Abstract
Anthocyanins are widespread water-soluble pigments in the plant kingdom. Anthocyanin accumulation is activated by the MYB-bHLH-WD40 (MBW) protein complex. In Arabidopsis, the R2R3-MYB transcription factor PAP1 activates anthocyanin biosynthesis. While prior research primarily focused on seedlings, seeds received limited attention. This study explores PAP1’s genome-wide target genes in anthocyanin biosynthesis in seeds. Our findings confirm that PAP1 is a positive regulator of anthocyanin biosynthesis in Arabidopsis seeds. PAP1 significantly increased anthocyanin content in developing and mature seeds in Arabidopsis. Transcriptome analysis at 12 days after pollination reveals the upregulation of numerous genes involved in anthocyanin accumulation in 35S:PAP1 developing seeds. Chromatin immunoprecipitation and dual luciferase reporter assays demonstrate PAP1’s direct promotion of ten key genes and indirect upregulation of TT8, TTG1, and eight key genes during seed maturation, thus enhancing seed anthocyanin accumulation. These findings enhance our understanding of PAP1’s novel role in regulating anthocyanin accumulation in Arabidopsis seeds.
1. Introduction
Anthocyanins, water-soluble pigments, fall under the flavonoids class of secondary metabolites, contributing red, purple, and blue hues to various fruits and vegetables [1]. These pigments play a role in attracting pollinators and seed dispersers agents [1,2]. Anthocyanin formation is influenced by environmental factors such as UV irradiation [3], temperature [4], drought [5], and nutrient deficiency [6]. Under adverse conditions, anthocyanin concentrations generally increase, indicating their involvement in biotic and abiotic stress responses [7,8,9]. Anthocyanins, with their antioxidant properties, also serve as vital micronutrients for humans, guarding against cardiovascular, neurodegenerative, metabolic diseases, and cancer [10]. Therefore, gaining a deeper understanding of anthocyanin accumulation and its regulatory mechanisms holds significant scientific and economic importance.
The anthocyanin biosynthetic pathway is a major branch of the general phenylpropanoid pathway that starts with phenylalanine (Phe) [11]. This pathway can be briefly divided into three parts: beginning steps of the general phenylpropanoid pathway, early steps of the flavonoid pathway, and late steps of the anthocyanin specific pathway. The expression of structural genes in the anthocyanin biosynthetic pathway primarily depends on the MYB-bHLH-WD40 (MBW) transcription complex, comprising MYB and basic helix-loop-helix (bHLH) transcription factors alongside WD40 proteins [12,13]. MYB proteins, one of the largest plant transcription factor families, play roles in cell differentiation [14], stress responses [15], metabolism [16], and development processes [17]. R2R3-MYB transcription factors, featuring an N-terminal conserved MYB domain and a C-terminal variable activation or repression domain, play a key role in determining the spatial and temporal patterns of anthocyanin accumulation [16,18]. ZmC1 (colorless1), the first discovered R2R3-MYB transcription factor, was reportedly essential for anthocyanin biosynthesis in maize aleurone tissues [19]. Several R2R3-MYB members act as positive regulators for anthocyanin biosynthetic genes, including PRODUCTION OF ANTHOCYANIN PIGMENTATION 1 (PAP1)/MYB75, PAP2/MYB90, MYB113, and MYB114 in Arabidopsis [20,21], MdMYB10 and MdMYB110a in apple [22], and MYB78 in canola [23]. Conversely, specific R2R3-MYB transcription factors repress anthocyanin biosynthesis [24,25], including MYB3, MYB4, and MYB6 in Arabidopsis [26] and MdMYB16, MdMYB17, and MdMYB111 in apple [27].
PAP1, also known as MYB75, functions as a key anthocyanin biosynthesis regulator [28,29]. In Arabidopsis seedlings, the pap1 loss-of-function mutant or PAP1 RNA interference plants displayed reduced anthocyanin induction [21,30]. Conversely, over-expression of PAP1 in the activation-tagged PAP1 gain-of-function mutant (pap1-D) resulted in hyperaccumulation of anthocyanins in leaves, roots, stems, and flowers [20]. For governing the anthocyanin biosynthetic pathway, PAP1 forms a complex with bHLH anthocyanin regulators GLABRA 3, ENHANCER OF GLABRA 3, and TRANSPARENT TESTA 8 (TT8), along with a WD40-repeat protein, TRANSPARENT TESTA GLABRA 1 (TTG1). This complex enhances the expression of late anthocyanin biosynthetic genes: dihydroflavonol-4-reducatse (DFR), leucoanthocyanidin dioxygenase (LDOX), and UDP-glucose: flavonoid-3-O-glucosyl-transferase (UF3GT) [11,21,28]. Further studies have demonstrated that the post-translational modification of MBW proteins modulates the MBW protein complex’s transcriptional activity. PAP1 degradation in the dark is mediated by the CONSTITUTIVE PHOTOMORPHOGENIC 1/SUPPRESSOR of PHYA-105 ubiquitin ligase [31], while phosphorylation by MAP KINASE 4 stabilizes PAP1, which is essential for light-induced anthocyanin accumulation [32]. Another post-translational modification hinders the formation of the MBW protein complex. The DNA-binding homeodomain ZIP transcription factor HAT1 interferes with the formation of the MBW protein complex by interacting with PAP1 and recruiting the TOPLESS corepressor to epigenetically modulate anthocyanin biosynthetic genes [33]. Furthermore, the phosphate starvation signaling pathway repressor SPX4 physically interacts with PAP1, disrupting the PAP1–TT8 interaction and impairing the transcriptional activation of the anthocyanin biosynthesis gene DFR [34]. A recent study reveals that PHYTOCHROME-INTERACTING Factor 4 competes with TT8 to bind PAP1, thus affecting the regulation of the MBW protein complex in anthocyanin biosynthesis [35].
These findings suggest that the MBW complex serves as a central regulatory hub for anthocyanin accumulation. Notably, PAP1 is a key activator in the anthocyanin biosynthesis pathway with conserved functions in various crops [11,21,28]. However, genome-wide targets of PAP1 in Arabidopsis seeds remain unexplored. In our study, we demonstrated that PAP1 enhances seed anthocyanin accumulation by upregulating some anthocyanin biosynthesis-related genes during Arabidopsis seed development. Our results offer new insights into PAP1’s regulatory role in Arabidopsis seed anthocyanin accumulation.
2. Results
2.1. Positive Correlation of PAP1 Levels and Anthocyanin Accumulation in Seeds
To validate PAP1’s role in anthocyanin accumulation in Arabidopsis seeds, we introduced the 35S:PAP1-6HA construct into Arabidopsis Columbia-0 (Col-0), yielding fifteen transgenic lines. We selected three independent 35S:PAP1 T3 homozygous transgenic lines with the highest PAP1 expression levels, 35S:PAP1 #1, 35S:PAP1 #3, and 35S:PAP1 #5, for subsequent experiments (Figure 1).
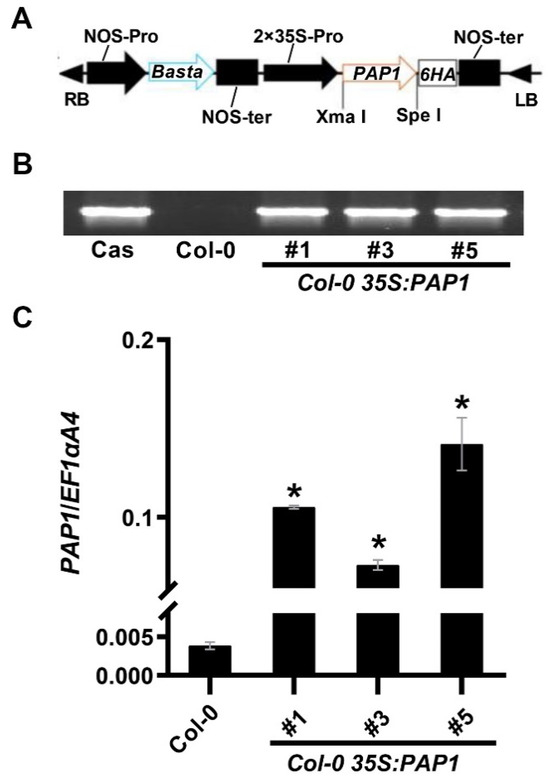
Figure 1.
