Fitness Trade-Offs between Phage and Antibiotic Sensitivity in Phage-Resistant Variants: Molecular Action and Insights into Clinical Applications for Phage Therapy
Abstract
:1. Introduction
2. Virology of Phages
3. Phage Therapy within the Context of Antimicrobial Resistance
4. Adverse Effects of Phage Resistance on Therapeutic Outcomes
5. Molecular Mechanisms of Fitness Trade-Offs between Phage and Antibiotic Resistance
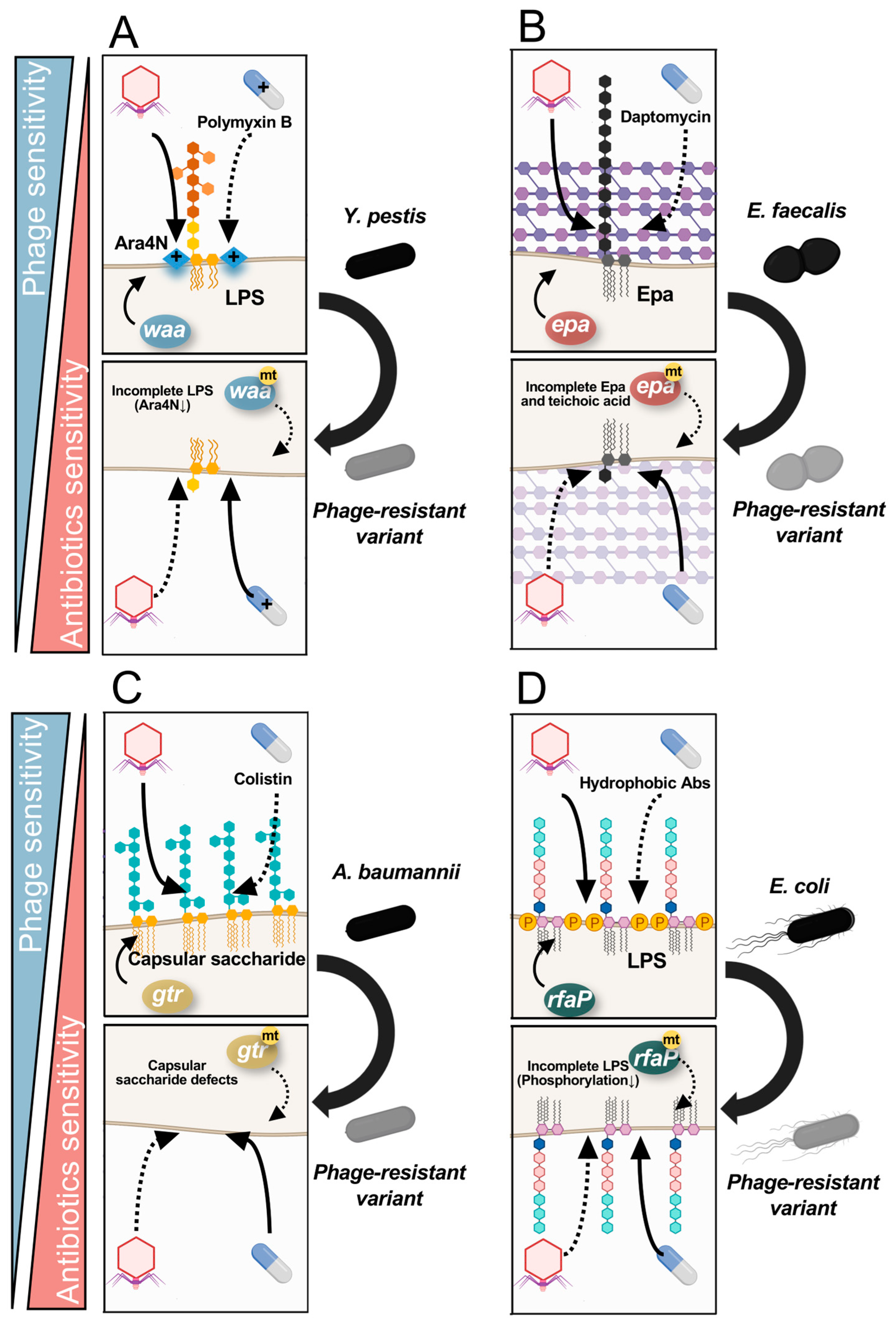
5.1. Fitness Trade-Offs via Saccharide
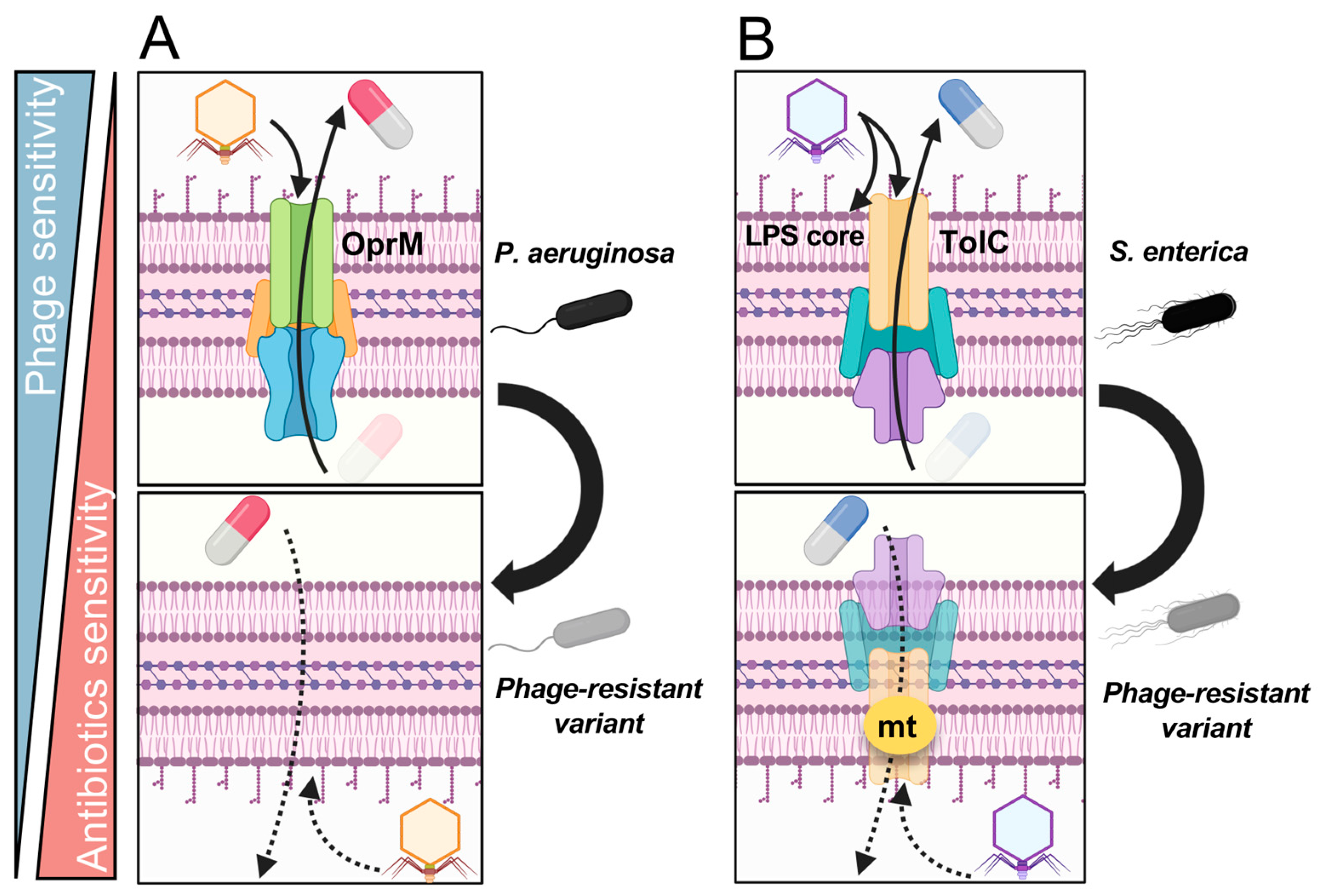
5.2. Fitness Trade-Offs via Drug Efflux Transporters
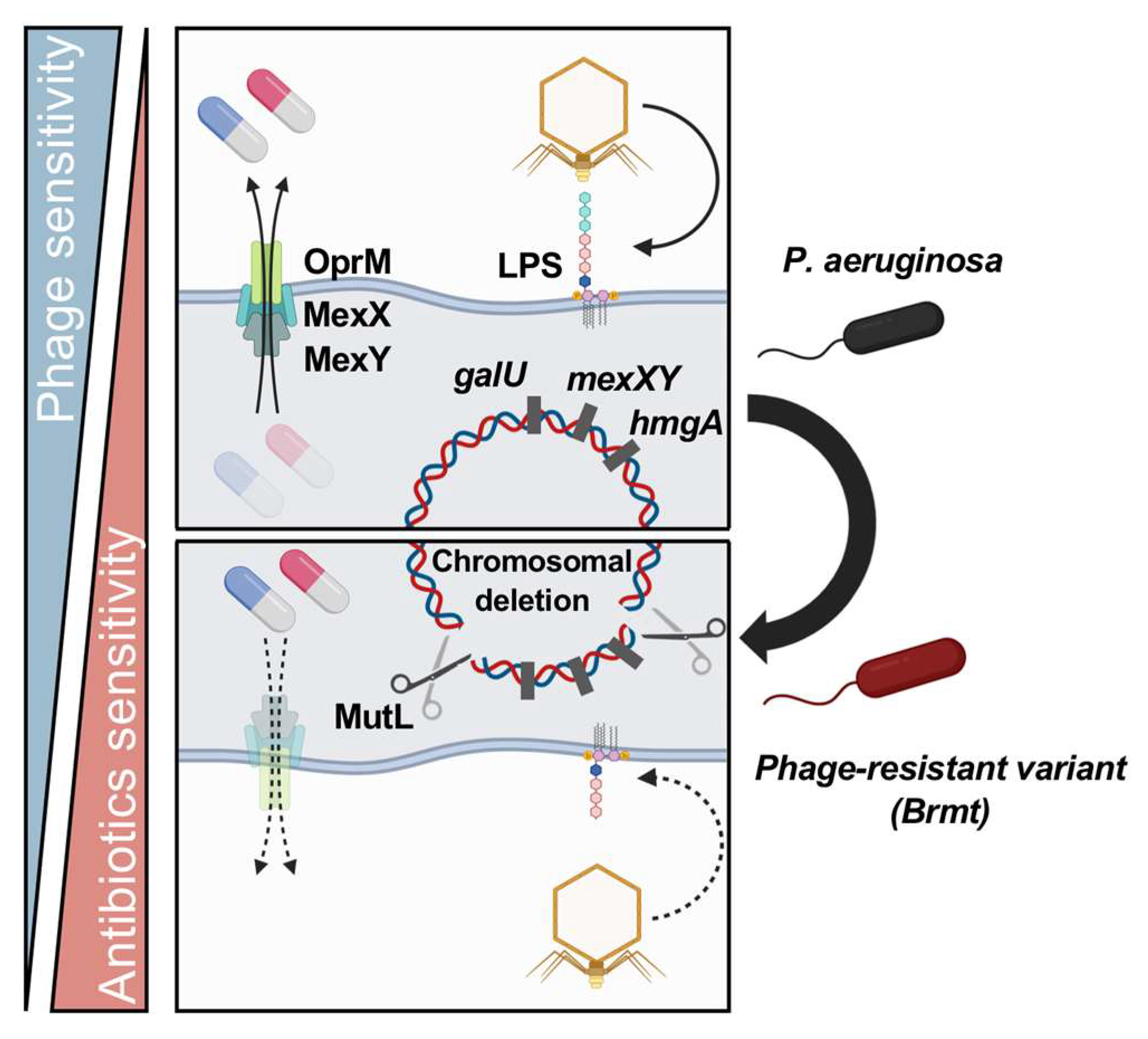
5.3. Fitness Trade-Offs via Large Chromosomal Deletions
6. Synergistic Interactions between Phages and Antibiotics in Phage Therapy
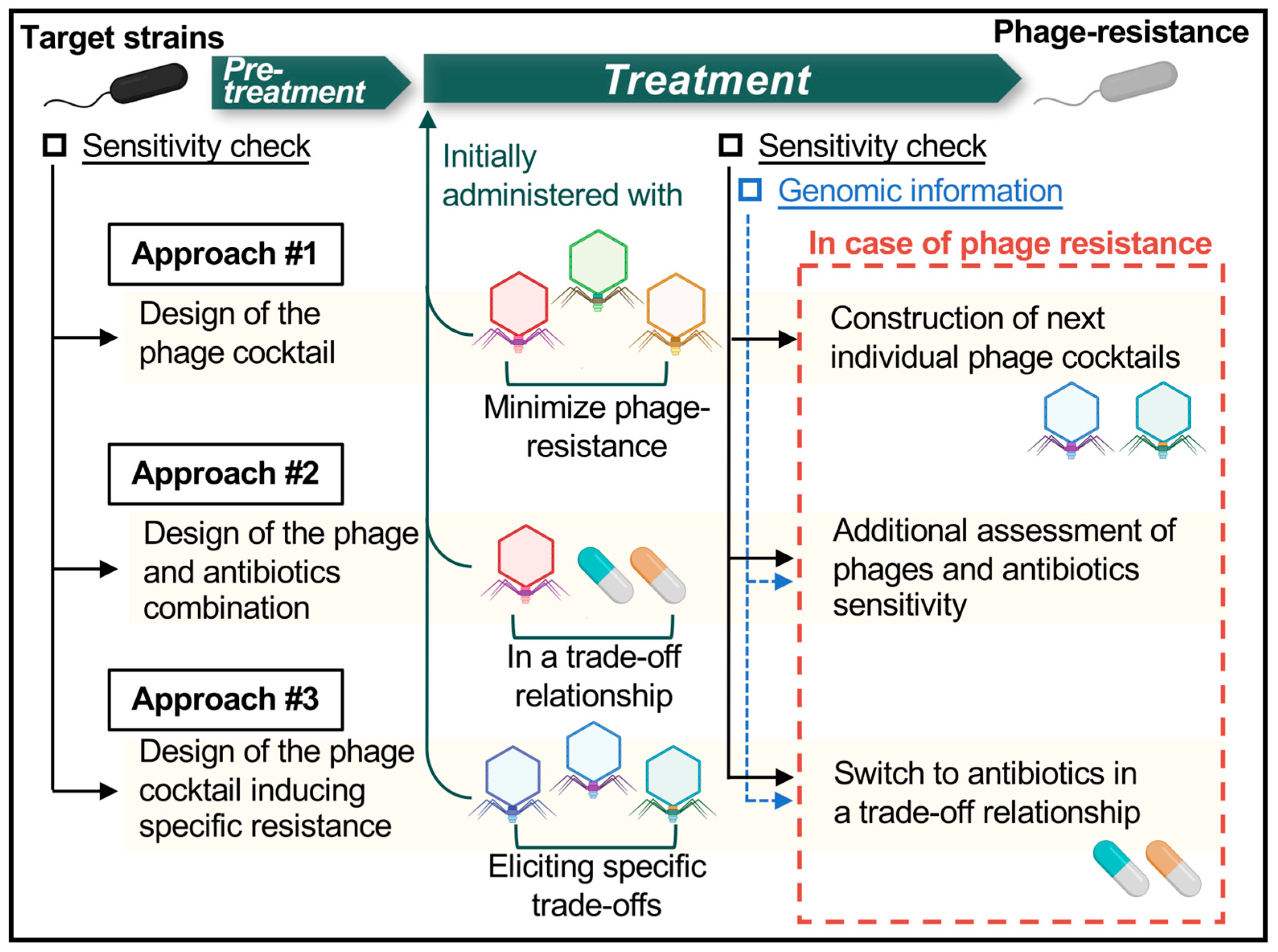
7. Future Perspectives in Phage Therapy
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Twort, F.W. An investigation on the nature of ultra-microscopic viruses. Lancet 1915, 186, 1241–1243. [Google Scholar] [CrossRef]
- D’Herelle, F. Sur un microbe invisible antagoniste des bacilles dysenteriques. C. R. Acad. Sci. 1917, 165, 373–375. (In French) [Google Scholar]
- Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. 1929. Bull. World Health Organ. 2001, 79, 780–790. [Google Scholar] [PubMed]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage therapy: From biological mechanisms to future directions. Cell 2023, 186, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef]
- Reardon, S. Phage therapy gets revitalized. Nature 2014, 510, 15–16. [Google Scholar] [CrossRef]
- Oromí-Bosch, A.; Antani, J.D.; Turner, P.E. Developing Phage Therapy That Overcomes the Evolution of Bacterial Resistance. Annu. Rev. Virol. 2023, 10, 503–524. [Google Scholar] [CrossRef]
- Hampton, H.G.; Watson, B.N.J.; Fineran, P.C. The arms race between bacteria and their phage foes. Nature 2020, 577, 327–336. [Google Scholar] [CrossRef]
- Oechslin, F. Resistance Development to Bacteriophages Occurring during Bacteriophage Therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef]
- Mangalea, M.R.; Duerkop, B.A. Fitness Trade-Offs Resulting from Bacteriophage Resistance Potentiate Synergistic Antibacterial Strategies. Infect. Immun. 2020, 88, e00926-19. [Google Scholar] [CrossRef]
- Mushegian, A.R. Are There 1031 Virus Particles on Earth, or More, or Fewer? J. Bacteriol. 2020, 202, e00052-20. [Google Scholar] [CrossRef] [PubMed]
