Plant Responses of Maize to Two formae speciales of Sporisorium reilianum Support Recent Fungal Host Jump
Abstract
1. Introduction
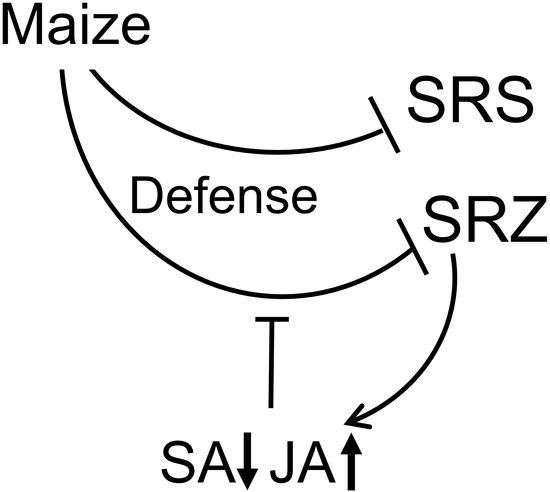
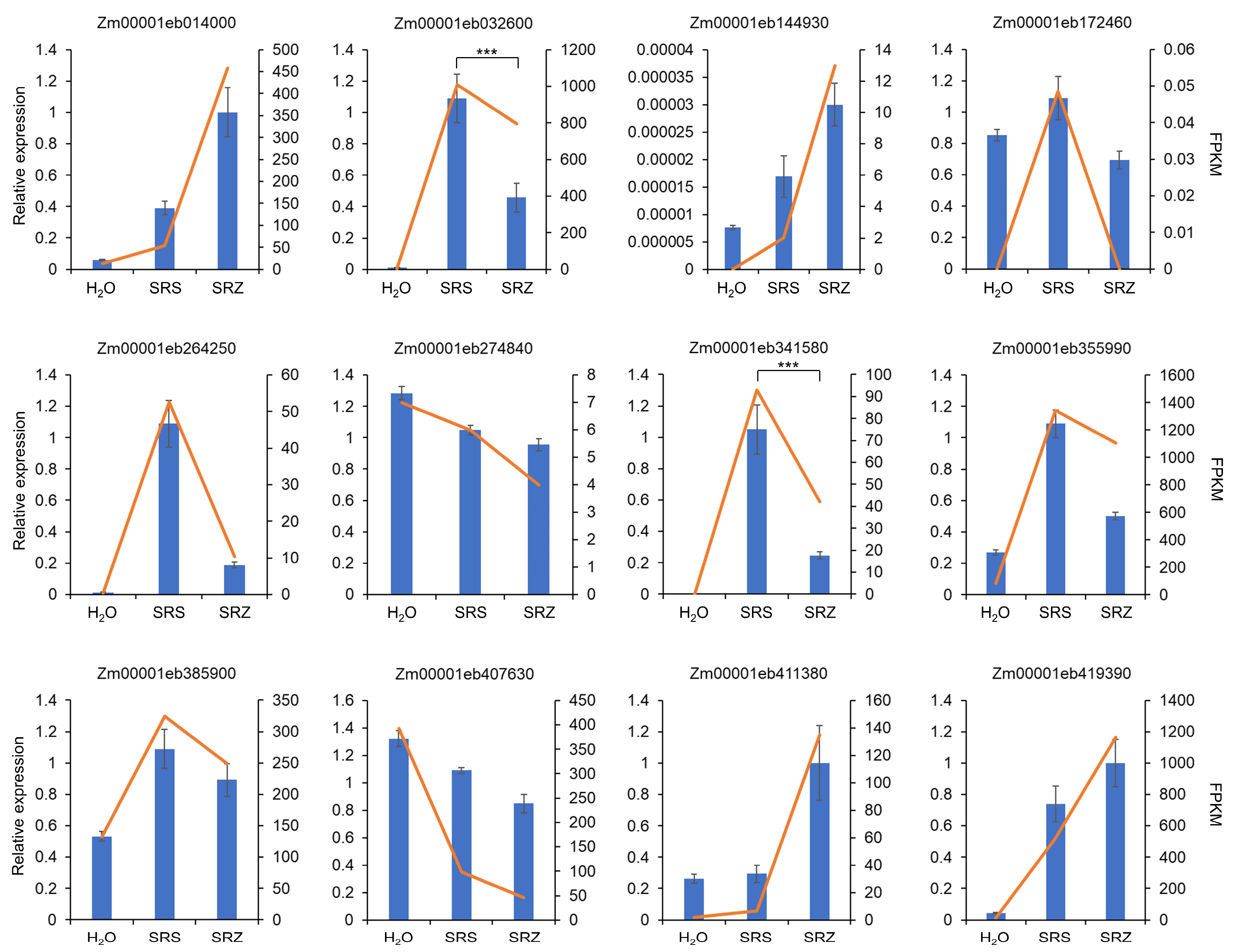
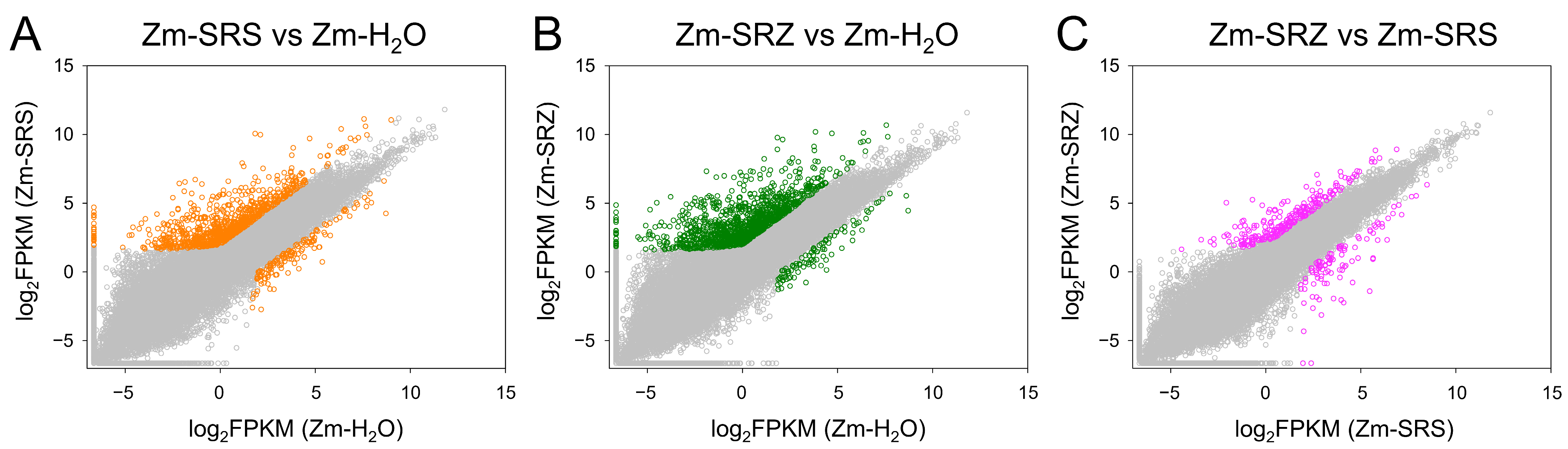
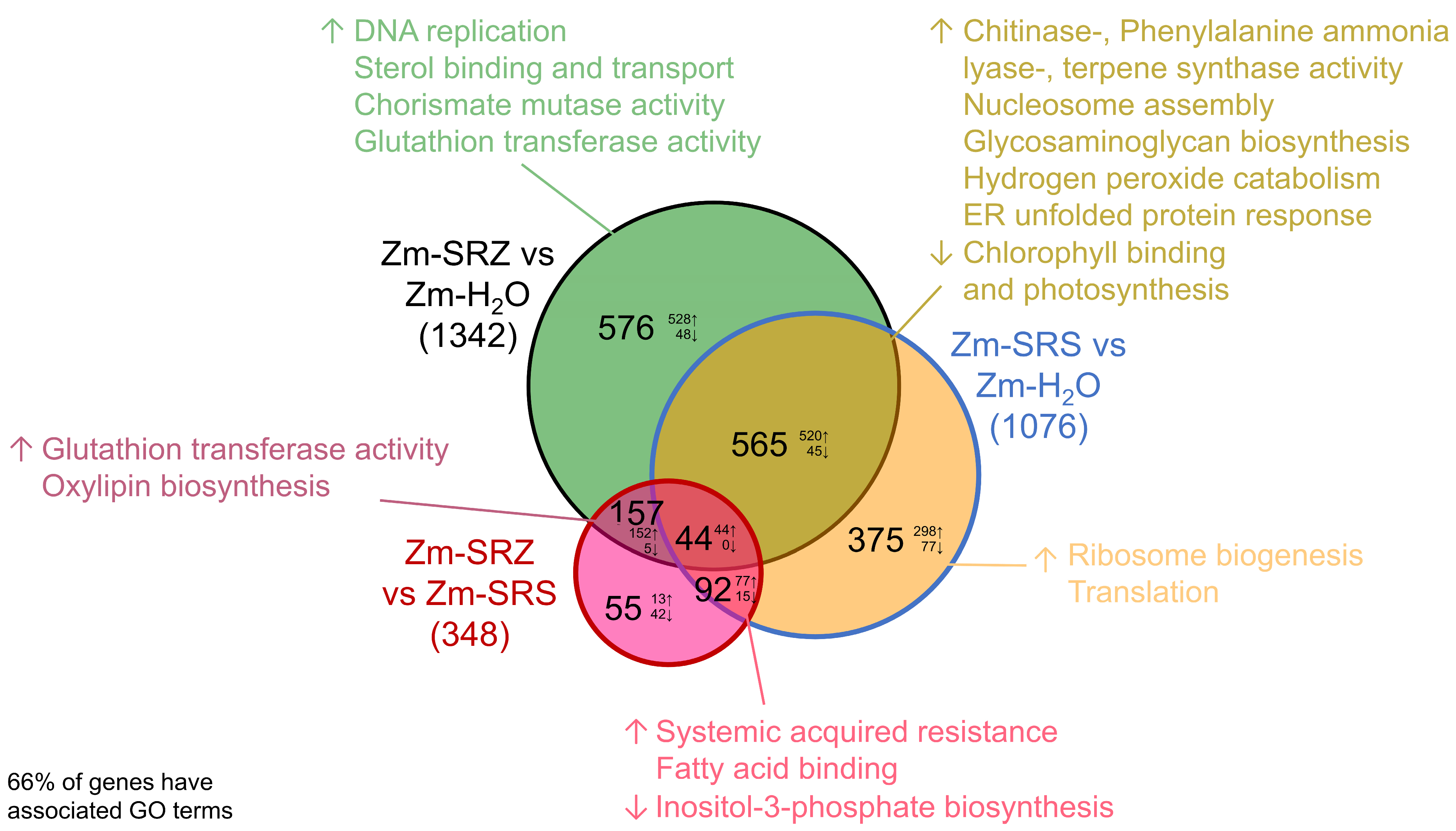
2. Results
3. Discussion
4. Materials and Methods
4.1. Experimental Design and Accession Numbers
4.2. Transcriptome Data Analysis
4.3. Gene Expression Validation Via Real-Time PCR
4.4. Phytohormone Measurements
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Urashima, S.; Igarashi, S.; Kato, H. Host range, mating type, and fertility of Pyricularia grisea from wheat in Brazil. Plant Dis. 1993, 77, 1211–1216. [Google Scholar] [CrossRef]
- Islam, M.T.; Croll, D.; Gladieux, P.; Soanes, D.M.; Persoons, A.; Bhattacharjee, P.; Hossain, M.S.; Gupta, D.R.; Rahman, M.M.; Mahboob, M.G.; et al. Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol. 2016, 14, 84. [Google Scholar]
- Malaker, P.K.; Barma, N.C.; Tiwari, T.P.; Collis, W.J.; Duveiller, E.; Singh, P.K.; Joshi, A.K.; Singh, R.P.; Braun, H.J.; Peterson, G.L.; et al. First report of wheat blast caused by Magnaporthe oryzae pathotype triticum in Bangladesh. Plant Dis. 2016, 100, 2330. [Google Scholar]
- Navaud, O.; Barbacci, A.; Taylor, A.; Clarkson, J.P.; Raffaele, S. Shifts in diversification rates and host jump frequencies shaped the diversity of host range among Sclerotiniaceae fungal plant pathogens. Mol. Ecol. 2018, 27, 1309–1323. [Google Scholar] [CrossRef] [PubMed]
- Begerow, D. Koevolution–Zur Bedeutung organismischer Interaktion für die Evolution. In Die Evolution des Lebendigen; Betz, O., Köhler, H.-R., Eds.; Attempto-Verlag: Tübingen, Germany, 2008; pp. 173–186. [Google Scholar]
- Munkacsi, A.B.; Stoxen, S.; May, G. Domestication of maize, sorghum, and sugarcane did not drive the divergence of their smut pathogens. Evolution 2007, 61, 388–403. [Google Scholar] [CrossRef] [PubMed]
