Abstract
The plant actin cytoskeleton is characterized by the basic properties of dynamic array, which plays a central role in numerous conserved processes that are required for diverse cellular functions. Here, we focus on how actins and actin-related proteins (ARPs), which represent two classical branches of a greatly diverse superfamily of ATPases, are involved in fundamental functions underlying signal regulation of plant growth and development. Moreover, we review the structure, assembly dynamics, and biological functions of filamentous actin (F-actin) from a molecular perspective. The various accessory proteins known as actin-binding proteins (ABPs) partner with F-actin to finely tune actin dynamics, often in response to various cell signaling pathways. Our understanding of the significance of the actin cytoskeleton in vital cellular activities has been furthered by comparison of conserved functions of actin filaments across different species combined with advanced microscopic techniques and experimental methods. We discuss the current model of the plant actin cytoskeleton, followed by examples of the signaling mechanisms under the supervision of F-actin related to cell morphogenesis, polar growth, and cytoplasmic streaming. Determination of the theoretical basis of how the cytoskeleton works is important in itself and is beneficial to future applications aimed at improving crop biomass and production efficiency.
1. Introduction
The optimization of cellular architecture is one of the most efficient means to advance crop productivity and stress tolerance. While plant cell growth and morphology are modulated by a number of factors, the actin cytoskeleton plays a decisive role in almost all plant developmental processes, including cargo trafficking, cellular motility, apical growth, cell wall formation, and cytoplasmic streaming [1,2,3,4,5,6]. In the cytoplasm, actin has different physiological functions depending on its form, as a monomer, oligomer, or polymer or as a complex with actin-binding proteins (ABPs). Polymeric and non-polymeric forms of actin maintain a dynamic balance in the cell. Actin molecules come together into a filament, a double helix polymer with a head-to-tail orientation that gives the filament a +/− molecular polarity. This polarity is crucial to the mechanism of actin assembly, with the barbed (or +) end of a filament always elongating much faster than the pointed (or –) end. Shortly after each actin monomer assembles into a filament, its bound ATP is hydrolyzed into ADP, such that the filament is a linear mix of ATP-actin and then ADP-actin. ATP-actin that associates at the barbed end and ADP-actin that dissociates from the pointed end are affected by ATP hydrolysis and phosphate dissociation. The overall mechanism at steady state denotes a dynamic equilibrium known as “treadmilling” [7,8]. The cell monitors both temporal and spatial actin dynamics, such as actin filament nucleation, movement, assembly, and disassembly. As such a basic part of cellular function, it is necessary to understand the intriguing roles of actin proteins in various cellular processes.
The actin-related proteins (ARPs) were discovered in eukaryotes in the 1990s. The ARPs are classified into various classes or subfamilies, which are extremely conserved across a wide range of eukaryotes, from yeast to plants and humans [9]. The amino acid sequence of ARPs differs by 20% to 60% compared to the canonical actin [10]. Nevertheless, the expression patterns of ARP genes do not correlate strongly with those observed for either actins or ABPs [11]. Actually, ARP classes assemble with other proteins to form stable hetero-multimeric protein complexes [12,13,14], so it is important to take into account how ARPs function biologically in light of their roles as parts of larger macromolecular machinery.
ABPs have evolved as partners that facilitate and manipulate the formation and turnover of actin filaments [2,7,8]. Genetic or physical interactions between actin and different ABPs can be placed into various signaling networks that participate in specific plant morphogenetic pathways that are supervised by the actin cytoskeleton. A combination of approaches has been used to predict the impact of ABPs on actin dynamics so that a deeper understanding of actin-mediated functions in plants can be achieved [15]. Beyond that, an increasing number of ABPs and microtubule-associated proteins (MAPs) have been identified as indispensable elements for sensing environmental signals and for the coordinated regulation of cytoskeleton reorganization [16,17,18,19]. Whether the functions of plant and animal ABPs are evolutionarily conserved has been the thrust of research in the field for a long time.
In this review, we summarize recent reports that have uncovered the functions of actin and ARPs during the growth and development of plants and highlight critical new genetic and biochemical evidence that plant ABPs coordinate filamentous actin (F-actin) formation to perform different functions. Additionally, these complex arrangements reveal crosstalk between the actin cytoskeleton and regulatory proteins within signaling networks in response to environmental change. Despite recent progress, research on actin in plants is rather fragmentary in comparison to our understanding of the animal actin cytoskeleton. We hope that this review provides a serviceable update on the functional properties of actin proteins in multiple plant species.
2. Identification and Annotation of Arabidopsis Actin Family
Actin, as a cytoskeletal protein, is encoded by a relatively diverse and ancient family of genes whose phylogeny began with the emergence of vascular plants [20]. We focus this part of the review on differential expression of genes within the actin family and their different functions as selective forces that preserve divergent members.
2.1. Tissue-Specific Expression of Arabidopsis Actin Genes
There are ten actin genes in Arabidopsis, eight of which are strongly expressed at certain times and locations during plant development [21]. Vegetative actins (ACT2, ACT7 and ACT8) are strongly expressed in the roots, stems, and leaves of germinating seedlings, young plants, and mature plants. In contrast, five other actin genes, namely ACT1, ACT3, ACT4, ACT11, and ACT12, are characterized by expression in pollen, ovules, and seeds [20]. Joint analysis of the phylogeny and tissue localization of the actin family proteins shows that the division of each member in the phylogenetic tree is distinct, while their gene expression patterns partially overlap (Figure 1A). The rate of divergence among actins is mild, with members of the actin family more closely related than members of the ARP family. This suggests that the evolution of respective regulatory elements with each protein may be more complex in comparison to that of their coding sequences. Given that the expression patterns of actin genes have remained consistent throughout lengthy evolutionary time periods, it is reasonable to consider selective constraint keeping the actin family so conserved.
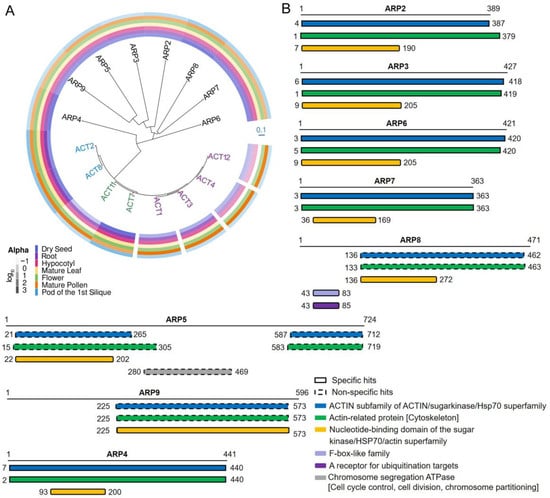
Figure 1.