Characterization of Col-0 35S:PAP1 lines: (A) Schematic diagram of the constitutive expression cassette of the PAP1 gene in the binary vector pGreen-35S-6HA used for plant transformation. RB, right border; LB, left border; NOS-pro, nopaline synthase promoter; NOS-ter, nopaline synthase terminator; Basta, glyphosate; 35S-pro, CaMV 35S promoter. (B) PCR-based DNA genotyping of Col-0 35S:PAP1 transgenic plants using specific primers for the 35S_P/PAP1_R. Cas, cassette. (C) Expression analysis of PAP1 in wild-type (Col-0) and three independent Col-0 35S:PAP1 transgenic plants using RT-qPCR. RNA samples were extracted from rosette leaves. Results were normalized against the expression of Arabidopsis EF1aA4 as the internal control. Values are means ± SD (n = 3). Asterisks (*) indicate a significant difference in gene expression in the transgenic plants of PAP1 compared with Col-0 plants (two-tailed paired Student’s t-test, p ≤ 0.05).
Compared to wild-type plants, the developing seeds of these three PAP1-over-expressing transgenic lines exhibited enhanced pigmentation at 10 and 12 days after pollination (DAP) as well as in mature seeds (Figure 2A–C). Additionally, the seedlings of the transgenic lines (35S:PAP1 #1, 35S:PAP1 #3, and 35S:PAP1 #5) displayed purple stems and petioles, while wild-type seedlings were green (Supplementary Figure S1).
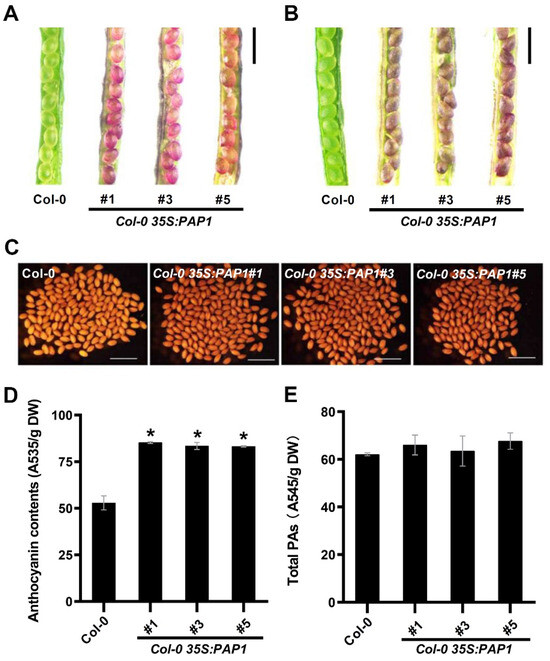
Figure 2.
Effect of PAP1 on the accumulation of anthocyanin and PAs in seeds: (A–C) Phenotypes of wild type (Col-0) and Col-0 35S:AtPAP1 immature seeds at 10 DAP (A), 12 DAP (B), and mature seeds (C). Scale bars = 1 mm. (D) Total anthocyanin contents in mature seeds of wild-type (Col-0) and Col-0 35S:PAP1. Asterisks (*) denote the statistically significant differences between the indicated samples (Student’s t-test, p ≤ 0.05). Values are means ± SD (n = 3). DW, dry weight. (E) Total PAs contents in mature seeds of wild-type (Col-0) and Col-0 35S:PAP1. Values are means ± SD (n = 3). DW, dry weight.
Furthermore, we measured the anthocyanin levels in mature seeds of the wild-type and three transgenic lines, revealing a significant increase in anthocyanin content in the transgenic lines compared to wild-type plants (Figure 2D). Proanthocyanidins (PAs) are a class of oligomeric or polymeric flavonoids. The intermediate compound dihydroflavonol can be further converted to anthocyanins or PAs through distinct branches of the flavonoid pathway [36]. Previous studies have shown that PAs accumulate in the seed coat and protect the embryo and endosperm [37]. Consequently, we assessed the PAs levels in mature seeds of both the wild-type and the three transgenic lines, revealing no significant difference in PAs content (Figure 2E). In summary, our findings suggested that PAP1 selectively regulates anthocyanin accumulation, but not PAs, in seeds during Arabidopsis seed development.
2.2. A Whole-Genome Analysis of Genes Associated with Seed Anthocyanin Accumulation
To elucidate the regulatory mechanism of PAP1 in seed anthocyanin accumulation, we performed RNA-Sequencing (RNA-Seq) analysis on developing seeds from the transgenic line 35S:PAP1 #5 and wild-type Col-0 plants at 12 DAP. The results identified 5174 differentially expressed genes (DEGs), with 4760 upregulated and 414 downregulated DEGs (Table 1). Among them, seventy-four upregulated genes (1.6%) and three downregulated genes (0.7%) were involved in flavonoid biosynthesis (Table 1). Additionally, 30% of upregulated genes participated in primary metabolic processes, including carbohydrate metabolism (12.8%), nucleic acid (5.2%), amino acid and protein (5.0%), cell wall (3.3%), and photosynthesis (2.3%) (Table 1). Notably, 780 upregulated genes (16.4%) and 95 downregulated genes (22.9%) were linked to stress/defense responses (Table 1). These findings highlight PAP1’s pivotal role in seed anthocyanin accumulation and other crucial physiological and biochemical processes.

Table 1.
Functional classification of differentially expressed genes (DEGs) in developing seeds between wild-type Col-0 and Col-0 35S:PAP1 #5 plants at 12 days after pollination (DAP).
2.3. Validation of Seed Anthocyanin Accumulation-Related Genes
To confirm the regulation of anthocyanin biosynthesis-related genes in developing 35S:PAP1 #5 seeds at 12 DAP and to identify target genes controlled by PAP1 in the seed anthocyanin biosynthetic pathway, we conducted quantitative real-time PCR (RT-qPCR) to compare expression patterns between 35S:PAP1 #5 and wild-type plants. We selected twenty highly upregulated genes, including two transcription factors (TT8 and TTG1) and eighteen structural genes (Table 2). The RT-qPCR results aligned with the RNA-seq data, confirming significant upregulation of these twenty genes (Figure 3 and Table 2). Notably, pivotal genes involved in Phe synthesis, such as arogenate dehydratase 5 (ADT5), and genes related to Phe metabolic pathways, such as cinnamate 4-hydroxylase (C4H) and 4-coumarate: CoA ligase 3 (4CL3), exhibited higher expression in 35S:PAP1 #5 compared to the wild-type seeds at 12 DAP (Figure 3). Additionally, genes associated with anthocyanin biosynthesis, like chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3’H), DFR, and anthocyanidin synthase (ANS), as well as genes involved in anthocyanin modification and transport, including flavonoid 3-O-glycosyltransferase (3GT), anthocyanin 5-O-glycosyltransferase (5GT), UF3GT, UDP-glycosyltransferases (UGT79B2 and UGT79B3), anthocyanin 3-O-glucoside-6″-O-acyltransferases (3AT1 and 3AT2), anthocyanidin 5-O-glucoside-6″-O-malonyltransferase (5MAT), and glutathione S-transferase 26 (GST26), were likewise upregulated in 35S:PAP1 #5 at 12 DAP (Figure 3). In summary, these findings underscore the role of PAP1 in enhancing seed anthocyanin accumulation by activating the expression of regulatory and structural genes involved in anthocyanin biosynthesis, modification, and transport.