- Halisky, P.M.; Smeltzer, D.G. Head smut. Disease of corn, sorghum and sudangrass established in California. Calif. Agric. 1961, 1, 10–12. [Google Scholar]
- Halisky, P.M. Physiologic specialization and genetics of the smut fungi III. Bot. Rev. 1965, 1, 114–150. [Google Scholar]
- Poloni, A.; Schirawski, J. Host specificity in Sporisorium reilianum is determined by distinct mechanisms in maize and sorghum. Mol. Plant Pathol. 2016, 17, 741–754. [Google Scholar] [CrossRef]
- Halisky, P.M. Head smut of Sorghum, Sudangrass, and Corn, Caused by Sphacelotheca reiliana (Kühn) Clint. Hilgardia 1963, 34, 287–304. [Google Scholar] [CrossRef][Green Version]
- Schweizer, G.; Münch, K.; Mannhaupt, G.; Schirawski, J.; Kahmann, R.; Dutheil, J.Y. Positively Selected Effector Genes and Their Contribution to Virulence in the Smut Fungus Sporisorium reilianum. Genome Biol. Evol. 2018, 10, 629–645. [Google Scholar] [CrossRef]
- Schirawski, J.; Mannhaupt, G.; Münch, K.; Brefort, T.; Schipper, K.; Doehlemann, G.; Di Stasio, M.; Rössel, N.; Mendoza-Mendoza, A.; Pester, D.; et al. Pathogenicity determinants in smut fungi revealed by genome comparison. Science 2010, 330, 1546–1548. [Google Scholar] [CrossRef] [PubMed]
- Schirawski, J.; Heinze, B.; Wagenknecht, M.; Kahmann, R. Mating type loci of Sporisorium reilianum: Novel pattern with three a and multiple b specificities. Eukaryot. Cell 2005, 4, 1317–1327. [Google Scholar] [CrossRef]
- Prom, L.K.; Perumal, R.; Erattaimuthu, S.R.; Erpelding, J.E.; Montes, N.; Odvody, G.N.; Greenwald, C.; Jin, Z.; Frederiksen, R.; Magill, C.W. Virulence and Molecular Genotyping Studies of Sporisorium reilianum Isolates in Sorghum. Plant Dis. 2011, 95, 523–529. [Google Scholar] [CrossRef]
- Zhang, F.; Ping, J.; Du, Z.; Cheng, Q.; Huang, Y. Identification of a new race of Sporisorium reilianum and characterization of the reaction of sorghum lines to four races of the head smut pathogen. J. Phytopathol. 2010, 159, 342–346. [Google Scholar] [CrossRef]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Lübberstedt, T.; Xia, X.C.; Tan, G.; Liu, X.; Melchinger, A.E. QTL mapping of resistance to Sporisorium reiliana in maize. Theor. Appl. Genet. 1999, 99, 593–598. [Google Scholar] [CrossRef]