Homology analysis of actins and ACTIN-RELATED PROTEINs (ARPs). (A) Phylogenetic relationships of actin proteins and ARPs. The protein sequences of Arabidopsis actins (AtACT1, AtACT2, AtACT3, AtACT4, AtACT7, AtACT8, AtACT11, and AtACT12) and ARPs (AtARP2, AtARP3, AtARP4, AtARP5, AtARP6, AtARP7, AtARP8, and AtARP9) are compared in a neighbor-joining (NBJ) tree. The branch length in the NBJ tree relays the degree of sequence divergence or genetic distance. Therefore, compared with actins, the accelerated rate of divergence among Arabidopsis ARPs is greater throughout the course of evolution. The expression patterns in different organs (dry seed, root, hypocotyl, mature leaf, flower, mature pollen, and pod of the first silique) of the ARPs and actins are shown in the color bars, which display the log10 expression value acquired the integration platform ePlant. (ePlant: Visualizing and Exploring Multiple Levels of Data for Hypothesis Generation in Plant Biology, http://bar.utoronto.ca/eplant (accessed on 14 September 2021)). (B) The search for conserved domains in the ARP protein sequences from NCBI (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/cdd (accessed on 14 September 2021)) is shown.
2.2. Subcellular Localization of Actin
The subcellular localization of each actin protein will be related to its function. Prediction of the subcellular localization of the sequences highly homologous to Arabidopsis ACT1 in other species of plants has been completed on the crop proteins with annotated locations (cropPAL, https://croppal.org/ (accessed on 27 February 2022)). A query of the database showed that actins may be present in the nucleus, cytoplasm, and plastids (Table S1). We specifically list the predicted subcellular localizations of different actin isoforms in Arabidopsis, and the data from different sources highlight the cytoskeleton, nucleus, and cytosol as the main sites of actin accumulation (Table S2).
However, many non-specific interactions between actin and different kinds of binding partners have been reported, fueling debate about the function of actin-nucleoprotein binding. The functions of actin in the nucleus are better understood in mammalian cells compared to plant cells. Although it is not clear how much actin is normally present in the nucleus, a few studies have revealed that actin could shuttle in and out of the nucleus [22,23]. In mammalian cells, members of the actin family take part in a variety of activities in the nucleus in multifarious forms, including monomers, short filaments, and novel oligomer forms [24]. Actin is involved in the export of mRNA transcripts at the nuclear pore complex (NPC) through binding to heterogeneous ribonucleoprotein (hnRNP) complexes [25]. Actin also maintains the stability of the nuclear structure by participating in the regulation of the nuclear lamina and the enhanced adult sensory threshold (EAST) endoskeleton [26]. Additionally, actin is one of the components of the chromatin remodeling complex [13], and actin can change or maintain the chromatin structure [27]. Moreover, super-resolution imaging and RNA-seq reveal that nuclear actin can promote the formation of a transcription factory for inducible genes, which can respond quickly to the external environment [28]. Many reports indicate that actin is present in the nucleus and has a variety of roles in mammalian cells in processes such as transcription, nucleocytoplasmic transport, DNA damage repair, chromatin remodeling, and nuclear structure stabilization [29,30,31,32,33,34,35,36,37,38,39].
Whether plant actin is in the nucleus has been a matter of concern. In fact, a specific form of actin in the nucleus has long been reported [40]. ACT2, ACT7, and ACT8 are distributed throughout the nucleoplasm in isolated nuclei of Arabidopsis, with ACT7 more concentrated in nuclear speckles than the other two [40]. ACT2 and ACT7 aggregate into different types of filamentous structures in Arabidopsis protoplasts and are distributed across the whole protoplast [41]. Moreover, flowering plants have developed a distinctive double-fertilization process accompanied by F-actin-based gamete nuclear migration systems [42]. In Arabidopsis root hairs, the migration direction of cell nuclei is closely linked to the perinuclear actin filaments [43]. On the other side, the actin cytoskeleton plays a crucial role in facilitating intercellular communication through plasmodesmata (PD) [44,45,46]. Supporting this idea, it has been demonstrated that actin and certain actin-associated proteins localize to PD [47,48].
2.3. Diverse Functions of Actin Isoforms in Plants
As one of the key components of the cytoskeleton, the monomeric actin plays an irreplaceable role. For example, misexpression of ACT1 leads to dwarfism and altered organ morphology in Arabidopsis. At the highest levels of transgenic overexpression of ACT1, a large number of actin filaments are polymerized, bundled, and reorganized [49], indicating that the abnormal expression of ACT1 in vegetative tissues affects the dynamic arrangement of actin and the orientation of actin-related proteins, disrupting the normal development of plants. Furthermore, misexpression of ACT1 alters the microfilament architecture due to inappropriate interaction of ACT1 with endogenous ABPs [50]. ACT2 is located around the chloroplast and participates in chloroplast photorelocation movement [51]. Similarly, a requirement for ACT7 in actin-dependent chloroplast clustering also has been shown [52].
Actin has been shown to be critical for tip growth in an array of plant models. Plant root hairs achieve tubular shapes through cell tip growth, which requires the coordinated activity of the actin cytoskeleton and endomembrane systems [53,54]. The actin network exhibits rapid turnover near the site of rapid cell expansion. Furthermore, fine actin filament bundles in the apex and subapex are essential for the expansion of the tip, which serve as pathways for the transportation of secretory vesicles to or from the plasma membrane (PM), ultimately aiding in the rapid growth of root hairs. By contrast, thick actin filament bundles in the apical and subapical regions inhibit the growth of root hairs [55]. ACT2 is a key factor in the correct selection of the bulge site on the epidermal cell and for tip growth of root hair development in Arabidopsis [56]. The act2-1 mutant generally contains transversely oriented actin filaments, whereas the wild type displays longitudinal actin filaments [57]. Both single-point and T-DNA insertion mutants of ACT2 affect the length of actin bundles or the polymerization of actin filaments in specific tissues [58,59]. The act7-1 and act7-4 mutant alleles show delayed and inefficient germination and roots with increased twisting along with wavy but retarded growth [60]. However, the adult act2-1, act4-1, and act7-1 mutant plants are robust, morphologically normal and completely fertile because of functional redundancy [61]. The expression of ACT7 is more relevant to epidermal cell differentiation, cell division, and root architecture, while ACT2 and ACT8 are crucial for regulating root hair tip growth [57]. In the pollen tube tip, the arrangement and dynamic distribution of the actin cytoskeleton are both the powerful motivators of growth and the organizer of cell polarity. ACT1, ACT3, ACT4, and ACT12 are involved in the dynamic arrangement of actin filaments in germinating pollen grains and tip-growing pollen tubes [62]. Differential regulation of vegetative actin genes and the diversity of actin isovariant sequences are essential for plant development.