Table 2.
Differentially expressed genes (DEGs) contributing to anthocyanin biosynthesis in the developing seeds between wild-type Col-0 and Col-0 35S:PAP1 #5 plants at 12 days after pollination (DAP).
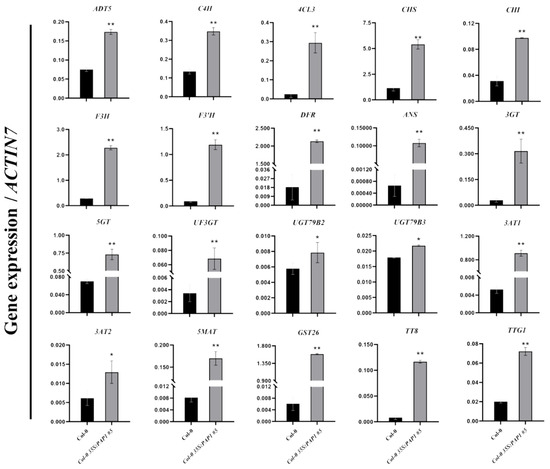
Figure 3.
RT-qPCR analysis of the expression of anthocyanin biosynthesis-related genes in the wild-type (Col-0) and Col-0 35S:PAP1 #5 developing seeds at 12 DAP. Results were normalized against the expression of Arabidopsis ACTIN7 as the internal control. Values are means ± SD (n = 3). ** p ≤ 0.01 and * p ≤ 0.05 indicate highly significant or significant differences in gene expression levels between wild-type (Col-0) and Col-0 35S:PAP1 #5 plants (two-tailed paired Student’s t-test).
2.4. PAP1 Promotes Anthocyanin Accumulation by Directly Activating the Expression of ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26 in Arabidopsis Developing Seeds
To investigate PAP1’s regulation of seed anthocyanin accumulation, we performed chromatin immunoprecipitation (ChIP) assays on developing siliques at 12 DAP from 35S:PAP1-6HA #5 plants. This allowed us to understand how PAP1 controls the transcription of target genes. From the twenty genes mentioned earlier, we selected ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26 due to their possession of PAP1 binding sites. The core binding motif of PAP1, identified as a 7 bp MYB-recognizing element (MRE) (ANCNNCC), was found in the promoter regions of these ten genes [34,63]. We designed primers to cover all possible MRE sites bound by PAP1 in the promoter regions of these genes. There are three MREs within the promoter of ADT5, five MREs in CHS, three MREs in F3H, two MREs in DFR, three MREs in ANS, three MREs in 3GT, two MREs in UGT79B2, two MREs in UGT79B3, three MREs in 5MAT, and four MREs in GST26 (Figure 4). The ChIP assay revealed that PAP1-6HA was associated with specific promoter regions: P3 of ADT5, P1 of CHS, P1 of F3H, P1 of DFR, P1 and P2 of ANS, P1 and P2 of 3GT, P2 of UGT79B2, P1 and P2 of UGT79B3, P3 of 5MAT, and P2 of GST26 (Figure 4). These results demonstrated that PAP1 directly binds to the promoter regions of ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26 to promote their expression.
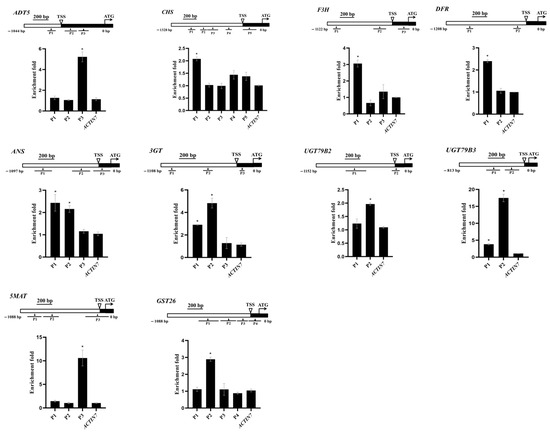
Figure 4.
PAP1 targets ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26 promoters and directly promotes their expressions in developing Arabidopsis seeds. Schematic diagrams show the promoter regions of ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26, while ChIP-qPCR assays show PAP1 binding to their promoter regions in the developing Arabidopsis siliques at 12 DAP. The transcriptional start site (TSS) and exons are represented by black boxes, whereas promoter regions are represented by white boxes. The triangle represents the MYB-recognizing element (MRE) site ANCNNCC and the black lines represented the DNA fragments amplified in ChIP assays for each gene. The enrichment fold of each fragment was calculated first by normalizing the amount of a target DNA fragment against a genomic fragment of Arabidopsis EF1aA4 as the internal control. Then, the value for Col-0 35S:PAP1 #5 was normalized against it for wild-type (Col-0) plants. The Arabidopsis ACTIN7 fragment was amplified as the negative control. Values are means ± SD (n = 3). Significant differences in comparison with the ACTIN7 fragment enrichment are indicated with asterisks (*) (two-tailed paired Student’s t-test, p ≤ 0.05).
Moreover, we further evaluated the positively regulatory function of PAP1 on the transcription of ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26 using a transient dual-luciferase reporter assay. We constructed effectors with or without the CDS of PAP1, and reporters containing firefly luciferase driven by the promoters of ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26 along with Renilla luciferase driven by the 35S promoter (Pro35S:REN) was used as an internal control (Figure 5A). After co-expression with effector and reporter in N. benthamiana leaves, the LUC/REN ratios of ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26 were markedly increased (Figure 5B), indicating that PAP1 directly activates their transcription in N. benthamiana leaves. In summary, our findings collectively indicate that PAP1 promotes anthocyanin accumulation by directly activating the expression of ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26, while also indirectly promoting the expression of C4H, 4CL3, CHI, F3′H, 5GT, UF3GT, 3AT1, 3AT2, TT8, and TTG1 during seed development in Arabidopsis.
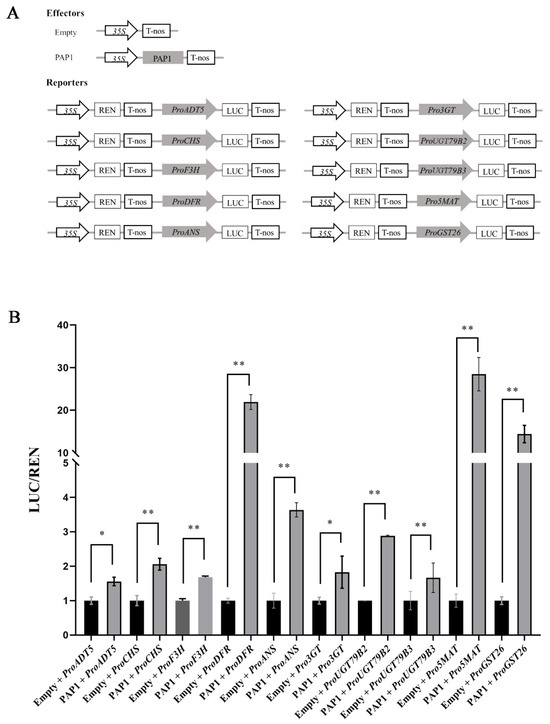
Figure 5.
PAP1 directly activates ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26 transcription in N. benthamiana leaves: (A) Schematic diagrams show the effectors with and without PAP1 and the reporters containing ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26 promoters. (B) Transient dual-luciferase reporter assay. The reporter constructs were transiently expressed in N. benthamiana leaf cells together with empty or PAP1 effector constructs. The expression level of Renilla (REN) was used as an internal control, and the LUC/REN represents the relative activity of ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26 promoters. Values are means ± SD (n = 6). Double asterisks (**) and asterisk (*) separately indicate highly significant (p ≤ 0.01) and significant differences (p ≤ 0.05), respectively, in the LUC/REN compared to PAP1 effector with an empty effector (two-tailed paired Student’s t-test).