- Parikka, K.J.; Le Romancer, M.; Wauters, N.; Jacquet, S. Deciphering the virus-to-prokaryote ratio (VPR): Insights into virus-host relationships in a variety of ecosystems. Biol. Rev. Camb. Philos. Soc. 2017, 92, 1081–1100. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Lang, S.; Demir, M.; Martin, A.; Jiang, L.; Zhang, X.; Duan, Y.; Gao, B.; Wisplinghoff, H.; Kasper, P.; Roderburg, C.; et al. Intestinal Virome Signature Associated with Severity of Nonalcoholic Fatty Liver Disease. Gastroenterology 2020, 159, 1839–1852. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, F.L.; Vlot, M.; de Jonge, P.A.; Dreesens, L.L.; Beaumont, H.J.E.; Lavigne, R.; Dutilh, B.E.; Brouns, S.J.J. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [CrossRef]
- Ackermann, H.W.; Prangishvili, D. Prokaryote viruses studied by electron microscopy. Arch. Virol. 2012, 157, 1843–1849. [Google Scholar] [CrossRef]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef]
- Abdelrahman, F.; Easwaran, M.; Daramola, O.I.; Ragab, S.; Lynch, S.; Oduselu, T.J.; Khan, F.M.; Ayobami, A.; Adnan, F.; Torrents, E.; et al. Phage-Encoded Endolysins. Antibiotics 2021, 10, 124. [Google Scholar] [CrossRef]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef]
- Khalil, R.K.; Skinner, C.; Patfield, S.; He, X. Phage-mediated Shiga toxin (Stx) horizontal gene transfer and expression in non-Shiga toxigenic Enterobacter and Escherichia coli strains. Pathog. Dis. 2016, 74, ftw037. [Google Scholar] [CrossRef]
- Kaiser, D.; Dworkin, M. Gene transfer to myxobacterium by Escherichia coli phage P1. Science 1975, 187, 653–654. [Google Scholar] [CrossRef] [PubMed]
- Ofir, G.; Sorek, R. Contemporary Phage Biology: From Classic Models to New Insights. Cell 2018, 172, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Tanji, Y.; Mizoguchi, K.; Akitsu, T.; Kijima, N.; Unno, H. Characterization of a virulent bacteriophage specific for Escherichia coli O157:H7 and analysis of its cellular receptor and two tail fiber genes. FEMS Microbiol. Lett. 2002, 211, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, J.; Suzuki, M.; Nishifuji, K.; Kato, S.I.; Miyata, R.; Nasukawa, T.; Yamaguchi, K.; Takemura-Uchiyama, I.; Ujihara, T.; Shimakura, H.; et al. Analyses of Short-Term Antagonistic Evolution of Pseudomonas aeruginosa Strain PAO1 and Phage KPP22 (Myoviridae Family, PB1-Like Virus Genus). Appl. Environ. Microbiol. 2016, 82, 4482–4491. [Google Scholar] [CrossRef] [PubMed]