The actin cytoskeleton is one of the important factors in plant cell expansion. Thinner and denser arrays of filamentous actin probably participate in expanding cell areas by means of delivering cargo, since the plant actin cytoskeleton is the primary backbone of cytoplasmic streaming [63,64,65]. In Arabidopsis roots, the epidermal and cortical cells have two patterns of rapid elongation: (1) elongating rapidly at the edge of the proximal meristem and transition zone [66,67] and (2) elongating at the boundary between the transition and elongation zones [68]. The second type of rapid cell elongation involves dynamic actin reorganization at the basal end of the transition zone, which is disrupted in the act7 and act2act8 mutants [69]. Actually, both the globular actin (G-actin) generated by expression of actin genes and the dynamic structure of the actin cytoskeleton regulated by the ACTIN-RELATED PROTEIN 2/3 (ARP2/3) complex have crucial effects on the second rapid cell elongation [69,70]. In Gossypium hirsutum, 15 GhACT genes are also differentially expressed in various tissues, among which GhACT1 is mainly expressed in fiber cells and significantly affects fiber elongation [71]. Additionally, the mutation of the actin gene Ligon lintless-1 (GhLi1) disrupts the normal arrangement of the actin cytoskeleton and affects cell elongation, which ultimately leads to formation of various distorted organs [72]. F-actin may act as a trajectory for vesicle movement, thereby regulating fiber cell elongation and secondary cell wall biosynthesis [72]. A general mechanism associated with fine F-actin formation could be involved in accumulating and retaining materials for cell expansion [4]. Another key factor in cell expansion is the auxin signaling pathway, but its association with the actin cytoskeleton remains unclear. During tissue culture, the speed of callus induction in the act7 mutant is slower under the effect of auxin compared with wild-type callus [73]. A high level of ACT7 protein may be induced by phytohormones to maintain the rapid growth of cell cultures; therefore, ACT7 is necessary for normal callus formation [73]. Additionally, Arabidopsis root meristem development is influenced by ACT7-mediated modulation of auxin-ethylene responses [74]. The unique functional properties of different actin genes may be the evolutionary result of the sessile lifestyle of plants for meeting the demands of various environmental challenges.
Actin also regulates various cellular events in response to external changes. Salt stress-induced microfilament assembly in Arabidopsis is an essential component of salt tolerance [75]. High external pH has no impact on microfilament stability in vitro, but it induces the depolymerization of microfilament in vivo, suggesting that alkaline stress may activate a signal that leads to the reorganization of microfilament [76]. Heat stress affects the organization of actin filaments in the subapex, leading to changes in vesicular transport and cell wall deposition processes [77]. Arabidopsis hypocotyl cells can respond to mechanical stimuli similar to those exerted by fungal and oomycete cells, in which actin microfilaments aggregate and reorganize toward the site of the indentation [78]. Moreover, remodeling of actin arrays features prominently during both early and late events associated with the innate immune response [79]. A summary of the responses of actin filaments under different conditions shows that actin is a key player in enabling plants to adapt and thrive in their ever-changing surroundings (Figure 2).
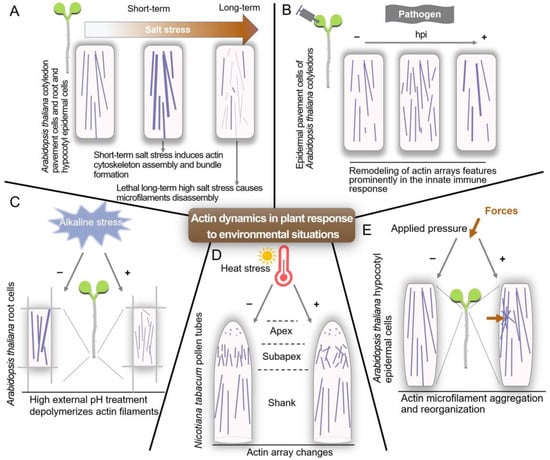
Figure 2.
Schematic model of actin dynamics in individual cell growth and plant development in response to environmental stresses. (A) Short-term salt stress can trigger the assembly and formation of microfilament bundles in Arabidopsis. However, the polymerization of microfilaments is inhibited in susceptible seedlings subjected to long-term or high salt stress. (B) Exposure to Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) induces two distinct changes in the arrangement of the cortical actin network. At 6 h post-inoculation (hpi), cells show an increase in the overall density of actin filament network. Later, ~24 hpi, a reduction in the number of individual filaments or an increase in the extent of actin filament bundling is obvious. (C) Under high external pH conditions, microfilament depolymerization is induced, which is associated with the inhibition of Arabidopsis root growth. (D) Damages caused by heat stress to actin filaments mainly concern the actin array present in the subapex, a region critical for regulating the organelle and vesicle distribution in the pollen tube apex. (E) The Arabidopsis hypocotyl epidermal cell exhibits actin microfilament aggregation in response to mechanical stimulation, and this applied pressure is utilized to simulate the attack by fungal or oomycete hyphae.
3. Structure and Evolution of the ARP Superfamily
ARPs are named based on their similarity to the canonical actin [10]. The expression patterns of AtARP5, AtARP6, and AtARP8 are distinct in seedlings, roots, leaves, flowers, and siliques, whereas expression of AtARP2 and AtARP3 was extremely low in all organs [9,11]. The AtARP4 and AtARP7 proteins were concentrated in flowers. Compared with members of the actin protein family, ARPs appear to be more divergent in their predicted structures (Figure 1A). The domains with known functions in the eight AtARP proteins are shown in Figure 1B. The AtARPs were characterized by the nucleotide-binding domain, which are like those mainly present in actin, nucleotide exchange factors, and HSP70 molecular chaperones. The nucleotide sits in a deep cleft generated between the two lobes of the nucleotide-binding domain (NBD), and the residues in the NBD are conserved [80,81,82,83,84,85]. Proteins in the actin and HSP70 superfamilies have functional activities that are regulated by allosteric effectors, which may influence the cleft closure. This conserved domain was prominent in all Arabidopsis ARPs, where the NBD location was a bit different. Overall, the conservation of individual domains in their respective clades may imply their significance for functional specificity.
There is a potential connection between ARP-based chromatin dynamics and the control of diverse developmental processes in plants [86]. ARP4 may exert its effect on plant architecture, floral senescence, flowering time, and fertility through adjustment of chromatin structure and changes in corresponding gene regulation [86]. ARP4 and ARP7 are likely to be involved in chromatin remodeling and transcriptional control during mitotic interphase [87]. ARP4- and ARP5-deficient plants are both hypersensitive to DNA-damaging agents, and a role for ARP5 in DNA repair was demonstrated [88]. ARP5-deficient plants display growth inhibition, morphological changes of individual cells, and abnormal organ development. Presumably, the functions of ARP5 are crucial to normal epigenetic control in Arabidopsis [88]. ARP5 and the chromatin-remodeler INO80 form a larger protein complex that has a synergistic effect in plant cell proliferation and response to replication stress [89]. At the same time, INO80 and ARP6 collaborate in embryogenesis and postembryonic plant development, while the synergy of ARP5 and ARP6 contributes to the maintenance of genomic stability [89]. Likewise, the arp7 mutant and ARP7 RNAi lines show abnormalities at different stages of plant growth and development due to the epigenetic impact of ARP7 complexes on chromatin-mediated regulation of gene expression [90,91].