3. Discussion
Anthocyanin accumulation is a dynamic phenomenon in various plant species. The transcription factor PAP1, an R2R3-MYB member, serves as a pivotal hub, integrating diverse internal and external stimuli affecting anthocyanin biosynthesis, with prior research mainly focused on seedling phenotypes [28,29]. However, information on the direct targets of the PAP1 in the regulation of anthocyanin accumulation, especially in seeds of Arabidopsis, remains limited. In this study, we identified new genes targeted by PAP1, directly or indirectly regulating anthocyanin accumulation at the genome-wide level in Arabidopsis seeds.
Numerous R2R3-MYB genes are known to positively or negatively regulate anthocyanin biosynthesis [24,64]. Previous reports indicated that two PAP1 over-expressing lines, the pap1-D mutant and the PAP1 cDNA over-expressing transgenic plant, exhibited similar anthocyanin accumulation in vegetative tissues but differed in seed color and accumulation patterns of anthocyanins and PAs in seeds [20,65]. The pap1-D mutant showed increased pigmentation in leaves, stems, and roots but no change in seed color [20,65]. In contrast, the PAP1 cDNA over-expressing plant displayed darker colors in all vegetative organs, including seeds [65]. Soluble anthocyanin levels increased in seeds of transgenic plants over-expressing PAP1 cDNA but remained unchanged in seeds of the pap1-D mutant [65]. Consistent with results from PAP1 cDNA over-expressing lines, our reverse genetic approach demonstrated that PAP1 over-expression modulates anthocyanin biosynthesis, leading to anthocyanin hyperaccumulation in both developing and mature seeds of Arabidopsis (Figure 2A–C). The increased anthocyanin content should be the reason for the darker color of mature seeds in transgenic lines (Figure 2C,D). Notably, total PAs content showed no difference between the wild-type and transgenic lines in mature seeds (Figure 2D). Successful PAs accumulation in Arabidopsis reportedly requires the cooperation of multiple genes [51]. Based on this observation and previous findings showing increased PAs content in the pap1-D mutant but decreased PAs content in PAP1 cDNA over-expressing plants in Arabidopsis seeds [65], it is reasonable to suggest that PAP1’s influence on accumulation of PAs is more complex than anthocyanin production in Arabidopsis seeds, likely due to various unknown factors. Further research is needed to understand the relationships between PAP1 and PAs in Arabidopsis seeds. Therefore, we speculate that PAP1 plays a specific positive role in anthocyanin accumulation, not PAs, during seed development in Arabidopsis seeds.
The regulation of gene expression involved in the anthocyanin biosynthetic pathway is largely coordinated by a complex network of interactions between transcription factors and their target genes [21,34]. The transcriptome analysis revealed that seventy-four upregulated genes (1.6%) and three downregulated genes (0.7%) in developing seeds of 35S:PAP1 #5 were related to flavonoid metabolism (Table 1 and Table S2). Additionally, a significant portion of all DEGs in developing seeds of 35S:PAP1 #5 (16.9%) were associated with stress/defense responses (Table 1), consistent with PAP1’s known role as a stress regulator [66,67]. Previous studies have discovered that anthocyanins play a crucial role in enhancing tolerance to biotic and abiotic stresses in vegetative tissues [8,9]. It is possible that PAP1 directly altered the expression patterns of these differentially expressed stress/defense-responsive genes (Supplementary Tables S2 and S3). Alternatively, the hyperaccumulation of anthocyanins in developing seeds may have indirectly regulated these stress/defense-responsive genes.
Multiple studies confirm PAP1’s potent impact on anthocyanin accumulation in Arabidopsis seedlings by specifically inducing anthocyanin-related gene expression. Tohge et al. [28], via microarray experiments showed upregulation of late anthocyanin biosynthetic genes, including glycosyltransferase genes UGT79B1 (At5g54060), UGT75C1 (At4g14090), and UGT78D2 (At5g17050), as well as early genes like 4CL, CHS, CHI, and F3′H in PAP1 over-expressing plants leaves. RNA gel blot analysis showed massive enhancement of the expression of genes across the entire phenylpropanoid pathway, including PAL1, CHS, DFR, and GST in 6-week-old pap1-D plants [20]. Our data corroborates this trend, as it shows a substantial upregulation of key genes throughout the entire anthocyanin biosynthetic pathway in developing seeds of 35S:PAP1 #5 (Figure 3). Additionally, molecular analyses reveal PAP1’s direct binding to the promoters of ten structural genes, including ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26, activating their transcription during anthocyanin biosynthesis in Arabidopsis seeds (Figure 4, Figure 5 and Figure 6). Notably, ADT plays a crucial role in converting arogenate into Phe during sucrose-induced anthocyanin biosynthesis in Arabidopsis. There are six ADT isoforms, ADT1 to ADT6, which redundantly regulate anthocyanin biosynthesis with varying contributions. Compared to other ADTs, when ADT4 or ADT5 were overexpressed, it results in Phe hyperaccumulation and a significant increase in anthocyanin content in Arabidopsis seedlings [38]. CHS catalyzes the initial step of the flavonoid pathway, condensing p-coumaroyl-CoA and malonyl-CoAs to produce tetrahydroxychalcone [42]. Over-expressing CHS enhances high light resistance by increasing anthocyanin synthesis [43]. Foliar application of CHS-specific dsRNAs and siRNAs resulted in an efficient downregulation of CHS and suppressed anthocyanin accumulation in Arabidopsis under anthocyanin biosynthesis-modulating conditions [44]. F3H hydroxylates flavanone to yield dihydrokaempferol [47]. Downregulating F3H in strawberries markedly reduces anthocyanin and moderately decrease flavonol content [68]. DFR reduces dihydroflavonol to leucoanthocyanidin, the initial reaction that leads to anthocyanin and proanthocyanidin biosynthesis. Pi starvation was found to destabilize the SPX4-PAP1 complex, allowing PAP1 to directly bind the DFR promoter, activating anthocyanin biosynthesis [34]. ANS, a pivotal enzyme in anthocyanin biosynthetic, converts colorless leucoanthocyanins into colored anthocyanidins. Arabidopsis ans mutants display reduced anthocyanin accumulation in hypocotyls [51] and rosette leaves [69]. 3GT, identified as flavonoid 3-O-glucosyltransferase, was predicted to participate in Arabidopsis’ anthocyanin biosynthesis [28]. UDP-glycosyltransferases (UGTs) catalyze the final step in anthocyanin biosynthesis, resulting in diverse anthocyanin molecules in Arabidopsis [11]. Two UGTs, UGT79B2 and UGT79B3, modified anthocyanins by adding UDP-rhamnose to cyanidin and 3-O-glucoside-cyanidin. Ectopic expression of UGT79B2/B3 significantly boosts anthocyanin levels, but ugt79b2/b3 double mutants exhibited reduced anthocyanin levels [5]. 5MAT encodes malonyl-CoA cyanidin 3,5-diglucoside transferase activity, thereby accelerating malonylated anthocyanin accumulation [57]. GST26, encoding an Arabidopsis glutathione S-transferase-like protein, is implicated in anthocyanin transport into vacuoles [58].
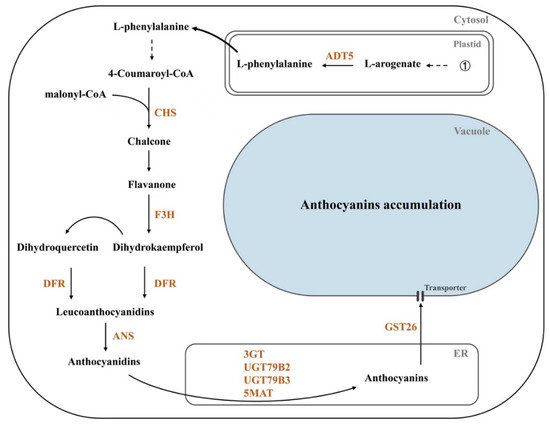
Figure 6.