- Xuan, G.; Dou, Q.; Kong, J.; Lin, H.; Wang, J. Pseudomonas aeruginosa Resists Phage Infection via Eavesdropping on Indole Signaling. Microbiol. Spectr. 2023, 11, e0391122. [Google Scholar] [CrossRef]
- Esteves, N.C.; Porwollik, S.; McClelland, M.; Scharf, B.E. The multi-drug efflux system AcrABZ-TolC is essential for infection of Salmonella Typhimurium by the flagellum-dependent bacteriophage Chi. J. Virol. 2021, 95, e00394-21. [Google Scholar] [CrossRef]
- Gao, D.; Ji, H.; Wang, L.; Li, X.; Hu, D.; Zhao, J.; Wang, S.; Tao, P.; Li, X.; Qian, P. Fitness Trade-Offs in Phage Cocktail-Resistant Salmonella enterica Serovar Enteritidis Results in Increased Antibiotic Susceptibility and Reduced Virulence. Microbiol. Spectr. 2022, 10, e0291422. [Google Scholar] [CrossRef]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef]
- Summers, W.C. Cholera and plague in India: The bacteriophage inquiry of 1927–1936. J. Hist. Med. Allied Sci. 1993, 48, 275–301. [Google Scholar] [CrossRef]
- Ho, K. Bacteriophage therapy for bacterial infections. Rekindling a memory from the pre-antibiotics era. Perspect. Biol. Med. 2001, 44, 1–16. [Google Scholar] [CrossRef]
- D’Herelle, F. Studies Upon Asiatic Cholera. Yale J. Biol. Med. 1929, 1, 195–219. [Google Scholar] [PubMed]
- Smith, J. The bacteriophage in the treatment of typhoid fever. Br. Med. J. 1924, 2, 47–49. [Google Scholar] [CrossRef] [PubMed]
- Hadley, P. The Twort-D’Herelle phenomenon: A critical review and presentation of a new conception (homogamic theory) of bacteriophage action. J. Infect. Dis. 1928, 42, 263–434. [Google Scholar] [CrossRef]
- Eaton, M.D.; Bayne-Jones, S. Bacteriophage therapy review of the principles and results of the use of bacteriophage in the treatment of infections. JAMA 1934, 103, 1769–1776. [Google Scholar] [CrossRef]
- Międzybrodzki, R.; Borysowski, J.; Weber-Dąbrowska, B.; Fortuna, W.; Letkiewicz, S.; Szufnarowski, K.; Pawełczyk, Z.; Rogóż, P.; Kłak, M.; Wojtasik, E.; et al. Clinical aspects of phage therapy. Adv. Virus Res. 2012, 83, 73–121. [Google Scholar] [PubMed]
- Fujiki, J.; Nakamura, T.; Nakamura, K.; Nishida, K.; Amano, Y.; Watanabe, Y.; Gondaira, S.; Usui, M.; Shimizu, M.; Miyanaga, K.; et al. Biological properties of Staphylococcus virus ΦSA012 for phage therapy. Sci. Rep. 2022, 12, 21297. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist. 2015, 1, 1–20. Available online: https://amr-review.org (accessed on 22 October 2023).