Functional investigations of ARP proteins show that they form stable hetero-multimeric protein complexes with other proteins [92]. For instance, ARP2/3 complexes are implicated in generating the actin arrays related to polarized growth [11,93]. In leaf pavement cells of Arabidopsis, the ARP2/3 complex covers the surface of organelles that can efficiently bind to actin filaments and the microtubule cytoskeleton, meaning that the function of ARP2/3 affects the structure of G- and F-actin during reconfigurations of the cytoskeleton [94]. ARP2/3 is intrinsically inactive [95], but its complex can be converted to a potent actin filament nucleator via interaction with nucleation promoting proteins, such as the Wiskott-Aldrich syndrome protein (WASP)/Scar homolog (WASH), the WASP homolog associated with actin, membranes and microtubules (WHAMM), and the junction-mediating regulator protein (JMY) [96]. ARP3/DISTORTED1 (DIS1) acts in amyloplast sedimentation through altering apparent local viscosity in the central columella cells and redistributes asymmetric auxin in the root tip by modification of PIN-FORMED (PIN) protein trafficking [97]. In plants, formin homologs and the ARP2/3 complex are the known actin nucleators. These two actin nucleation systems, the Arabidopsis thaliana FORMIN HOMOLOGY 1 (FH1) and the ARP2/3 complex subunit 5 (ARPC5), have complementary or parallel functions in terms of some aspects of cell morphogenesis [98]. The ARP2/3 complex, together with FH1, mediates actin patch formation, thereby contributing to host cellular defenses and penetration resistance against fungal invasion [99]. The ARP2/3 complex precisely regulates guard cell actin remodeling and stomatal movement in order to ensure an appropriate stomatal aperture in response to environmental challenges [100]. Additionally, the ARP2/3 complex can participate in mitochondrial-associated calcium signaling pathway to respond to salt stress [101]. In brief, most of our understanding of distinct ARP functions is based on how various ARP-containing complexes work, but our knowledge of their functional characteristics in plants is still in its infancy. It would be interesting to explore whether there are differences between animal and plant ARP features.
4. Role of ABPs: An Accurate Network Controller of Plant Actin Dynamics
Under normal conditions, about 95% of the actin cytoskeleton is composed of monomeric actin in a state of readiness [102,103]. The actin monomer is a core part of the microfilament skeleton and can receive signals or be regulated by actin monomer binding proteins, so that the actin cytoskeleton can quickly and accurately respond to changes in the external environment. Concurrent with the evolution of actin, certain specific ABPs also gradually developed [104]. Different ABPs not only regulate actin activity, but also play an important role in promoting nucleation, which controls the assembly and formation of microfilaments (Table 1). ABPs are divided into two categories according to their function: one category keeps the balance of an actin monomer pool through affecting the dynamics and assembly of the unit structure of microfilament; the other category can arrange the microfilament into more complex structures [105]. Here, we highlight the main classes of ABP found in plant cells and suggest their likely mechanism of action, as far as possible, based on both in vitro or in vivo studies.

Table 1.
Biochemical properties of representative plant ABPs in actin cytoskeleton.
4.1. Fimbrin
Fimbrins possess the actin-binding domain (ABD) composed of two tandem calponin-homology (CH) domains. Each fimbrin contains two ABDs, enabling it to crosslink actin filaments as a monomer and generate high-order actin structures [128,129]. Arabidopsis has five FIMBRIN genes, with FIMBRIN1 (FIM1) and FIM5 being crucial for maintaining the polarity of actin bundles in pollen tubes and playing a role in pollen development [120,121,130]. FIM4 acts in coordination with FIM5 to organize and maintain normal actin architecture in pollen tubes and pollen grains, thus fulfilling double fertilization in Arabidopsis [131]. Additionally, fimbrin-dependent cross-linking plays a significant role in creating robust microfilament bundles that facilitate cytoplasmic streaming. Fimbrin serves to safeguard these bundles from severing and depolymerizing agents [121,132]. Due to the distinct tissue expression patterns and biochemical activities observed in Arabidopsis fimbrins, it is believed that they function in fulfilling diverse actin-based physiological cellular processes [133,134].
4.2. Formin
Formin is a type of actin nucleation factor that has been implicated in the formation of linear actin bundles [135]. Formin proteins are distinguished by the presence of FH1 and FH2 domains, which have the ability to nucleate actin assembly from actin or actin-profilin complexes [135]. The formins have been implicated in many actin-based cellular processes in plants, including root growth, polarized pollen tube growth, cell division, cytokinesis, cell morphogenesis, and plant defense [136,137,138,139,140,141]. In Arabidopsis, there are a total of 11 class I formins and 10 class II formins. Among these, the class I formins possess a distinctive transmembrane domain at their N-terminus, allowing them to specifically target membranes [142]. During the ontogeny of root cells, FH1 undergoes relocation between membrane compartments and forms associations with PD [143]. FH2 regulates PD permeability by anchoring actin filaments to PD, which is crucial for normal intercellular trafficking [144]. In addition, rice formin protein OsFH13 is a putative traffic protein from the PM to the chloroplast membrane and bridges the actin cytoskeleton and light signaling [145]. A central mechanism in plant immune signaling suggests that rapid actin remodeling occurs through the nanoclustering of formin integrated into the PM [146,147]. In short, formins might have a shared role at cell–cell junctions in plant.
4.3. Capping Protein
Highly conserved homologs of capping protein (CP) are present in almost all eukaryotic cells, including higher plants, fungi, and various cells and tissues in vertebrates [127]. In vitro, CP, as a heterodimeric protein complex, binds tightly to the barbed ends of actin filaments, effectively preventing the addition or loss of actin subunits [127]. As actin polymerization occurs, AtCP is an efficient nucleator of actin filament formation from monomers and shortens the delay period prior to actin assembly. The activity of AtCP is not affected by calcium, but it does exhibit moderate sensitivity to the signaling lipid phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] [127]. In fact, CP is a phosphatidic acid biosensor and converter of fluxes of membrane-signaling phospholipids into dynamic changes in the actin cytoskeleton [148]. During plant innate immunity, the negative regulation of CP by phosphatidic acid plays a vital role in actin remodeling, and CP also act as an intermediary in ROS signaling to the actin cytoskeleton [149,150]. Moreover, AtC α and β subunits (i.e., AtCPA and AtCPB) show distinct expression patterns in vivo, and the downregulation of AtCPB leads to enhance thermotolerance in plants following exposure to heat shock stress [151].