A simplified scheme shows that PAP1 directly regulates the expression of structural genes that control the accumulation of seed anthocyanin in Arabidopsis. ①: Shikimic pathway; ADT: arogenate dehydratase; CHS: chalcone synthase; F3H: flavanone 3-hydroxylase; DFR: dihydroflavonol-4-reducatse; ANS: anthocyanidin synthase; 3GT: flavonoid 3-O-glycosyltransferase; UGTs: UDP-glycosyltransferases; 5MAT: anthocyanidin 5-O-glucoside-6″-O-malonyltransferase; GST: glutathione S-transferase; ER: endoplasmic reticulum.
However, we observed that PAP1 indirectly regulated eight upregulated structural genes essential for anthocyanin biosynthesis in Arabidopsis seeds. These genes are C4H, 4CL3, CHI, F3′H, 5GT, UF3GT, 3AT1, and 3AT2. C4H and 4CL are the second and third enzymes in the phenylpropanoid pathway, converting Phe to p-coumaroyl CoA [39,40,41]. CHI uses tetrahydroxychalcone to produce naringenin, and its mutation fails to accumulate anthocyanins [45,46]. F3′H hydroxylates dihydrokaempferol into dihydroquercetin [48,49]. 5GT encodes anthocyanin 5-O-glucosyltransferase, and UF3GT converts cyanidin 3-O-glucoside to cyanidin 3-O-xylosyl glucoside [28,55]. 3AT1 and 3AT2 encode coumaroyl CoA-cyanidin 3-O-glucose transferase and have redundant functions in anthocyanin stability [56].
Furthermore, we demonstrated that PAP1 indirectly enhanced the expression of two regulatory genes, TT8 and TTG1, during seed anthocyanin biosynthesis (Figure 3). This activation is instrumental in accelerating anthocyanin production in seeds (Figure 2). The feedback mechanisms between the MYB and bHLH components of the MBW activation complex play a pivotal role in flavonoid regulation. TT8, a bHLH transcription factor, regulates its own expression via a positive feedback loop through an MBW complex, contributing to anthocyanin and PA biosynthesis regulation [60,70]. TTG1, encoding a WD40 repeat transcription factor, collaborates with TT8 and TT2 (encoding MYB123) to mediate anthocyanin pigment production in developing seeds [59]. Our data suggests that PAP1, an R2R3-MYB factor, upregulates TT8 and TTG1 expression within the MBW complex for anthocyanin gene regulation, with the hierarchical regulation of TT8 and TTG1 expression requiring further investigation.
PAP1 expression has the potential to significantly enhance anthocyanin accumulation in many plant species, resulting in a dark purple color in various plant organs [20,21,65]. It has been demonstrated that the increased anthocyanin accumulation confers plants with enhanced tolerance to abiotic stress [66,67]. Thus, there is an interesting question needing to be investigated: do the enhanced anthocyanin levels in seeds enhance abiotic stress tolerance? Previous studies indicated that the anthocyanins present in seed extracts could serve as defense molecules against abiotic stresses such as UVB radiation, drought, and low or high temperatures [71]. Recently, some researchers have observed an association between seed dormancy and seed color [72,73]. These findings suggested that PAP1 may be a valuable candidate gene that is associated with seed dormancy and germination under stress due to its increased anthocyanin content.
In summary, our study reveals that the R2R3-MYB transcription factor PAP1 directly activates the expression of ten anthocyanin biosynthetic pathway-related structural genes, ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26, during seed development (Figure 6). This makes PAP1 a promising target for genetic manipulation to enhance seed anthocyanin levels, improving seed anthocyanin quality.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
The Arabidopsis thaliana ecotype Col-0 served as the wild-type control. As previously stated [74], the plants were grown in a 16 h light/8 h dark cycle at 22 °C in a growth chamber with overhead light at 160 μmol m−2 s−1.
4.2. Plasmid Construction and Transgenic Plants Generation
Specific primers were designed from the full-length coding sequence (CDS) of PAP1 (At1g56650) without a stop codon from the TAIR database. To create 35S:PAP1-6HA, the CDS of PAP1 (without the stop codon) was amplified using PAP1_F and PAP1_R primers (Supplemental Table S1). PCR products were digested using XmaI and SpeI, then ligated into the pGreen-35S-6HA binary vector to create an in-frame fusion of PAP1-6HA under the 35S promoter.
The 35S:PAP1-6HA vector was transformed into Arabidopsis Col-0 using the Agrobacterium-tumefaciens-mediated floral dip method [75]. 35S:PAP1 transgenic plants were selected based on their soil using Basta, and successful transformation was confirmed through DNA genotyping until homozygous T3 transgenic progenies were obtained.
4.3. Phenotypic Observation of Seeds Color and Seed Size
To analyze seed color and seed size, the immature (10 and 12 DAP) and mature seeds of three Arabidopsis transgenic lines were randomly selected from major inflorescences and photographed using a SZ61 stereomicroscope (Olympus, Tokyo, Japan).
4.4. Determination of Anthocyanin and PAs Content
Anthocyanin measurement followed the protocol by Li et al. [76] with minor adjustments. Seeds were briefly frozen in liquid nitrogen, ground in a mortar, and approximately 5 mg of seed powder was placed in a 10 mL graduated test tube and incubated overnight at 4 °C in 3 mL methanol solution with 1% (v/v) HCl. After a 60 min incubation at 75 °C and cooling to room temperature, the sample was centrifuged for 15 min at 1500 rpm (HC-3018R, Zonkia, Anhui, China). The supernatant was mixed with 2 mL of distilled water and an equal volume of chloroform, followed by centrifugation (1500 rpm, 15 min, HC-3018R, Zonkia, Anhui, China). The supernatant’s absorbance was measured at 535 nm using a spectrophotometer (V-1200, Mapada, Shanghai, China) and the anthocyanin content was then normalized to the dry seed weight.
PAs extraction, adapted from Kitamura et al. [77], involved grinding mature seeds (10 mg) and mixing the powder with 1.5 mL of 70% (v/v) acetone extraction buffer containing 5.26 mM Na2S2O5. This mixture was sonicated using an ultrasonic bath (SB-5200 DT, Scientz, Ningbo, China) for 20 min at room temperature and then centrifuged at 1500 rpm for 15 min (HC-3018R, Zonkia, Anhui, China). The supernatant was dried and resuspended in HCl:butanol:70% acetone (2:10:3). The resulting absorbance was measured at 545 nm using an Infinite M200 PRO (Tecan). Following this, the solution was heated at 95 °C for 60 min and the absorbance at 545 nm was recorded again. The soluble PA fraction was calculated by subtracting the initial absorbance from the final one. The pellet obtained after 70% acetone extraction was dried via evaporation, resuspended in the HCl/butanol solution, and hydrolyzed to determine the insoluble PAs. Three independent biological replicates were conducted, each with three technical repetitions.
4.5. RNA-Seq Analysis
The RNA-seq experiment utilized developing seeds at 12 DAP from the major inflorescences of wild-type (Col-0) and Col-0 35S:PAP1 #5 over-expressing plants. Three independent biological replicates from each genotype were sequenced using BGI-Tech (Shenzhen, China), following the standard protocol (http://bgitechsolutions.com/sequencing/45 (accessed on 18 November 2019)). DEGs were identified using |log2 ratios| ≥ 1 and a false discovery rate (FDR) of ≤0.05 (Supplemental Tables S2 and S3). DEGs underwent functional classification based on the biological process category of Arabidopsis Gene Ontology (http://www.geneontology.com (accessed on 6 February 2020)).