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails to Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2017, 61, e00954-17. [Google Scholar] [CrossRef]
- Aslam, S.; Lampley, E.; Wooten, D.; Karris, M.; Benson, C.; Strathdee, S.; Schooley, R.T. Lessons Learned From the First 10 Consecutive Cases of Intravenous Bacteriophage Therapy to Treat Multidrug-Resistant Bacterial Infections at a Single Center in the United States. Open Forum Infect. Dis. 2020, 7, ofaa389. [Google Scholar] [CrossRef]
- Pirnay, J.P.; Verbeken, G.; Ceyssens, P.J.; Huys, I.; De Vos, D.; Ameloot, C.; Fauconnier, A. The Magistral Phage. Viruses 2018, 10, 64. [Google Scholar] [CrossRef]
- Verbeken, G.; Pirnay, J.P. European regulatory aspects of phage therapy: Magistral phage preparations. Curr. Opin. Virol. 2022, 52, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.; Hawkins, C.H.; Anggård, E.E.; Harper, D.R. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 2009, 34, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.A.; Sultana, S.; Reuteler, G.; Moine, D.; Descombes, P.; Charton, F.; Bourdin, G.; McCallin, S.; Ngom-Bru, C.; Neville, T.; et al. Oral Phage Therapy of Acute Bacterial Diarrhea with Two Coliphage Preparations: A Randomized Trial in Children from Bangladesh. eBioMedicine 2016, 4, 124–137. [Google Scholar] [CrossRef]
- Jault, P.; Leclerc, T.; Jennes, S.; Pirnay, J.P.; Que, Y.A.; Resch, G.; Rousseau, A.F.; Ravat, F.; Carsin, H.; Le Floch, R.; et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): A randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 2019, 19, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Zhvania, P.; Hoyle, N.S.; Nadareishvili, L.; Nizharadze, D.; Kutateladze, M. Phage Therapy in a 16-Year-Old Boy with Netherton Syndrome. Front. Med. 2017, 4, 94. [Google Scholar] [CrossRef] [PubMed]
- Zaldastanishvili, E.; Leshkasheli, L.; Dadiani, M.; Nadareishvili, L.; Askilashvili, L.; Kvatadze, N.; Goderdzishvili, M.; Kutateladze, M.; Balarjishvili, N. Phage Therapy Experience at the Eliava Phage Therapy Center: Three Cases of Bacterial Persistence. Viruses 2021, 13, 1901. [Google Scholar] [CrossRef]
- Bao, J.; Wu, N.; Zeng, Y.; Chen, L.; Li, L.; Yang, L.; Zhang, Y.; Guo, M.; Li, L.; Li, J.; et al. Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae. Emerg. Microbes Infect. 2020, 9, 771–774. [Google Scholar] [CrossRef]
- Aslam, S.; Courtwright, A.M.; Koval, C.; Lehman, S.M.; Morales, S.; Furr, C.L.; Rosas, F.; Brownstein, M.J.; Fackler, J.R.; Sisson, B.M.; et al. Early clinical experience of bacteriophage therapy in 3 lung transplant recipients. Am. J. Transplant. 2019, 19, 2631–2639. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Freeman, K.G.; Nguyen, J.A.; Bahadirli-Talbott, A.; Smith, B.E.; Wu, A.E.; Ong, A.S.; Lin, C.T.; Ruppel, L.C.; Parrish, N.M.; et al. Potent antibody-mediated neutralization limits bacteriophage treatment of a pulmonary Mycobacterium abscessus infection. Nat. Med. 2021, 27, 1357–1361. [Google Scholar] [CrossRef]
- Blasco, L.; López-Hernández, I.; Rodríguez-Fernández, M.; Pérez-Florido, J.; Casimiro-Soriguer, C.S.; Djebara, S.; Merabishvili, M.; Pirnay, J.P.; Rodríguez-Baño, J.; Tomás, M.; et al. Case report: Analysis of phage therapy failure in a patient with a Pseudomonas aeruginosa prosthetic vascular graft infection. Front. Med. 2023, 10, 1199657. [Google Scholar] [CrossRef]
- Guerrero-Bustamante, C.A.; Dedrick, R.M.; Garlena, R.A.; Russell, D.A.; Hatfull, G.F. Toward a Phage Cocktail for Tuberculosis: Susceptibility and Tuberculocidal Action of Mycobacteriophages against Diverse Mycobacterium tuberculosis Strains. mBio 2021, 12, e00973-21. [Google Scholar] [CrossRef] [PubMed]
- Naknaen, A.; Samernate, T.; Wannasrichan, W.; Surachat, K.; Nonejuie, P.; Chaikeeratisak, V. Combination of genetically diverse Pseudomonas phages enhances the cocktail efficiency against bacteria. Sci. Rep. 2023, 13, 8921. [Google Scholar] [CrossRef]
- Zavascki, A.P.; Goldani, L.Z.; Li, J.; Nation, R.L. Polymyxin B for the treatment of multidrug-resistant pathogens: A critical review. J. Antimicrob. Chemother. 2007, 60, 1206–1215. [Google Scholar] [CrossRef]
- Velkov, T.; Thompson, P.E.; Nation, R.L.; Li, J. Structure—Activity relationships of polymyxin antibiotics. J. Med. Chem. 2010, 53, 1898–1916. [Google Scholar] [CrossRef]
- Rebeil, R.; Ernst, R.K.; Gowen, B.B.; Miller, S.I.; Hinnebusch, B.J. Variation in lipid A structure in the pathogenic yersiniae. Mol. Microbiol. 2004, 52, 1363–1373. [Google Scholar] [CrossRef] [PubMed]
- Nizet, V. Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr. Issues Mol. Biol. 2006, 8, 11–26. [Google Scholar] [PubMed]
- Anisimov, A.P.; Dentovskaya, S.V.; Titareva, G.M.; Bakhteeva, I.V.; Shaikhutdinova, R.Z.; Balakhonov, S.V.; Lindner, B.; Kocharova, N.A.; Senchenkova, S.N.; Holst, O.; et al. Intraspecies and temperature-dependent variations in susceptibility of Yersinia pestis to the bactericidal action of serum and to polymyxin B. Infect. Immun. 2005, 73, 7324–7331. [Google Scholar] [CrossRef]
- Knirel, Y.A.; Lindner, B.; Vinogradov, E.V.; Kocharova, N.A.; Senchenkova, S.N.; Shaikhutdinova, R.Z.; Dentovskaya, S.V.; Fursova, N.K.; Bakhteeva, I.V.; Titareva, G.M.; et al. Temperature-dependent variations and intraspecies diversity of the structure of the lipopolysaccharide of Yersinia pestis. Biochemistry 2005, 44, 1731–1743. [Google Scholar] [CrossRef]
- Anisimov, A.P.; Dentovskaya, S.V.; Kondakova, A.N.; Lindner, B.; Shaikhutdinova, R.Z.; Kocharova, N.A.; Senchenkova, S.Y.N.; Knirel, Y.A. Yersinia pestis lipopolysaccharide in host-pathogen interactions. In The Challenge of Highly Pathogenic Microorganisms: Mechanisms of Virulence and Novel Medical Countermeasures London; Springer: New York, NY, USA, 2010; pp. 77–87. [Google Scholar]
- Filippov, A.A.; Sergueev, K.V.; He, Y.; Huang, X.Z.; Gnade, B.T.; Mueller, A.J.; Fernandez-Prada, C.M.; Nikolich, M.P. Bacteriophage-resistant mutants in Yersinia pestis: Identification of phage receptors and attenuation for mice. PLoS ONE 2011, 6, e25486. [Google Scholar] [CrossRef]