- Poloni, A.; Garde, R.; Dittiger, L.D.; Heidrich, T.; Müller, C.; Drechsler, F.; Zhao, Y.; Mazumdar, T.; Schirawski, J. Transcriptome Analysis Reveals Contrasting Plant Responses of Sorghum bicolor upon Colonization by Two Formae Speciales of Sporisorium reilianum. Int. J. Mol. Sci. 2022, 23, 8864. [Google Scholar] [CrossRef]
- Hufford, M.B.; Seetharam, A.S.; Woodhouse, M.R.; Chougule, K.M.; Ou, S.; Liu, J.; Ricci, W.A.; Guo, T.; Olson, A.; Qiu, Y.; et al. De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. Science 2021, 373, 655–662. [Google Scholar] [CrossRef]
- Zuther, K.; Kahnt, J.; Utermark, J.; Imkampe, J.; Uhse, S.; Schirawski, J. Host specificity of Sporisorium reilianum is tightly linked to generation of the phytoalexin luteolinidin by Sorghum bicolor. Mol. Plant Microbe Interact. 2012, 25, 1230–1237. [Google Scholar] [CrossRef]
- Doehlemann, G.; Wahl, R.; Vranes, M.; de Vries, R.P.; Kämper, J.; Kahmann, R. Establishment of compatibility in the Ustilago maydis/maize pathosystem. J. Plant Physiol. 2008, 165, 29–40. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Kahmann, R. Manipulation of Phytohormone Pathways by Effectors of Filamentous Plant Pathogens. Front. Plant Sci. 2019, 10, 822. [Google Scholar] [CrossRef]
- Grant, M.R.; Jones, J.D. Hormone (dis)harmony moulds plant health and disease. Science 2009, 324, 750–752. [Google Scholar] [CrossRef]
- Djamei, A.; Schipper, K.; Rabe, F.; Ghosh, A.; Vincon, V.; Kahnt, J.; Osorio, S.; Tohge, T.; Fernie, A.R.; Feussner, I.; et al. Metabolic priming by a secreted fungal effector. Nature 2011, 478, 395–398. [Google Scholar] [CrossRef]
- Darino, M.; Chia, K.S.; Marques, J.; Aleksza, D.; Soto-Jiménez, L.M.; Saado, I.; Uhse, S.; Borg, M.; Betz, R.; Bindics, J.; et al. Ustilago maydis effector Jsi1 interacts with Topless corepressor, hijacking plant jasmonate/ethylene signaling. New Phytol. 2021, 229, 3393–3407. [Google Scholar] [CrossRef] [PubMed]