4.6. RNA Extraction and RT-qPCR Analysis
Total RNA extraction from 12 DAP developing seeds was performed using the SteadyPure Plant RNA Extraction Kit (Accurate Biology, Changsha, China), followed by cDNA reverse transcription (TransGen, Beijing, China). RT-qPCR analysis was conducted with three independent biological replicates using SYBR Green Master Mix (Cofitt, Hongkong, China) on the QuantStudioTM 7 Flex Real-Time System (Thermo Fisher Scientific, Waltham, MA, USA). The Arabidopsis house-keeping gene EF1αA4 served as the internal control, and relative expression values of the target genes were calculated via normalization against EF1αA4 using a modified double-delta method [78]. The RT-qPCR primer details are listed in Supplemental Table S4.
4.7. ChIP-qPCR Assay
The ChIP-qPCR assay followed a previously described protocol [79]. Developing siliques (3−5 g) at 12 DAP were harvested from both wild-type (Col-0) and Col-0 35S:PAP1 #5 over-expressing plants. The samples underwent triple ddH2O washes and were cross-linked using 1% (v/v) formaldehyde (37 mL) under vacuum on ice for 15 min. Crosslinking was terminated by adding 2.5 mL of 2 M glycine. After being ground in liquid nitrogen, nuclear protein was separately extracted using sucrose-based buffers containing 0.4, 0.25, and 1.7 M sucrose. Chromatins were isolated, and DNA was sheared into 200–700 bp fragments through sonication with ultrasonic cell disruptors (Scientz-IID, Scientz, Ningbo, China). After centrifugation at 4 °C for 5 min at 12,000 rpm (Sorval LegendTM Micro 17, Thermo Fisher Scientific, Waltham, MA, USA), the chromatin remained in the upper aqueous phase. PAP1-6HA chromatin DNA was immunoprecipitated overnight using anti-HA magnetic beads (Thermo, USA) at 4 °C. The beads were washed and collected using a magnetic rack, and the immune complexes were eluted twice. Subsequently, the complexes were eluted and reversely crosslinked at 65 °C for 10 h in 5 M NaCl. Proteins were digested with 0.5 M EDTA, 1 M Tris-HCl (pH 6.5), and 3 mL of proteinase K (10 mg/mL) at 45 °C for 1 h. The DNA fragments were extracted using a Phenol/chloroform/isoamyl alcohol solution (25:24:1, pH > 7) and stored at −80 °C. The relative enrichment of each fragment was assessed via RT-qPCR. Each experiment involved three biological replicates with three technical replicates per biological replicate. Arabidopsis EF1αA4 and ACTIN7 served as the internal reference and negative control, respectively. The ChIP-qPCR assay primer details are provided in Supplemental Table S5.
4.8. Transient Dual-Luciferase Reporter Analysis
The PAP1 CDS was amplified and cloned into pGreenII 62-SK under the 35S promoter to form effector constructs. The effector constructs without PAP1 served as the empty control. Promoters for ADT5, CHS, F3H, DFR, ANS, 3GT, UGT79B2, UGT79B3, 5MAT, and GST26 were separately cloned into pGreenII 0800-LUC [80] to form reporter constructs. A. tumefaciens strain GV3101 was transformed with all the constructs along with pSoup-P19 (Weidi Biotechnology, Shanghai, China). Effector and reporter constructs were mixed in a buffer comprising 10 mM MgCl2, 10 mM MES-KOH (pH 5.8), and 10 μM acetosyringone in a 1:1 ratio and then injected into the young leaves of 4-week-old N. benthamiana. The infiltrated plants were cultured in a climate incubator (RXD-1000D-LED, Prandt, Ningbo, China) with a light/dark 16:8 h photoperiod cycle at 22 °C for 72 h. The firefly luciferase (LUC) and Renilla luciferase (REN, an internal control) activities were assessed using a dual-luciferase reporter assay kit (YEASEN, Shanghai, China) on a multifunctional enzyme label instrument (Spark®, Tecan, Männedorf, Switzerland). Six independent biological samples were examined. The primers for the dual-luc assay are provided in Supplemental Table S6.
4.9. Statistical Analysis
This study used a completely randomized design. Data were expressed as mean and standard deviation and analyzed using one-way analysis of variance (ANOVA) via SPSS software (version 17.0, SPSS Inc., Chicago, IL, USA). Significant differences were determined using a two-tailed paired Student’s t-test at the 0.05 significance level.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms242216049/s1.
Author Contributions
M.C. conceived the project and designed the experiments; Y.G., D.L. and T.L. performed most of the experiments and the data analyses; Y.L., J.L., M.H. and X.C. provided technical assistance; Z.L. advised on the data analyses; Y.G. and D.L. wrote the draft of the manuscript; M.C. reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Fundamental Research Funds for the Central Universities (grant No. 2452023090), the Key Research and Development Program of Shaanxi Province (grant No. 2022NY-158), and the Yang Ling Seed Industry Innovation Center (grants No. K3031122024 and K3031123009).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Chittka, L.; Raine, N.E. Recognition of flowers by pollinators. Curr. Opin. Plant Biol. 2006, 9, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Zoratti, L.; Sarala, M.; Carvalho, E.; Karppinen, K.; Martens, S.; Giongo, L.; Häggman, H.; Jaakola, L. Monochromatic light increases anthocyanin content during fruit development in bilberry. BMC Plant Biol. 2014, 14, 377. [Google Scholar] [CrossRef]
- Kim, S.; Hwang, G.; Lee, S.; Zhu, J.Y.; Paik, I.; Nguyen, T.T.; Kim, J.; Oh, E. High ambient temperature represses anthocyanin biosynthesis through degradation of HY5. Front. Plant Sci. 2017, 8, 1787. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef]
- Liang, J.; He, J. Protective role of anthocyanins in plants under low nitrogen stress. Biochem. Biophys. Res. Commun. 2018, 498, 946–953. [Google Scholar] [CrossRef]
- Gutha, L.R.; Casassa, L.F.; Harbertson, J.F.; Naidu, R.A. Modulation of flavonoid biosynthetic pathway genes and anthocyanins due to virus infection in grapevine (Vitis vinifera L.) leaves. BMC Plant Biol. 2010, 10, 187. [Google Scholar] [CrossRef]
- Ahmed, N.U.; Park, J.I.; Jung, H.J.; Hur, Y.; Nou, I.S. Anthocyanin biosynthesis for cold and freezing stress tolerance and desirable color in Brassica rapa. Funct. Integr. Genom. 2015, 15, 383–394. [Google Scholar] [CrossRef]
- Kim, J.; Lee, W.J.; Vu, T.T.; Jeong, C.Y.; Hong, S.W.; Lee, H. High accumulation of anthocyanins via the ectopic expression of AtDFR confers significant salt stress tolerance in Brassica napus L. Plant Cell Rep. 2017, 36, 1215–1224. [Google Scholar] [CrossRef]
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A comprehensive review of their chemical properties and health effects on cardiovascular and neurodegenerative diseases. Molecules 2020, 25, 3809. [Google Scholar] [CrossRef]
- Shi, M.Z.; Xie, D.Y. Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Pat. Biotechnol. 2014, 8, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, N.A.; Glover, B.J. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Xie, S.; Lei, Y.; Chen, H.; Li, J.; Chen, H.; Zhang, Z. R2R3-MYB transcription factors regulate anthocyanin biosynthesis in grapevine vegetative tissues. Front. Plant Sci. 2020, 11, 527. [Google Scholar] [CrossRef]