4.4. Villin
Villins regulate actin by promoting actin-bundling [152]. These actin-bundling proteins crosslink adjacent actin to bundle several parallel actin filaments [153]. In Arabidopsis, there are five VILLINs divided into three functionally distinct groups [154]. Even when plant cells receive signals that lead to the depolymerization of actin arrays, such as actin-depolymerizing factor (ADF)-mediated actin filament depolymerization, VILLIN1 maintains the cable network by regulating actin filament bundle formation and stability [115]. Nevertheless, ADF7 inhibits VILLIN1 to modulate F-actin dynamics in root hair formation in response to osmotic stress [155]. VILLIN3 phosphorylation by MITOGEN-ACTIVATED PROTEIN KINASE 3/6 (MPK3/6) regulates actin remodeling to trigger stomatal defense in Arabidopsis [156]. Besides bundling F-actin, VILLIN3 can sever actin filaments in a Ca2+-dependent manner and promote bundle turnover [154]. Similarly, the regulation of long axial and short apical actin bundles by VILLIN4 is important for cell growth and cytoplasmic streaming in root hairs, which is also influenced by Ca2+ signaling [6]. VILLIN4 plays a crucial role in regulating the dynamics of PIN2 in response to 2,3,5-triiodobenzoic acid, supporting the important role of actin dynamics in the mechanism of auxin transport [157]. In addition, the unifying mechanism behind the VILLIN2- and VILLIN5-mediated regulation of actin dynamics in the apical dome may be that VILLIN proteins sever the actin filaments at the apex of pollen tubes in conjunction with an apically focused Ca2+ gradient [114,158]. These actin bundles are essential for cell expansion during directional organ growth [159]. Given that VILLINs perform multiple actions as Ca2+-responsive and F-actin regulatory proteins, it will be challenging to investigate how Ca2+ signaling affects the actin dynamics through the integration of VILLINs activity.
4.5. LIM Domain-Containing Protein
LIM domain-containing proteins (LIMs), a class of LIM domain proteins related to animal Cys-rich proteins, have been reported to initiate the formation of actin bundles, a significant assembly of the higher-order cytoskeleton [125,126]. In the six Arabidopsis thaliana LIMs, WLIM1, WLIM2a, and WLIM2b are widely expressed, whereas PLIM2a, PLIM2b, and PLIM2c are predominantly expressed in pollen [160]. Furthermore, the balance between PLIM2a/PLIM2b and FIMBRIN5 (FIM5) is essential to maintain the proper organization and normal bundling of longitudinal actin bundles in pollen tubes [122]. LIMs contribute to the regulation of actin bundling in virtually all plant cells as a highly specialized ABP family [160]. In lily pollen tubes, LlLIM1 plays a rational role in integrating endomembrane trafficking with a pH- and calcium-sensitive manner [125].
4.6. Myosin
The actin cytoskeleton and relevant motor proteins underlie the movement of plant organelles and multiple materials within plant cells. Two myosin families, VIII and XI, drive cytoplasmic streaming, organize actin, and regulate cell expansion [161]. Myosin VIII (four genes in A. thaliana) in the roots of maize and cress localizes on the PD and at newly formed cell plates [162]. Myosin VIII may participate in intercellular transport through PD [163]. Enzymatically, myosin VIII can generate or sense tension, and this tension makes the PD better for cargo transference and junction formation between the endoplasmic reticulum and the PM [45,164]. Among the 13 A. thaliana myosin XI isoforms, myosin XI1, XI2, and XIK generate force that enables the buckling and straightening of actin filaments and bundles, as well as facilitate actin filament turnover [165]. Moreover, myosin XI functions in vesicle exocytosis and cellulose production at the cytoskeleton-PM-cell wall nexus [166]. Specifically, myosin XIK associates with secretory vesicles earlier than the exocyst, and it is likely responsible for recruiting or stabilizing the exocyst at the PM tethering site to facilitate vesicle tethering [167]. Myosin XIG regulates the meshwork F-actin movement via myosin functions that are distinct from organelle movement [42]. The four functional domains of myosins enable them to glide along actin filaments using energy from ATP hydrolysis and then to transport organelles or protein complexes. It is the close relationship between myosins and cytoplasmic streaming that plays an important role in the polar growth by affecting directional tip elongation of typically long cells, such as root hairs, pollen tubes, and moss protonemata [168,169,170]. Moreover, myosin function in cell division is distinct from driving cytoplasmic streaming [171]. Because organelle traffic and cytoplasmic flow occur in both growing and fully differentiated cells, the functional allocation between the molecular motor- and the membrane receptor-dependent pathway needs to be further confirmed while these processes facilitate intracellular homeostasis [3].
4.7. Profilin
Profilin (PRF) has a profound influence on the organization of the actin cytoskeleton mainly through sequestering actin monomers to maintain the level of polymerizable actin monomers. PRF can bind to proteins containing proline-rich sequences and phospholipids, which further drives PRF to bind actin monomers in multiple ways. The conserved domain in PRFs is the PRF-actin interacting region (PAINR), which plays vital roles in the binding process [172,173]. PRFs, in direct or indirect interaction with membranes, transmit information between the actin cytoskeleton and the PM via PHOSPHATIDYLINOSITOL 4, 5-BISPHOSPHATE (PIP2) [118,174]. PRFs can affect fiber growth in cotton, cell shape maintenance in Arabidopsis, as well as flowering time in tobacco [175]. In addition to influencing various aspects of cellular development, PRFs also positively contribute to management of stresses, such as salinity stress [175,176]. PRFs may coordinate with formins during actin polymerization. The polyproline tract sequences located in FH1 enable the profilin-actin complex to bind FH1 [177,178]. During plant cell expansion, PRF1 coordinates the stochastic dynamic of actin filaments by modulating formin-mediated actin nucleation and assembly [179]. AtPRF3 is an atypical isoform of profilin found in Arabidopsis, and it possesses an N-terminal extension that results in protein oligomerization and hampers the formin-mediated actin assembly [180]. Furthermore, AtPRF3 regulates the immune responses triggered by pathogen associated molecular patterns (PAMPs), which in turn also influence the degradation of AtPRF3 [180]. In pollen germination, PRF4 and PRF5 control vesicle movement and polarity establishment by facilitating FH5-mediated actin polymerization and strengthening the interaction between FH5 and actin filaments [181].
4.8. Cyclase-Associated Protein
Another class of proteins that can bind actin monomers is the cyclase-associated proteins (CAPs), which perform multiple functions due to their two distinct domains that bind adenylyl cyclase and G-actin at the N- or C-terminus, respectively [182]. In contrast to plant PRFs, CAP1 is capable of directly accelerating the nucleotide exchange on G-actin, even without the role of ADF/cofilin [183]. It has been speculated that coupling between ADF/cofilin-mediated depolymerization of actin filaments and PRF-mediated assembly of ATP-G-actin needs a CAP protein as a key intermediate [123]. CAP is also involved in the process of regulating cell expansion by the actin cytoskeleton in Arabidopsis. Both the number and size of leaf cells are altered through overexpression of CAP, which causes a reduction in organ size [184]. During normal pollen tube growth, CAP1 is an abundant cellular protein that acts in concert with PRF and ADF to enhance actin turnover and ADP-G-actin nucleotide exchange in vitro. The change of apical actin polymerization has differential effects on various regions of pollen tubes by altering the actin cytoskeleton [185]. In addition, a defect of CAP1 alters the developmental tendency of multiple cell types, such as meandering roots and curling inflorescences [186].
4.9. Actin-Depolymerizing Factor
The ADF family contains well-characterized ABPs that can change orientation and arrangement of actin filaments through binding monomeric actin or modifying filamentous actin [187,188]. The diverse tissue expression patterns of the 11 Arabidopsis ADFs suggest that they have evolved different physiological characteristics [189].