- Bindics, J.; Khan, M.; Uhse, S.; Kogelmann, B.; Baggely, L.; Reumann, D.; Ingole, K.D.; Stirnberg, A.; Rybecky, A.; Darino, M.; et al. Many ways to TOPLESS-manipulation of plant auxin signalling by a cluster of fungal effectors. New Phytol. 2022, 236, 1455–1470. [Google Scholar] [CrossRef] [PubMed]
- Ghareeb, H.; Zhao, Y.; Schirawski, J. Sporisorium reilianum possesses a pool of effector proteins that modulate virulence on maize. Mol. Plant Pathol. 2019, 20, 124–136. [Google Scholar] [CrossRef]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Deng, C.; Leonard, A.; Cahill, J.; Lv, M.; Li, Y.; Thatcher, S.; Li, X.; Zhao, X.; Du, W.; Li, Z.; et al. The RppC-AvrRppC NLR-effector interaction mediates the resistance to southern corn rust in maize. Mol. Plant 2022, 15, 904–912. [Google Scholar] [CrossRef]
- Cheng, Y.; Li, X.; Jiang, H.; Ma, W.; Miao, W.; Yamada, T.; Zhang, M. Systematic analysis and comparison of nucleotide-binding site disease resistance genes in maize. FEBS J. 2012, 279, 2431–2443. [Google Scholar] [CrossRef]
- Zhang, X.; Fernandes, S.B.; Kaiser, C.; Adhikari, P.; Brown, P.J.; Mideros, S.X.; Jamann, T.M. Conserved defense responses between maize and sorghum to Exserohilum turcicum. BMC Plant Biol. 2020, 20, 67. [Google Scholar] [CrossRef]
- OmicsBox-Bioinformatics Made Easy, version 3.0.30; BioBam Bioinformatics: Valencia, Spain, 2019.
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. (Version FastQC 0.11.8). Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 4 October 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2010, 30, 2114–2120. [Google Scholar]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2014, 31, 166–169. [Google Scholar] [PubMed]
- Tarazona, S.; Furio-Tari, P.; Turra, D.; Di Pietro, A.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [PubMed]
