and Vasiliy Akimkin. 2023. "CRISPR/Cas9 Landscape: Current State and Future Perspectives" International Journal of Molecular Sciences 24, no. 22: 16077. https://doi.org/10.3390/ijms242216077
APA StyleTyumentseva, M., Tyumentsev, A., & Akimkin, V. (2023). CRISPR/Cas9 Landscape: Current State and Future Perspectives. International Journal of Molecular Sciences, 24(22), 16077. https://doi.org/10.3390/ijms242216077