In pollen tubes of Arabidopsis, ADF7 evolved to promote turnover of longitudinal actin cables through severing actin filaments that might occur in the actin fringe [190,191]. ADF10 arranges filamentous actin around the pore of the mature pollen grain in the developing gametophyte [192] and promotes the circulation and arrangement of the apical actin filament to regulate vesicle trafficking and pollen tube growth [193]. In Nicotiana tabacum, overexpression of NtADF1 in elongating pollen tubes disrupts the actin cytoskeleton and causes a decrease in growth rate [194]. In Physcomitrium patens, F-actin organization is altered with loss of ADF function, resulting in an inhibition of tip growth [195]. Phospho-regulation at serine 6 is a requisite for effects of ADF on polarized growth [195]. Beyond that, an altered expression level of ADF1 or ADF9 affects F-actin organization, flowering time, and cell expansion in Arabidopsis [196]. ADF9 is an actin-bundling protein whose activity is adjusted by pH conditions and antagonizes ADF1 activity through reducing its ability to potentiate F-actin depolymerization [110]. ADF9 is a novel photoperiod-dependent early flowering repressor, which is regulated by CONSTANS (CO)- and FLOWERING LOCUS C (FLC)-related networks [196,197]. ADF4 plays a role in the process of modulating actin filament turnover, and the adf4 mutant displays altered cytoskeletal arrays and morphologies of hypocotyl and epidermal cells [198].
The functions of ADF family proteins are implicated in response of plants to environmental stimuli, but the evidence is indirect or limited. ADF4 has been identified as a signaling component that can transport the Pseudomonas syringae effector proteins Avirulence protein Pseudomonas phaseolicola B (AvrPphB), and RESISTANT TO PSEUDOMONAS SYRINGAE 5 (RPS5) around the PM by mediating rearrangement of the actin cytoskeleton, which aids in the subsequent identification of cargo [199]. Inhibition of ADF4 activity during innate immune signaling can modulate actin dynamics so as to implement PAMP-triggered immunity (PTI)-related response strategies in Arabidopsis [200]. ADF4 is a physiological substrate of CALCIUM-DEPENDENT PROTEIN KINASE 3 (CPK3), and phosphorylation of ADF4 by CPK3 controls actin cytoskeletal organization associated with pattern-triggered immunity [201]. During the response to abiotic stress, ADF5 may be involved in the ABA signaling pathway and regulate the actin cytoskeleton to affect stomatal movement in response to drought stress [202]. Under a low-temperature environment, C-REPEAT/DRE BINDING FACTORs (CBFs) can bind to the CRT/DRE DNA regulatory element of the ADF5 promoter, which activates the expression of ADF5 and modulates the actin cytoskeleton dynamics [203]. The opposing and diverse biochemical properties of plant ADFs are caused by evolutionary changes in key amino acids [106]. The regulatory mechanism of ADF activity in the organization of F-actin and the correlation between ADF expression and nuclear function should be investigated in more detail.
5. Signals and Pathways Regulating the Actin Cytoskeleton
Many direct correlations between the actin cytoskeleton reorganization and signal transduction exist in plants, including polarized growth, Ca2+ homeostasis, cytoplasmic streaming, and responses to extracellular stimulus (Figure 3). A focus of research in this area will make it easier to analyze the evolutionary direction of actin function.
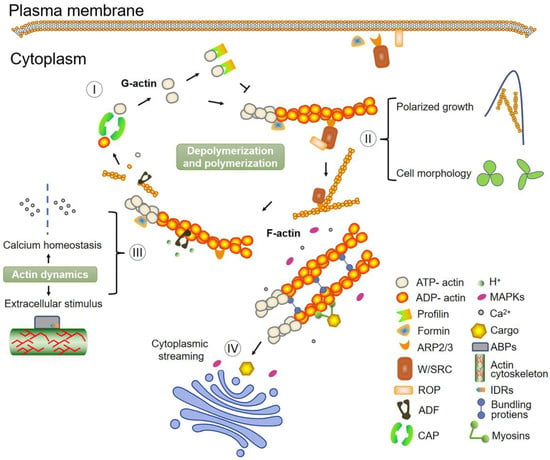
Figure 3.
Fundamental model depicting actin dynamics and various processes that involve the actin filaments. (I) The conversion from ADP-actin to ATP-actin is accomplished by CAP and is a strategic step that guarantees the polymerization cycle. The nucleation factors, like ARP2/3 and the formins, order the polymerization of the actin monomers (G-actin). Profilin can also bind actin monomers to maintain the actin monomer pool by inhibiting the polymerization of actin filaments. Generated actin filaments (F-actin) are bundled at different angles via binding proteins or crosslinkers. Any depolymerized actin filaments will re-enter the assembly system. (II–IV) The dynamic changes of the actin cytoskeleton participate in plant cell development through the functional association of different signaling pathways. Filamentous actin arrays are associated with plant cell growth, and the activity of ABPs is essential for proper cell morphogenesis. Cytoplasmic streaming plays a vital role in the transportation of materials necessary for tip elongation. Meanwhile, actin reorganization during cellular processes, such as polar growth and cytoplasmic streaming, is frequently correlated with calcium influxes or intracellular calcium gradients.
The RHO-RELATED GTPases (ROPs) is a family of small signaling GTPases in plants that can transmit specific signals to the actin cytoskeleton in response to intracellular and extracellular signals [204,205]. Fine actin architecture and ROPs are localized in the tip area of apical cells. Multidimensional cell expansion during early stages of tissue development may be regulated by a related mechanism involving ROP signaling-dependent formation of cortical F-actin [206,207]. Downstream of the ROP and phosphoinositide (PI) signaling pathway, Ca2+ concentration and pH level modulate ABPs, which interact with phospholipids and several critical proteins in a phosphorylation-dependent manner [208,209,210]. Moreover, activity of ARP2/3 is positively regulated by the WASP FAMILY VERPROLIN HOMOLOGOUS PROTEIN (WAVE) complex that may be a ROP2 effector complex [211]. The readjustment of actin filament dynamics under the WAVE-ARP2/3 pathway can guide the polar growth of early stage trichomes [211]. These two complexes function in nucleating actin filaments, which ultimately leads to changes in cell morphology.
Reactive oxygen species (ROS) not only serve as significant stressors for cellular components but also play diverse roles in cell physiology [212]. There are the crucial connections between ROS signaling and actin cytoskeleton [213]. ARP2/3-mediated actin dynamics are essential for stomatal movement in response to ABA-induced ROS signaling [100,214,215]. Actin arrays in guard cells of respiratory burst oxidase homologues D/F (rbohD/F) mutants treated with ABA fail to reorganize, whereas applying H2O2 to rbohD/F mutants recapitulates actin remodeling in the absence of ABA [215]. PHOSPHOLIPASE Dα1 (PLDα1)-generated phosphatidic acid (PA) functions upstream and binds directly to RBOHD, resulting in the production of ROS during ABA signaling and subsequent stomatal closure [216]. In addition, exogenous ROS treatment induces the accumulation of actin filaments in leaf epidermal cells. During plant innate immunity, it has been found that CP is essential for transducing RBOHD/ROS signaling to facilitate actin remodeling in response to flg22, a 22-amino-acid epitope derived from flagellin [150]. Since PA regulates actin remodeling through CP, there is a negative feedback regulation by CP or the actin cytoskeleton to modulate ROS production elicited by flg22 [217].