- Tarazona, S.; Garcia-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [PubMed]
- Al-Shahrour, F.; Díaz-Uriarte, R.; Dopazo, J. Fatigo: A web tool for finding significant associations of gene ontology terms with groups of genes. Bioinformatics 2004, 20, 578–580. [Google Scholar] [PubMed]
- Lin, Y.; Zhang, C.; Lan, H.; Gao, S.; Liu, H.; Liu, J.; Cao, M.; Pan, G.; Rong, T.; Zhang, S. Validation of Potential Reference Genes for qPCR in Maize across Abiotic Stresses, Hormone Treatments, and Tissue Types. PLoS ONE 2014, 9, e95445. [Google Scholar] [CrossRef]
- Gallinger, J.; Zikeli, K.; Zimmermann, M.R.; Görg, L.M.; Mithöfer, A.; Reichelt, M.; Seemüller, E.; Gross, J.; Furch, A.C. Specialized 16SrX phytoplasmas induce diverse morphological and physiological changes in their respective fruit crops. PLoS Pathog. 2004, 17, e1009459. [Google Scholar] [CrossRef]

| Sample | Total Reads | Uniquely Mapped Reads (% 1) | Unmapped Reads (% 1) |
|---|---|---|---|
| Zm-H2O | 51,290,554 | 45,811,710 (89.3) | 5,478,844 (10.7) |
| Zm-SRS | 69,785,329 | 61,663,182 (88.4) | 8,122,147 (11.6) |
| Zm-SRZ | 40,313,666 | 35,178,627 (87.3) | 5,135,039 (12.7) |
| Primer | Description | Sequence | Primer Efficiency (%) |
|---|---|---|---|
| oLD227 | FP_Zm00001eb341580 | GAACTCGCCTCAAGACTACC | 92.6 |
| oLD228 | RP_Zm00001eb341580 | TACTTCTCCGCGAACTGC | |
| oLD231 | FP_Zm00001eb274840 | ACCTACAACAGCCTGATGG | 95.8 |
| oLD232 | RP_Zm00001eb274840 | GCAGAACCCGTTTATGACC | |
| oLD235 | FP_Zm00001eb144930 | TTCCATCTGATTCGATCGAG | 93.3 |
| oLD236 | RP_Zm00001eb144930 | CACATTATTATTGGGAAACCAAC | |
| oSC105 | FP_Zm00001eb014000 | GCGTCAGGCAGTTCAACTTC | 91.0 |
| oSC106 | RP_Zm00001eb014000 | CCTTGGCGATCTCGTCCTTC | |
| oSC113 | FP_Zm00001eb355990 | ACGCCAAGAAGGTGATCCTC | 96.7 |
| oSC114 | RP_Zm00001eb355990 | CGACGATGTCGACGAAGATG | |
| oSC115 | FP_Zm00001eb411380 | TGGAGGCTGCCTTAAATGAC | 99.3 |
| oSC116 | RP_Zm00001eb411380 | TGTAGCGCGTGCAGTTATTG | |
| oSC117 | FP_Zm00001eb032600 | GCCAGGACTTCTACGACATC | 91.1 |