- Canfield, G.S.; Duerkop, B.A. Molecular mechanisms of enterococcal-bacteriophage interactions and implications for human health. Curr. Opin. Microbiol. 2020, 56, 38–44. [Google Scholar] [CrossRef]
- Ho, K.; Huo, W.; Pas, S.; Dao, R.; Palmer, K.L. Loss-of-Function Mutations in epaR Confer Resistance to ϕNPV1 Infection in Enterococcus faecalis OG1RF. Antimicrob. Agents Chemother. 2018, 62, e00758-18. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Johnson, C.N.; Luong, P.; Hullahalli, K.; McBride, S.W.; Schubert, A.M.; Palmer, K.L.; Carlson, P.E., Jr.; Duerkop, B.A. Bacteriophage Resistance Alters Antibiotic-Mediated Intestinal Expansion of Enterococci. Infect. Immun. 2019, 87, e00085-19. [Google Scholar] [CrossRef] [PubMed]
- Dale, J.L.; Cagnazzo, J.; Phan, C.Q.; Barnes, A.M.; Dunny, G.M. Multiple roles for Enterococcus faecalis glycosyltransferases in biofilm-associated antibiotic resistance, cell envelope integrity, and conjugative transfer. Antimicrob. Agents Chemother. 2015, 59, 4094–4105. [Google Scholar] [CrossRef]
- Canfield, G.S.; Chatterjee, A.; Espinosa, J.; Mangalea, M.R.; Sheriff, E.K.; Keidan, M.; McBride, S.W.; McCollister, B.D.; Hang, H.C.; Duerkop, B.A. Lytic bacteriophages facilitate antibiotic sensitization of Enterococcus faecium. Antimicrob. Agents Chemother. 2023, 65, e00143-21. [Google Scholar] [CrossRef] [PubMed]
- Guerardel, Y.; Sadovskaya, I.; Maes, E.; Furlan, S.; Chapot-Chartier, M.P.; Mesnage, S.; Rigottier-Gois, L.; Serror, P. Complete Structure of the Enterococcal Polysaccharide Antigen (EPA) of Vancomycin-Resistant Enterococcus faecalis V583 Reveals that EPA Decorations Are Teichoic Acids Covalently Linked to a Rhamnopolysaccharide Backbone. mBio 2020, 11, e00277-20. [Google Scholar] [CrossRef]
- Gaupp, R.; Lei, S.; Reed, J.M.; Peisker, H.; Boyle-Vavra, S.; Bayer, A.S.; Bischoff, M.; Herrmann, M.; Daum, R.S.; Powers, R.; et al. Staphylococcus aureus metabolic adaptations during the transition from a daptomycin susceptibility phenotype to a daptomycin nonsusceptibility phenotype. Antimicrob. Agents Chemother. 2015, 59, 4226–4238. [Google Scholar] [CrossRef]
- Bertsche, U.; Weidenmaier, C.; Kuehner, D.; Yang, S.J.; Baur, S.; Wanner, S.; Francois, P.; Schrenzel, J.; Yeaman, M.R.; Bayer, A.S. Correlation of daptomycin resistance in a clinical Staphylococcus aureus strain with increased cell wall teichoic acid production and D-alanylation. Antimicrob. Agents Chemother. 2011, 55, 3922–3928. [Google Scholar] [CrossRef]
- Mechler, L.; Bonetti, E.J.; Reichert, S.; Flötenmeyer, M.; Schrenzel, J.; Bertram, R.; François, P.; Götz, F. Daptomycin Tolerance in the Staphylococcus aureus pitA6 Mutant Is Due to Upregulation of the dlt Operon. Antimicrob. Agents Chemother. 2016, 60, 2684–2691. [Google Scholar] [CrossRef]
- Mello, S.S.; Van Tyne, D.; Lebreton, F.; Silva, S.Q.; Nogueira, M.C.L.; Gilmore, M.S.; Camargo, I. A mutation in the glycosyltransferase gene lafB causes daptomycin hypersusceptibility in Enterococcus faecium. J. Antimicrob. Chemother. 2020, 75, 36–45. [Google Scholar] [CrossRef]
- Popova, A.V.; Lavysh, D.G.; Klimuk, E.I.; Edelstein, M.V.; Bogun, A.G.; Shneider, M.M.; Goncharov, A.E.; Leonov, S.V.; Severinov, K.V. Novel Fri1-like Viruses Infecting Acinetobacter baumannii-vB_AbaP_AS11 and vB_AbaP_AS12-Characterization, Comparative Genomic Analysis, and Host-Recognition Strategy. Viruses 2017, 9, 188. [Google Scholar] [CrossRef]
- Gordillo Altamirano, F.; Forsyth, J.H.; Patwa, R.; Kostoulias, X.; Trim, M.; Subedi, D.; Archer, S.K.; Morris, F.C.; Oliveira, C.; Kielty, L.; et al. Bacteriophage-resistant Acinetobacter baumannii are resensitized to antimicrobials. Nat. Microbiol. 2021, 6, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Loh, B.; Gordillo Altamirano, F.; Yu, Y.; Hua, X.; Leptihn, S. Colistin-phage combinations decrease antibiotic resistance in Acinetobacter baumannii via changes in envelope architecture. Emerg. Microbes Infect. 2021, 10, 2205–2219. [Google Scholar] [CrossRef] [PubMed]
- Yethon, J.A.; Heinrichs, D.E.; Monteiro, M.A.; Perry, M.B.; Whitfield, C. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J. Biol. Chem. 1998, 273, 26310–26316. [Google Scholar] [CrossRef] [PubMed]
- Pagnout, C.; Sohm, B.; Razafitianamaharavo, A.; Caillet, C.; Offroy, M.; Leduc, M.; Gendre, H.; Jomini, S.; Beaussart, A.; Bauda, P.; et al. Pleiotropic effects of rfa-gene mutations on Escherichia coli envelope properties. Sci. Rep. 2019, 9, 9696. [Google Scholar] [CrossRef] [PubMed]
- McGee, L.W.; Barhoush, Y.; Shima, R.; Hennessy, M. Phage-resistant mutations impact bacteria susceptibility to future phage infections and antibiotic response. Ecol. Evol. 2023, 13, e9712. [Google Scholar] [CrossRef]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef]
- Gurney, J.; Pradier, L.; Griffin, J.S.; Gougat-Barbera, C.; Chan, B.K.; Turner, P.E.; Kaltz, O.; Hochberg, M.E. Phage steering of antibiotic-resistance evolution in the bacterial pathogen, Pseudomonas aeruginosa. Evol. Med. Public Health 2020, 2020, 148–157. [Google Scholar] [CrossRef]
- Burmeister, A.R.; Fortier, A.; Roush, C.; Lessing, A.J.; Bender, R.G.; Barahman, R.; Grant, R.; Chan, B.K.; Turner, P.E. Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 11207–11216. [Google Scholar] [CrossRef]
- Shen, M.; Zhang, H.; Shen, W.; Zou, Z.; Lu, S.; Li, G.; He, X.; Agnello, M.; Shi, W.; Hu, F.; et al. Pseudomonas aeruginosa MutL promotes large chromosomal deletions through non-homologous end joining to prevent bacteriophage predation. Nucleic Acids Res. 2018, 46, 4505–4514. [Google Scholar] [CrossRef]
- Le, S.; Yao, X.; Lu, S.; Tan, Y.; Rao, X.; Li, M.; Jin, X.; Wang, J.; Zhao, Y.; Wu, N.C.; et al. Chromosomal DNA deletion confers phage resistance to Pseudomonas aeruginosa. Sci. Rep. 2014, 4, 4738. [Google Scholar] [CrossRef]