- Baumann, K.; Perez-Rodriguez, M.; Bradley, D.; Venail, J.; Bailey, P.; Jin, H.; Koes, R.; Roberts, K.; Martin, C. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 2007, 134, 1691–1701. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Li, S.; Zheng, Y.; Duan, H.; Xiao, F.; Zhuang, Y.; He, J.; Wu, G.; Zhao, S.; Zhou, H.; et al. SUMOylation of MYB30 enhances salt tolerance by elevating alternative respiration via transcriptionally upregulating AOX1a in Arabidopsis. Plant J. 2020, 102, 1157–1171. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Gao, Z.; Wang, J.; Huang, Y.; Chen, Y.; Li, J.; Lv, M.; Wang, J.; Luo, M.; Zuo, K. Cotton fiber elongation requires the transcription factor GhMYB212 to regulate sucrose transportation into expanding fibers. N. Phytol. 2019, 222, 864–881. [Google Scholar] [CrossRef]
- Costantini, L.; Malacarne, G.; Lorenzi, S.; Troggio, M.; Mattivi, F.; Moser, C.; Grando, M.S. New candidate genes for the fine regulation of the colour of grapes. J. Exp. Bot. 2015, 66, 4427–4440. [Google Scholar] [CrossRef]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Petersont, P.A.; Saedler, H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. Embo J. 1987, 6, 3353–3558. [Google Scholar] [CrossRef]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2000, 12, 2383–2393. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Chagné, D.; Lin-Wang, K.; Espley, R.V.; Volz, R.K.; How, N.M.; Rouse, S.; Brendolise, C.; Carlisle, C.M.; Kumar, S.; De Silva, N.; et al. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 2013, 161, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Niu, F.; Liu, W.Z.; Yang, B.; Zhang, J.; Ma, J.; Cheng, H.; Han, F.; Jiang, Y.Q. Identification, cloning and characterization of R2R3-MYB gene family in canola (Brassica napus L.) identify a novel member modulating ROS accumulation and hypersensitive-like cell death. DNA Res. 2016, 23, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Bioch. 2019, 136, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Constabel, C.P. MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Cominelli, E.; Bailey, P.; Parr, A.; Mehrtens, F.; Jones, J.; Tonelli, C.; Weisshaar, B.; Martin, C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. Embo J. 2000, 19, 6150–6161. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Micheletti, D.; Palmer, J.; Volz, R.; Lozano, L.; Espley, R.; Hellens, R.P.; Chagnè, D.; Rowan, D.D.; Troggio, M.; et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 2011, 34, 1176–1190. [Google Scholar] [CrossRef]
- Tohge, T.; Nishiyama, Y.; Hirai, M.Y.; Yano, M.; Nakajima, J.; Awazuhara, M.; Inoue, E.; Takahashi, H.; Goodenowe, D.B.; Kitayama, M.; et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005, 42, 218–235. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Fan, K.; Li, Z.; Jia, Q.; Lin, W.; Zhang, Y. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) negatively regulates anthocyanin accumulation by inhibiting PAP1 transcription in Arabidopsis seedlings. Plant Sci. 2021, 303, 110788. [Google Scholar] [CrossRef]
- Teng, S.; Keurentjes, J.; Bentsink, L.; Koornneef, M.; Smeekens, S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005, 139, 1840–1852. [Google Scholar] [CrossRef]
- Maier, A.; Schrader, A.; Kokkelink, L.; Falke, C.; Welter, B.; Iniesto, E.; Rubio, V.; Uhrig, J.F.; Hülskamp, M.; Hoecker, U. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 2013, 74, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, W.; Gao, J.; Yin, K.; Wang, R.; Wang, C.; Petersen, M.; Mundy, J.; Qiu, J.L. MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell 2016, 28, 2866–2883. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Tan, W.; Yang, H.; Zhang, L.; Li, T.; Liu, B.; Zhang, D.; Lin, H. Regulation of anthocyanin accumulation via MYB75/HAT1/TPL-mediated transcriptional repression. PLoS Genet. 2019, 15, e1007993. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhang, X.; Li, L.; Sun, Z.; Li, J.; Chen, X.; Hong, G. SPX4 interacts with both PHR1 and PAP1 to regulate critical steps in phosphorus-status-dependent anthocyanin biosynthesis. N. Phytol. 2021, 230, 205–217. [Google Scholar] [CrossRef]
- Qin, J.; Zhao, C.; Wang, S.; Gao, N.; Wang, X.; Na, X.; Wang, X.; Bi, Y. PIF4-PAP1 interaction affects MYB-bHLH-WD40 complex formation and anthocyanin accumulation in Arabidopsis. J. Plant Physiol. 2022, 268, 153558. [Google Scholar] [CrossRef]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef]
- Debeaujon, I.; Nesi, N.; Perez, P.; Devic, M.; Grandjean, O.; Caboche, M.; Lepiniec, L. Proanthocyanidin-accumulating cells in Arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell 2003, 15, 2514–2531. [Google Scholar] [CrossRef]
- Chen, Q.; Man, C.; Li, D.; Tan, H.; Xie, Y.; Huang, J. Arogenate dehydratase isoforms differentially regulate anthocyanin biosynthesis in Arabidopsis thaliana. Mol. Plant 2016, 9, 1609–1619. [Google Scholar] [CrossRef]
- Schilmiller, A.L.; Stout, J.; Weng, J.K.; Humphreys, J.; Ruegger, M.O.; Chapple, C. Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 2009, 60, 771–782. [Google Scholar] [CrossRef]
- Kim, J.I.; Hidalgo-Shrestha, C.; Bonawitz, N.D.; Franke, R.B.; Chapple, C. Spatio-temporal control of phenylpropanoid biosynthesis by inducible complementation of a cinnamate 4-hydroxylase mutant. J. Exp. Bot. 2021, 72, 3061–3073. [Google Scholar] [CrossRef]
- Li, Y.; Kim, J.I.; Pysh, L.; Chapple, C. Four isoforms of Arabidopsis 4-coumarate: CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 2015, 169, 2409–2421. [Google Scholar] [CrossRef]
- Bharti, A.K.; Khurana, J.P. Molecular characterization of transparent testa (tt) mutants of Arabidopsis thaliana (ecotype Estland) impaired in flavonoid biosynthetic pathway. Plant Sci. 2003, 165, 1321–1332. [Google Scholar] [CrossRef]
- Zhang, X.H.; Zheng, X.T.; Sun, B.Y.; Peng, C.L.; Chow, W.S. Over-expression of the CHS gene enhances resistance of Arabidopsis leaves to high light. Environ. Exp. Bot. 2018, 154, 33–43. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Ogneva, Z.V.; Kalachev, A.V.; Dubrovina, A.S. External dsRNA downregulates anthocyanin biosynthesis-related genes and affects anthocyanin accumulation in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 22, 6749. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Yin, Q.; Wu, R.; Zheng, G.; Liu, J.; Dixon, R.A.; Pang, Y. Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 7165–7179. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, L.; Irani, N.G.; Koo, A.J.K.; Bohorquez-Restrepo, A.; Howe, G.A.; Grotewold, E. A chemical complementation approach reveals genes and interactions of flavonoids with other pathways. Plant J. 2013, 74, 383–397. [Google Scholar] [CrossRef]
- Dai, M.; Kang, X.; Wang, Y.; Huang, S.; Guo, Y.; Wang, R.; Chao, N.; Liu, L. Functional characterization of Flavanone 3-Hydroxylase (F3H) and its role in anthocyanin and flavonoid biosynthesis in mulberry. Molecules 2022, 27, 3341. [Google Scholar] [CrossRef]