| oSC118 | RP_Zm00001eb032600 | GGCAGAAGGTGACTTGGTAG | |
| oSC121 | FP_Zm00001eb172460 | CATGGCCGTCATCACATGAG | 99.6 |
| oSC122 | RP_Zm00001eb172460 | AGTTGGTGCAGCGATGAG | |
| oSC123 | FP_Zm00001eb264250 | ACTACCCGCTTATGGTCTCC | 92.9 |
| oSC124 | RP_Zm00001eb264250 | ACTACTCCACGGGCAAACTC | |
| oSC127 | FP_Zm00001eb419390 | TGATACTCGTCGGCACTCTG | 91.0 |
| oSC128 | RP_Zm00001eb419390 | CGTTGACCGACACGTCATTG | |
| oSC129 | FP_Zm00001eb407630 | ATGCTGGCACGGAGTACAAG | 97.6 |
| oSC130 | RP_Zm00001eb407630 | TCCGTAAGCGCGTTTGTTGG | |
| oSC131 | FP_Zm00001eb173410 | CCATCACTGCCACACAGAAAAC | 93.7 |
| oSC132 | RP_Zm00001eb173410 | AGGAACACGGAAGGACATACCAG | |
| oSC133 | FP_Zm00001eb385900 | TGGGCCTACTGGTCTTACTACTGA | 93.5 |
| oSC134 | RP_Zm00001eb385900 | ACATACCCACGCTTCAGATCCT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dittiger, L.D.; Chaudhary, S.; Furch, A.C.U.; Mithöfer, A.; Schirawski, J. Plant Responses of Maize to Two formae speciales of Sporisorium reilianum Support Recent Fungal Host Jump. Int. J. Mol. Sci. 2023, 24, 15604. https://doi.org/10.3390/ijms242115604
Dittiger LD, Chaudhary S, Furch ACU, Mithöfer A, Schirawski J. Plant Responses of Maize to Two formae speciales of Sporisorium reilianum Support Recent Fungal Host Jump. International Journal of Molecular Sciences. 2023; 24(21):15604. https://doi.org/10.3390/ijms242115604
Chicago/Turabian StyleDittiger, Lukas Dorian, Shivam Chaudhary, Alexandra Charlotte Ursula Furch, Axel Mithöfer, and Jan Schirawski. 2023. "Plant Responses of Maize to Two formae speciales of Sporisorium reilianum Support Recent Fungal Host Jump" International Journal of Molecular Sciences 24, no. 21: 15604. https://doi.org/10.3390/ijms242115604
APA StyleDittiger, L. D., Chaudhary, S., Furch, A. C. U., Mithöfer, A., & Schirawski, J. (2023). Plant Responses of Maize to Two formae speciales of Sporisorium reilianum Support Recent Fungal Host Jump. International Journal of Molecular Sciences, 24(21), 15604. https://doi.org/10.3390/ijms242115604