The plant hormone cytokinin (CK) plays pivotal roles in plant development and throughout plant life. Root system architecture induced by CK involves the reorganization of actin filament [218]. In particularly, the transition from the transition zone (TZ) to the elongation/differentiation zone (EDZ) is an important mode of rapid cell elongation in epidermal and cortical cells of Arabidopsis roots [69]. During this time, CK promotes actin bundling and the resultant cell elongation through activating the ARABIDOPSIS HISTIDINE KINASE 3/4 (AHK3/4)-ARABIDOPSIS RESPONSE REGULATOR 2 (ARR2) pathway [69]. Additionally, the impact of CK on the actin cytoskeleton could lead to changes in trafficking rates and paths for endomembrane compartments, which could affect the distribution of defense-related cargo and result in altered defense signaling [219]. Thus, the modulation of the cellular cytoskeleton and trafficking could potentially serve as a mechanism that executes downstream responses of CK signaling [219].
The plant chloroplast and nucleus change position in response to light. Plant cells have seemingly evolved distinct mechanisms to regulate actin organization, which is necessary for driving the movements of these organelles [220]. In Arabidopsis leaf cells, blue-light-dependent nuclear positioning is regulated by the blue light receptor PHOTOTROPIN 2 (PHOT2)-dependent reorganization of the actin cytoskeleton [221]. The chloroplast-actin filaments (cp-actin filaments) emerge from the chloroplast edge and display rapid turnover. When chloroplast movement is induced by blue light, the cp-actin filaments undergo relocalization to the leading edge of chloroplasts both before and during photorelocation and are regulated by PHOT1 and PHOT2 [222]. THRUMIN1 plays a key role in connecting phototropin photoreceptor activity at the PM with actin-dependent chloroplast movements [223,224]. Moreover, the spatial reorganization of F-actin is also affected by red light in water plants, where chloroplast movements are closely linked with cytoplasmic streaming [225]. In Physcomitrella patens, KINESIN-LIKE PROTEIN 1/2 (KAC1/2) mediate the actin-dependent chloroplast light avoidance response [226]. Therefore, actin-based mechanism is important for light signaling-directed organelles movement.
MITOGEN-ACTIVATED PROTEIN KINASEs (MAPKs) signal transduction is involved in nearly all regulation of crucial cellular processes. The cross-talk between actin dynamics and MAPK signaling exists in response to environment stimuli and cell shape and morphogenesis in plants. In alfalfa, STRESS-INDUCED MAPK (SIMK) and STRESS-ACTIVATED MAP KINASE (SAMK) are involved in responses to osmotic, cold, and heat stress. They are activated when the actin cytoskeleton is disrupted through treatment with microfilament depolymerising drugs [227,228]. MAPK signaling pathways may be the key sensor for balancing the intracellular forces used for controlling cellular architecture [229]. In Papaver rhoeas, self-fertilization and consequent inbreeding rely on the specific self-recognition of pollen regulated by self-incompatibility (SI) pathways. SI-induced tip growth inhibition may be accomplished through an alteration of F-actin by a Ca2+-dependent signaling cascade [230,231]. In addition, Ca2+ can modulate ABP activity by directly binding them or indirectly through Ca2+-stimulated protein kinases, such as calcium-dependent protein kinases (CDPKs) [208,209]. On the other hand, actin dynamics can regulate the activity of Ca2+-permeable channels to maintain Ca2+ homeostasis [232].
Intrinsically disordered regions (IDRs) are present in a majority of ABPs and affect the activity of ABPs indirectly by altering conformational changes during complex protein assembly [233]. In a sense, IDRs can create diverse and flexible interactions between ABPs and the actin cytoskeleton to influence various signal transduction events [233,234,235]. The distinctive characteristics of IDR-mediated plant actin remodeling would enrich our comprehension of the structure–function relationships involved in actin assembly [236]. In summary, the actin cytoskeleton can be thought of as a ubiquitous downstream signal effector that participates in plant individual growth and system development (Figure 4). The further understanding of key links between such signaling cascades and actin dynamics is a top priority.
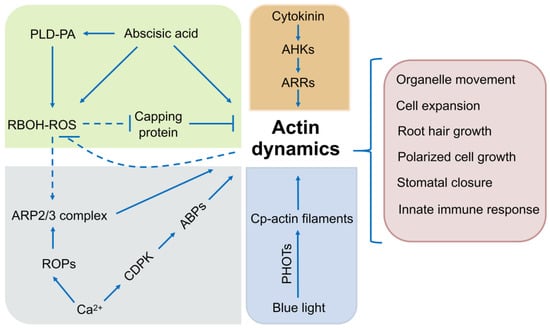
Figure 4.
Potential connections between the actin cytoskeleton and signaling molecules, involving ROS signaling, Ca2+ signaling, light signaling, cytokinin signaling, and ROP signaling. This scheme is based on published research data discussed in this review. Solid lines, direct interaction; dashed line, potential interaction; arrows, activation of signaling cascades; bars, inhibitory effect; PLD, PHOSPHOLIPASE D; PA, phosphatidic acid; RBOH, RESPIRATORY BURST OXIDASE HOMOLOGUE; CP, chloroplast; AHK, ARABIDOPSIS HISTIDINE KINASE; ARR, ARABIDOPSIS RESPONSE REGULATOR; ARP2/3, ACTIN-RELATED PROTEIN 2/3; ROP, RHO-RELATED GTPase; CDPK, calcium-dependent protein kinase; ABP, actin-binding protein.
6. Support for In-Depth Investigation on Plant Actin
Actin is one of the most highly conserved proteins across multiple kingdoms of life, with an 80% sequence conservation at the gene level between humans and the yeast Saccharomyces cerevisiae [104]. In mammalian cells, nuclear actin can regulate the transcription of induced genes by enhancing the aggregation of RNA polymerase II [28]. Actin genes in plant species have been identified by in silico analysis and other research approaches [237,238]. Whether it is the application of 3D culture systems based on organoids or the extension of super-resolution microscopy through fluorescence observation [239], animal cell materials seem to be more convenient for use of these new technologies and methods compared with plant cells. The further development of cryo-electron microscopy has made it possible to more accurately explore the structure of filamentous actin [240]. Using Zea mays pollen and rabbit skeletal muscle actin filaments, single-molecule magnetic tweezer and structural data analyses have found that plant actin filaments are more stable compared with animal actin filaments [241]. Differences in structure may determine the many functional differences between mammalian and plant actins that arose during their evolution. Therefore, it is a long process to further understand the dynamic regulation of actin in multiple plant cellular functions.