- Sanz-García, F.; Hernando-Amado, S.; Martínez, J.L. Mutation-Driven Evolution of Pseudomonas aeruginosa in the Presence of either Ceftazidime or Ceftazidime-Avibactam. Antimicrob. Agents Chemother. 2018, 62, e01379-18. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Amado, S.; Sanz-García, F.; Martínez, J.L. Rapid and robust evolution of collateral sensitivity in Pseudomonas aeruginosa antibiotic-resistant mutants. Sci. Adv. 2020, 6, eaba5493. [Google Scholar] [CrossRef]
- Nakamura, K.; Fujiki, J.; Nakamura, T.; Furusawa, T.; Gondaira, S.; Usui, M.; Higuchi, H.; Tamura, Y.; Iwano, H. Fluctuating Bacteriophage-induced galU Deficiency Region is Involved in Trade-off Effects on the Phage and Fluoroquinolone Sensitivity in Pseudomonas aeruginosa. Virus Res. 2021, 306, 198596. [Google Scholar] [CrossRef]
- Koderi Valappil, S.; Shetty, P.; Deim, Z.; Terhes, G.; Urbán, E.; Váczi, S.; Patai, R.; Polgár, T.; Pertics, B.Z.; Schneider, G.; et al. Survival Comes at a Cost: A Coevolution of Phage and Its Host Leads to Phage Resistance and Antibiotic Sensitivity of Pseudomonas aeruginosa Multidrug Resistant Strains. Front. Microbiol. 2021, 12, 783722. [Google Scholar] [CrossRef]
- Menon, N.D.; Penziner, S.; Montaño, E.T.; Zurich, R.; Pride, D.T.; Nair, B.G.; Kumar, G.B.; Nizet, V. Increased Innate Immune Susceptibility in Hyperpigmented Bacteriophage-Resistant Mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2022, 66, e0023922. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Willett, J.L.E.; Nguyen, U.T.; Monogue, B.; Palmer, K.L.; Dunny, G.M.; Duerkop, B.A. Parallel Genomics Uncover Novel Enterococcal-Bacteriophage Interactions. mBio 2020, 11, e03120-19. [Google Scholar] [CrossRef]
- Van Nieuwenhuyse, B.; Van der Linden, D.; Chatzis, O.; Lood, C.; Wagemans, J.; Lavigne, R.; Schroven, K.; Paeshuyse, J.; de Magnée, C.; Sokal, E.; et al. Bacteriophage-antibiotic combination therapy against extensively drug-resistant Pseudomonas aeruginosa infection to allow liver transplantation in a toddler. Nat. Commun. 2022, 13, 5725. [Google Scholar] [CrossRef] [PubMed]
- Law, N.; Logan, C.; Yung, G.; Furr, C.L.; Lehman, S.M.; Morales, S.; Rosas, F.; Gaidamaka, A.; Bilinsky, I.; Grint, P.; et al. Successful adjunctive use of bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa infection in a cystic fibrosis patient. Infection 2019, 47, 665–668. [Google Scholar] [CrossRef]
- Chan, B.K.; Turner, P.E.; Kim, S.; Mojibian, H.R.; Elefteriades, J.A.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 2018, 60–66. [Google Scholar] [CrossRef]
- Nir-Paz, R.; Gelman, D.; Khouri, A.; Sisson, B.M.; Fackler, J.; Alkalay-Oren, S.; Khalifa, L.; Rimon, A.; Yerushalmy, O.; Bader, R.; et al. Successful Treatment of Antibiotic-resistant, Poly-microbial Bone Infection with Bacteriophages and Antibiotics Combination. Clin. Infect. Dis. 2019, 69, 2015–2018. [Google Scholar] [CrossRef]
- Willy, C.; Bugert, J.J.; Classen, A.Y.; Deng, L.; Düchting, A.; Gross, J.; Hammerl, J.A.; Korf, I.H.E.; Kühn, C.; Lieberknecht-Jouy, S.; et al. Phage Therapy in Germany-Update 2023. Viruses 2023, 15, 588. [Google Scholar] [CrossRef] [PubMed]
- Rubalskii, E.; Ruemke, S.; Salmoukas, C.; Boyle, E.C.; Warnecke, G.; Tudorache, I.; Shrestha, M.; Schmitto, J.D.; Martens, A.; Rojas, S.V.; et al. Bacteriophage Therapy for Critical Infections Related to Cardiothoracic Surgery. Antibiotics 2020, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Borin, J.M.; Lee, J.J.; Gerbino, K.R.; Meyer, J.R. Comparison of bacterial suppression by phage cocktails, dual-receptor generalists, and coevolutionarily trained phages. Evol. Appl. 2023, 16, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Burrowes, B.H.; Molineux, I.J.; Fralick, J.A. Directed in Vitro Evolution of Therapeutic Bacteriophages: The Appelmans Protocol. Viruses 2019, 11, 241. [Google Scholar] [CrossRef]
- Mitsunaka, S.; Yamazaki, K.; Pramono, A.K.; Ikeuchi, M.; Kitao, T.; Ohara, N.; Kubori, T.; Nagai, H.; Ando, H. Synthetic engineering and biological containment of bacteriophages. Proc. Natl. Acad. Sci. USA 2022, 119, e2206739119. [Google Scholar] [CrossRef]
- Kiga, K.; Tan, X.E.; Ibarra-Chávez, R.; Watanabe, S.; Aiba, Y.; Sato’o, Y.; Li, F.Y.; Sasahara, T.; Cui, B.; Kawauchi, M.; et al. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat. Commun. 2020, 11, 2934. [Google Scholar] [CrossRef]
- Sanmukh, S.G.; Admella, J.; Moya-Andérico, L.; Fehér, T.; Arévalo-Jaimes, B.V.; Blanco-Cabra, N.; Torrents, E. Accessing the In Vivo Efficiency of Clinically Isolated Phages against Uropathogenic and Invasive Biofilm-Forming Escherichia coli Strains for Phage Therapy. Cells 2023, 12, 344. [Google Scholar] [CrossRef]
| Target Bacteria | Disease | Phage and Dose | Trial Design and Treatment Method | Pre-Phage Sensitivity | Post-Phage Sensitivity | Phage Resistance Analysis | Outcome | Ref. |
|---|---|---|---|---|---|---|---|---|
| P. aeruginosa | Otitis | 6 phages (109 PFUs) Single dose —intra-aural administration | Treatment group —12 individuals received phages Placebo group —12 individuals received glycerol-PBS buffer Double blind | Checked | Not checked | Not performed | Significant differences in terms of clinical improvement scores and levels of P. aeruginosa were observed between the phage-treated group and the placebo control. No adverse effects. | [42] |
| E. coli | Diarrhea | 11 T4-like phages (3.6 × 108 PFUs) or ColiProteus (1.4 × 109) Three times/day for 4 days (12 doses) —oral administration without gastric neutralization | Treatment group —39 individuals received 11 T4-like phages —40 individuals received ColiProteus Placebo group —41 individuals received rehydration solution Double blind | Not checked | Checked | Not performed | There were no indications of phage efficacy on diarrhea parameters in the treatment group, and as a result, the trial was terminated early. No adverse effects. | [43] |