- Shih, C.H.; Chu, I.K.; Yip, W.K.; Lo, C. Differential expression of two Flavonoid 3′-Hydroxylase cDNAs involved in biosynthesis of anthocyanin pigments and 3-Deoxyanthocyanidin phytoalexins in sorghum. Plant Cell Physiol. 2006, 47, 1412–1419. [Google Scholar] [CrossRef]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Rosales-Mendoza, S.; Zheng, D.; Lygin, A.V.; Korban, S.S. Ectopic expression of apple F3′H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiol. 2010, 153, 806–820. [Google Scholar] [CrossRef]
- Appelhagen, I.; Jahns, O.; Bartelniewoehner, L.; Sagasser, M.; Weisshaar, B.; Stracke, R. Leucoanthocyanidin dioxygenase in Arabidopsis thaliana: Characterization of mutant alleles and regulation by MYB–BHLH–TTG1 transcription factor complexes. Gene 2011, 484, 61–68. [Google Scholar] [CrossRef]
- Abrahams, S.; Lee, E.; Walker, A.R.; Tanner, G.J.; Larkin, P.J.; Ashton, A.R. The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J. 2003, 35, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, W.; Hu, Y.; Zhang, Y.; Sun, J.; Guo, X.; Lu, H.; Yang, Y.; Fang, C.; Niu, X.; et al. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 2019, 99, 359–378. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, P.; Ma, X.; Zhao, W.; Li, M.; Yao, S.; Liu, Y.; Gao, L.; Xia, T. Exploration of the substrate diversity of leucoanthocyanidin reductases. J. Agric. Food Chem. 2020, 68, 3903–3911. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Dixon, R.A.; Duan, C. A role for ascorbate conjugates of (+)-catechin in proanthocyanidin polymerization. Nat. Commun. 2022, 13, 3425. [Google Scholar] [CrossRef]
- Yonekura-Sakakibara, K.; Fukushima, A.; Nakabayashi, R.; Hanada, K.; Matsuda, F.; Sugawara, S.; Inoue, E.; Kuromori, T.; Ito, T.; Shinozaki, K.; et al. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 2012, 69, 154–167. [Google Scholar] [CrossRef]
- Luo, J.; Nishiyama, Y.; Fuell, C.; Taguchi, G.; Elliott, K.; Hill, L.; Tanaka, Y.; Kitayama, M.; Yamazaki, M.; Bailey, P.; et al. Convergent evolution in the BAHD family of acyl transferases: Identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 2007, 50, 678–695. [Google Scholar] [CrossRef]
- D’Auria, J.C.; Reichelt, M.; Luck, K.; Svatoš, A.; Gershenzon, J. Identification and characterization of the BAHD acyltransferase malonyl CoA: Anthocyanidin 5-O-glucoside-6″-O-malonyltransferase (At5MAT) in Arabidopsis thaliana. FEBS Lett. 2007, 581, 872–878. [Google Scholar] [CrossRef]
- Kitamura, S.; Shikazono, N.; Tanaka, A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 2004, 37, 104–114. [Google Scholar] [CrossRef]
- Nesi, N.; Debeaujon, I.; Jond, C.; Pelletier, G.; Caboche, M.; Lepiniec, L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 2000, 12, 1863–1878. [Google Scholar] [CrossRef]
- Baudry, A.; Caboche, M.; Lepiniec, L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2006, 46, 768–779. [Google Scholar] [CrossRef]
- Walker, A.R.; Davison, P.A.; Bolognesi-Winfield, A.C.; James, C.M.; Srinivasan, N.; Blundell, T.L.; Esch, J.J.; Marks, M.D.; Gray, J.C. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 Repeat Protein. Plant Cell 1999, 11, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, H.; Wang, Y.; Guan, S.; Wang, F.; Tang, J.; Zhang, R.; Xie, L.; Lu, Y. Characterization of the cis elements in the proximal promoter regions of the anthocyanin pathway genes reveals a common regulatory logic that governs pathway regulation. J. Exp. Bot. 2015, 66, 3775–3789. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yu, W.; Xu, J.; Lu, X.; Liu, Y. Anthocyanin biosynthesis induced by MYB transcription factors in plants. Int. J. Mol. Sci. 2022, 23, 11701. [Google Scholar] [CrossRef]
- Tohge, T.; Matsui, K.; Ohme-Takagi, M.; Yamazaki, M.; Saito, K. Enhanced radical scavenging activity of genetically modified Arabidopsis seeds. Biotechnol. Lett. 2005, 27, 297–303. [Google Scholar] [CrossRef]
- Lee, W.J.; Jeong, C.Y.; Kwon, J.; Kien, V.V.; Lee, D.; Hong, S.W.; Lee, H. Drastic anthocyanin increase in response to PAP1 overexpression in fls1 knockout mutant confers enhanced osmotic stress tolerance in Arabidopsis thaliana. Plant Cell Rep. 2016, 35, 2369–2379. [Google Scholar] [CrossRef]
- Zheng, T.; Lu, X.; Yang, F.; Zhang, D. Synergetic modulation of plant cadmium tolerance via MYB75-mediated ROS homeostasis and transcriptional regulation. Plant Cell Rep. 2022, 41, 1515–1530. [Google Scholar] [CrossRef]
- Jiang, F.; Wang, J.Y.; Jia, H.F.; Jia, W.S.; Wang, H.Q.; Xiao, M. RNAi-mediated silencing of the flavanone 3-hydroxylase gene and its effect on flavonoid biosynthesis in strawberry Fruit. J. Plant Growth Regul. 2013, 32, 182–190. [Google Scholar] [CrossRef]
- Zheng, X.T.; Chen, Y.L.; Zhang, X.H.; Cai, M.L.; Yu, Z.C.; Peng, C.L. ANS-deficient Arabidopsis is sensitive to high light due to impaired anthocyanin photoprotection. Funct. Plant Biol. 2019, 46, 756–765. [Google Scholar] [CrossRef]
- Xu, W.; Grain, D.; Gourrierec, J.L.; Harscoët, E.; Berger, A.; Jauvion, V.; Scagnelli, A.; Berger, N.; Bidzinski, P.; Kelemen, Z.; et al. Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. N. Phytol. 2013, 198, 59–70. [Google Scholar] [CrossRef]
- Chalker-Scott, L. Environmental significance of anthocyanins in plant stress responses. Photochem. Photobiol. 1999, 70, 1–9. [Google Scholar] [CrossRef]
- Gu, X.Y.; Foley, M.E.; Horvath, D.P.; Anderson, J.V.; Feng, J.; Zhang, L.; Mowry, C.R.; Ye, H.; Suttle, J.C.; Kadowaki, K.; et al. Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice. Genetics 2011, 189, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Li, X.; Jia, J.; Yuan, G.; Chen, S.; Qi, D.; Cheng, L.; Liu, G. bHLH92 from sheepgrass acts as a negative regulator of anthocyanin/proanthocyandin accumulation and influences seed dormancy. J. Exp. Bot. 2019, 70, 269–284. [Google Scholar] [CrossRef]
- Li, D.; Jin, C.; Duan, S.; Zhu, Y.; Qi, S.; Liu, K.; Gao, C.; Ma, H.; Zhang, M.; Liao, Y.; et al. MYB89 transcription factor represses seed oil accumulation. Plant Physiol. 2017, 173, 1211–1225. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wu, H.; Ding, Q.; Li, H.; Li, Z.; Ding, J.; Li, Y. The heterologous expression of Arabidopsis PAP2 induces anthocyanin accumulation and inhibits plant growth in tomato. Funct. Integr. Genom. 2018, 18, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, S.; Matsuda, F.; Tohge, T.; Yonekura-Sakakibara, K.; Yamazaki, M.; Saito, K.; Narumi, I. Metabolic profiling and cytological analysis of proanthocyanidins in immature seeds of Arabidopsis thaliana flavonoid accumulation mutants. Plant J. 2010, 62, 549–559. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nuleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef]
- Bowler, C.; Benvenuto, G.; Laflamme, P.; Molino, D.; Probst, A.V.; Tariq, M.; Paszkowski, J. Chromatin techniques for plant cells. Plant J. 2004, 39, 776–789. [Google Scholar] [CrossRef]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).