| P. aeruginosa | Burn wound infection | 12 phages (2 × 107 PFUs were expected but 200–2000 PFUs were observed) Once/day for 7 days (7 doses) —topical administration | Treatment group —12 individuals received phages Placebo group —13 individuals received standard of care (1% sulfadiazine silver) Blind | Not checked | Checked | Not performed | Phage treatment resulted in a slower reduction in bacterial load in burn wounds compared to the standard of care. The administered phage dose was significantly lower than the expected dose, leading to the trial being halted. | [44] |
| Case | Year | Institution and Country | Target Bacteria | Disease | Patient | Phages | Dose and Treatment | Occurrence of Phage Resistance | Impact of Phage Resistance | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2016 | University of California, San Diego, USA | A. baumannii | Necrotizing pancreatitis | Male, 68-years-old | Phage cocktail (ΦPC, 4 phages, Therapeutic dose was not applicable -used for intracavitary wash) Phage cocktail (ΦIV, 4 phages, 5 × 109 PFUs) | ΦPC: percutaneous catheters drained the pseudocyst cavity, the biliary cavity, and a third intra-abdominal cavity for 18 weeks ΦIV: intravenous administration for 16 weeks | TP3, which was isolated 8 days after the initial administration of phage cocktails (ΦPC and ΦIV), exhibited resistance to both of the cocktails | Alternative personalized phage cocktails were constructed against resistant variants. | [38,39] |
| 2 | 2016 | Eliava Phage Therapy Center, Georgia | S. aureus | Netherton syndrome | Male, 16-years-old | Staphylococcus phage Sb1 and Pyo phage cocktail contains phages active against S. aureus | Oral administration of both 10 mL once a day and spray (Pyo) and cream (Sb1) for 40 days in total, next second 3 month period, phages for 2 weeks, with 2 week breaks in between treatment courses | After 3 months of treatment, phage sensitivity profiles showed resistance to Pyo phage cocktail | Alternative personalized phage cocktails were constructed against resistant variants. | [45] |
| 3 | 2017 | Eliava Phage Therapy Center, Georgia | P. aeruginosa | Cystic fibrosis | Male, 43-years-old | Pyo and Intesti phage cocktails (both of which contain phages active against P. aeruginosa) | 8 mL taken orally once a day and 2 mL via nebulizer | Throughout the initial trimester of treatment for 3 months, the patient’s P. aeruginosa strains gradually developed resistance to Pyo and Intesti phage cocktails | Alternative personalized phage cocktails were constructed against resistant variants. | [46] |
| 4 | 2019 | Shanghai Public Health Clinical Center, China | K. pneumoniae | Urinary tract infection | Female, 63-years-old | Phage cocktail I (5 phages, 5 × 108 PFUs/mL of each phage) Phage cocktail II (5 phages, active against phage-resistant variants against cocktail I) | Phage cocktail I: 10 mL bladder irrigation, daily for 5 days Phage cocktail II: 50 mL bladder irrigation, daily for 5 days | In two rounds of phage therapy (1st using phage cocktail I, 2nd using phage cocktail II), phage-resistant mutants developed within days in both of rounds | As reemergence of phage-resistant was repeated, novel therapeutic strategies are needed. Eventually, by combination of phage and antibiotics, the patient was cured. | [47] |
| 5 | Reported in 2020 | University of California, San Diego, USA | P. aeruginosa | Lower respiratory tract infections | Male, 67-years-old | Phage cocktail AB-PA01 (4 phages, 4 × 109 PFUs/mL) | Intravenous administration every 6 h and nebulized every 12 h for 4 weeks. | One isolate resistant to AB-PA01 appeared after cessation of AB-PA01 | A personalized approach was required to identify new phages active against the resistant isolates. | [39,48] |
| 6 | Reported in 2020 | University of California, San Diego, USA | P. aeruginosa | Ventricular assist device infection | Male, 82-years-old | Phage cocktail SDSU1 and 2 (2 phages, respectively, 2 × 105 PFUs/mL), | Intravenous administration every 8 h for 6 weeks, +1-time intraoperative dose followed by one phage alone for 10 days and followed by SDSU2 IV every 12 h for 3 weeks | During phage therapy, phage resistance developed. | Additional new phages, in a personalized treatment approach, were required to account for resistant isolates. | [39] |
| 7 | Reported in 2021 | Johns Hopkins University, USA | M. abscessus | Refractory abscessus lung disease | N/A, 81-years-old | Phage cocktail (3 phages, 2 × 109 PFUs) | Intravenous administration twice daily for 6 month | Post-treatment M. abscessus isolates demonstrated emergence of phage resistance against one phage in the cocktail | Although phage resistance was observed, phage therapy failed mainly due to potent antibody-mediated neutralization. | [49] |
| 8 | 2021 | Not specified, Spain | P. aeruginosa | Prosthetic vascular graft infection | Male, N/A | Phage cocktail (3 phages, 107 PFUs/mL) | Intravenous administration of 70 mL phage cocktail, once a day in a 6-h infusion for 1 week | The post-phage therapy isolate exhibited phage resistance | Failed to eradicate the infection by the phage cocktail with ceftazidime-avibactam. After phage therapy, a proximal vascular prosthesis replacement combined with antibiotic therapy was performed, and patient remains asymptomatic. | [50] |
| Case | Year | Institution and Country | Target Bacteria | Disease | Patient | Phages | Combined Antibiotics | Outcomes | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2015 | Hannover Medical School, Germany | S. aureus E. faecium P. aeruginosa | Prosthetic infection after aortic arch replacement | Male, 52-years-old | Phage cocktail (CH1 against S. aureus, Enf1 against E. faecium, PA5 and PA10 against P. aeruginosa, 108 PFUs/mL), topical administration via drainage and intraoperatively. One oral administration | Gentamicin and daptomycin (with drainage), cefepime, daptomycin, linezolid, tobramycin (iv) | S. aureus, E. faecium and P. aeruginosa were not detected after phage and antibiotic treatment. | [93] |
| 2 | 2016 | Yale University, USA | P. aeruginosa | Chronic infection in aortic Dacron graft with aorto-cutaneous fistula. | Male, 80-years-old | Phage OMKO1 (108 PFUs), topical administration | Ceftazidime | Clinically successful. Phage revealed bactericidal activity while also eliciting the emergence of phage resistance, which correlated with a resensitization to the antibiotic. | [90] |
| 3 | 2016 | Hannover Medical School, Germany | K. pneumoniae | Lung infection during drug-induced immuno-suppression after heart transplantation | Male, 40-years-old | Phage cocktail (KPV811 and KPV15 against K. pneumoniae, 108 PFUs/mL), Inhalation and topical administration via nasogastric tube | Ceftazidime, linezolid, avibactam, meropenem, cotrimoxazole, tobramycin (iv), colistin (inhalation) | K. pneumoniae was not detected in bronchial lavage after phage and antibiotic treatment. | [93] |
| 4 | 2017 | Hannover Medical School, Germany | S. aureus | Chronic vascular graft infection after aortic arch replacement. Replacement | Male, 59-years-old | Phage CH1 (109 PFUs/mL), topical administration via drainage | Rifampicin, flucloxacillin (iv) | S. aureus was not detected after phage and antibiotic treatment. | [93] |
| 5 | 2017 | Hannover Medical School, Germany | S. aureus | Fulminant pleural empyema after left-ventricular assist device implantation | Male, 62-years-old | Phage CH1 (109 PFUs/mL), topical administration via drainage | Daptomycin (iv) | Although S. aureus was not detected after phage and antibiotic treatment. The patient died due to transplant failure after 20 months of transplantation. | [93] |
| 6 | 2017 | Hannover Medical School, Germany | S. aureus | Chronic left-ventricular assist device infection | Male, 51-years-old | Phage cocktail (Sa30, CH1, SCH11 and SCH111, 109 PFUs/mL). Topical administration via drainage, intranasal and oral administration | Daptomycin (iv) | The level of S. aureus was reduced to 10−2 in the drainage fluid and eradicated completely from nose and throat. Occurrence of phage-resistant variants was not observed throughout phage therapy. Nevertheless, the patient died 1.5 months after. | [93] |
| 7 | 2018 | Saint-Luc University Hospital, Belgium | P. aeruginosa | Sepsis after liver transplantation | Male, toddler | Phage cocktail, BFC1, containing one S. aureus phage (ISP) and two P. aeruginosa phages (PNM and 14-1), intravenous, intralesional and intra-abdominal administration | Vancomycin, gentamycin, colistin, aztreonam, cotrimoxazole, fluconazole | The occurrence of bacterial phage resistance did not result in therapeutic failure and in vitro phage-antibiotic synergies were clearly observed. | [88] |
| 8 | 2018 | Hannover Medical School, Germany | E. coli | Sternal wall healing disorder after mitral valve replacement and aortocoronary bypass | Female, 66-years-old | Phage cocktail (ECD7 and V18, 4 × 1010 PFUs/mL), intraoperatively administration mixed with fibrin glue | Clindamycin (Oral) | Phage mixture with fibrin glue allowed for the sustained release of phages to infected sites. The wound completely healed and E. coli was no longer detected after phage administration. | [93] |
| 9 | 2018 | Hannover Medical School, Germany | P. aeruginosa | Sternal wound abscesses after double lung transplantation | Male, 13-years old | Phage cocktail (PA5 and PA10, 4 × 1010 PFUs/mL), intraoperatively administration mixed with fibrin glue | Colistin, ceftazidime, avibactam (iv) | Phage mixture with fibrin glue allowed for the sustained release of phages to infected sites. The wound completely healed and P. aeruginosa was no longer detected after phage administration. | [93] |
| 10 | Reported in 2019 | University of California, San Diego, USA | P. aeruginosa | Cystic fibrosis | Female, 26-years-old | Phage cocktail (AB-PA01, 4 phages), intravenous administration | Azithromycin, ciprofloxacin, doripenem, piperacillin-tazobactam, vancomycin | Clinical success. Although a phage-resistant isolate temporarily emerged, it did not pose a therapeutic problem. This is likely an effect of the combination of antimicrobial agents. | [39,89] |
| 11 | Reported in 2019 | Hadassah-Hebrew University Medical Center, Israel | A. baumannii K. pneumoniae | Trauma-related left tibial infection | Male, 42-years-old | Phage cocktail (AbKT21phi3 against A. baumannii and KpKT21phi1 against K. pneumoniae), intravenous administration | Meropenem, colistin | Clinically successful and the patient’s leg did not have to be amputated. No phage-resistant variants emerged in the patient, likely because the combination of antibiotics and phages was very effective. | [91] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujiki, J.; Nakamura, K.; Nakamura, T.; Iwano, H. Fitness Trade-Offs between Phage and Antibiotic Sensitivity in Phage-Resistant Variants: Molecular Action and Insights into Clinical Applications for Phage Therapy. Int. J. Mol. Sci. 2023, 24, 15628. https://doi.org/10.3390/ijms242115628
Fujiki J, Nakamura K, Nakamura T, Iwano H. Fitness Trade-Offs between Phage and Antibiotic Sensitivity in Phage-Resistant Variants: Molecular Action and Insights into Clinical Applications for Phage Therapy. International Journal of Molecular Sciences. 2023; 24(21):15628. https://doi.org/10.3390/ijms242115628
Chicago/Turabian StyleFujiki, Jumpei, Keisuke Nakamura, Tomohiro Nakamura, and Hidetomo Iwano. 2023. "Fitness Trade-Offs between Phage and Antibiotic Sensitivity in Phage-Resistant Variants: Molecular Action and Insights into Clinical Applications for Phage Therapy" International Journal of Molecular Sciences 24, no. 21: 15628. https://doi.org/10.3390/ijms242115628
APA StyleFujiki, J., Nakamura, K., Nakamura, T., & Iwano, H. (2023). Fitness Trade-Offs between Phage and Antibiotic Sensitivity in Phage-Resistant Variants: Molecular Action and Insights into Clinical Applications for Phage Therapy. International Journal of Molecular Sciences, 24(21), 15628. https://doi.org/10.3390/ijms242115628