The cytoskeleton is an influential factor in the dynamics of subcellular membranes, organelle movement, membrane trafficking, and cellular morphogenesis [242,243,244]. However, not all proteins identified in animal systems to control membrane dynamics and the cytoskeleton operate similarly in plants, especially those proteins that act as membrane-actin adaptors. The mechanism of action of the actin-binding proteins in the NETWORKED (NET) superfamily represents an important field for evaluating plant actin cytoskeleton-endomembrane interactions [245]. In addition, different stages of autophagosome formation require a functional actin network as a support for vesicle trafficking and membrane fusion, which has been demonstrated in animal and yeast cells [246,247,248]. The function of the ARP2/3 complex in plant and mammalian cells is overall different [124,249]. In plants, the ARP2/3 complex and the related activators, the SCAR/WAVE and AtEH/Pan1 complexes, can inhibit autophagosome biogenesis when mutated, resulting in decreased abiotic stress resistance [250,251,252]. Although some autophagosome-actin associations have been summarized into detailed models [253], it remains to be confirmed whether the function of actin in autophagosome formation is independent or irreplaceable. Indeed, various mechanisms of action involving the actin cytoskeleton are conserved among organisms, but there are some notable exceptions, both in signaling components and in physiologically relevant links. Many actin-binding/regulatory proteins involved in apoptotic signaling pathways in mammalian cells have not been identified in yeast and do not exist in the Arabidopsis genome database [254]. Admittedly, it may be of considerable interest to investigate whether proteins with similar sequences or activities also perform similar functions across species.
The ability to fluorescently label proteins has allowed the increasingly accurate examination of the dynamic distribution of the actin cytoskeleton in living plants [15,255]. Meanwhile, advanced fluorescence microscopy has provided super-resolution approaches that have improved the spatial and temporal resolution of plant cell dynamics [256,257]. A combination of high-speed F-actin co-sedimentation assay and total internal reflection fluorescence microscopy (TIRFM) imaging technology effectively achieves direct visual analysis of actin filament severing [258]. For example, ACTIN-INTERACTING PROTEIN 1 (AIP1) and ADF synergistically regulate the turnover of actin filaments within the growth zone of pollen tubes, which pushes forward construction of the unique apical actin structure at the pollen tube tip [258]. Additionally, to comprehensively understand the cellular functions of actin organization and dynamics, some parameters have been created to quantify the organization of actin filaments, such as slope, filament density, and skewness [259,260]. From this perspective, the technological and inquisitive advances hold great promise for grasping the intricate details related to the actin cytoskeleton regulatory network at a molecular scale.
Action potentials are fundamental for facilitating long-distance signaling in plants and animals, and their functional linkage with the actin cytoskeleton associated with membranes is of utmost importance [261]. The actin cytoskeleton functions downstream of action potentials, which are accomplished through excitable membranes. Action potentials directly affect the lipid bilayer phospholipids and the proteins embedded within the PM. These include ion channels and transporters, as well as transmembrane proteins that directly regulate actin polymerization, such as formins [136,262,263,264,265]. In the root apex transition zone, the cells with the highest rates of electric spikes are the ones that assemble dense F-actin meshworks through formin activities. These meshworks play a critical role in supporting endocytosis and the recycling of endocytic vesicles [262,264,265]. Besides formins, myosin VIII may be relevant for the propagation of the action potentials [266,267]. The plant-specific actin-based endocytic motor is involved in the myosin-actin-based gating of plant plasmodesmata, which is important for the transmission of action potentials between cells [268,269,270]. Action potentials regulate various aspects of plant life [261]. Precise connections between the inherent bioelectricity of eukaryotic membranes and the actin cytoskeleton are still under-investigated, and studies in this field serve as a fundamental pillar in our pursuit to comprehend actin-related cellular life activities.
7. The Coordinating Effect of Microtubules and Microfilaments
Microtubules were originally viewed as a network with separate functions distinct from actin filaments. However, microfilaments and microtubules have repeatedly been found to work together in yeast and animal cells [271]. The dynamic interaction between actin filaments and microtubules in Arabidopsis has been confirmed by ultra-clear images derived from confocal microscopy [272,273], suggesting that there must exist indirect or direct connections. ABPs and microtubule-associated proteins (MAPs) are major players determining the spatiotemporal dynamics of microfilaments and microtubules and act as sensory hinges that converge different signals to regulate plant cytoskeletal behavior. For instance, the actin binding protein NETWORKED 3C (NET3C) interacts with two microtubule binding proteins, IQ67-DOMAIN 2 (IQD2) and KINESIN LIGHT CHAIN-RELATED PROTEIN 1 (KLCR1), to form a novel module for the organization and maintenance of endoplasmic reticulum (ER) morphology and cytoskeletal structure [274]. Members of the MICROTUBULE DESTABILIZING PROTEIN (MDP) family, MDP25 and MDP18, are mediated by Ca2+ signaling and affect microfilaments and microtubules during pollen tube growth [5,275,276,277]. MDP25-mediated actin dynamics are associated with MDP25 function under salt stress [278]. In addition, phragmoplasts are responsible for plant cytokinesis during cell expansion, and this structure is composed of microtubules, actin filaments, and membrane vesicles, all of which support the formation and expansion of the cell plate [279,280]. Cell plate formation is delayed by the breakdown of actin filaments, a process that may be linked to interruption of the initial phragmoplast microtubules [281]. In terms of polar growth, microtubules can direct the localization of actin nucleation factors, and the resulting actin filaments further focus the microtubules [282]. This process creates a positive feedback loop that brings actin polymerization and cell expansion together at a proper location to support sustainable polar growth [282].
8. Conclusions and Future Outlines
This review provides a wide discussion in regard to how actins, ARPs, and ABPs work together to drive cellular functions. Understanding of the multiple ways that F-actin influences the regulation of various cellular functions is one of the ultimate research goals in this field. The first further inquiry is finding out what higher-order structures are established by actin monomers and how the dynamic balance of the entire actin filament is maintained. Then, the numerous ABPs, including novel ABPs that have not yet been discovered, should be studied, including how microfilaments function and integrate into complex signal transduction processes. Because the components related to the actin cytoskeleton are highly conserved across many species, we can deduce functions through the subtle differences between sequences and work out similarities and connections to cellular components. Meanwhile, the development of super-resolution technologies and experimental strategies can be used to reveal the specific roles of the actin cytoskeleton in vital activities.
In plants, the signaling between actin and microtubules occurs all the time for many cellular processes. Many specific proteins ensure that the pivotal mechanisms that regulate microtubule–actin interactions operate. The proteins are conserved across diverse eukaryotes, which can serve as a major direction for follow-up investigations. It is also of interest to figure out what coordination between regulatory proteins modulate actin and microtubule dynamics under physiological conditions and how plants respond to extracellular challenges by harmonizing the activities and levels of ABPs and MAPs. These questions seem to point to a number of mechanisms underlying the control of MAPs and ABPs in cytoskeletal organization and dynamics. Finally, the newest 3D in vivo imaging combined with diverse fluorescent biosensors can be used to explore the above problems [273,283,284,285] and to address the gaps in our understanding of functional microtubule–actin interactions. Further dissection of cytoskeleton-related signaling mechanisms should enable plant improvement, supporting the development of sustainable and enhanced crops.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms242015480/s1.
Author Contributions
G.Y. and T.Y. conceived and designed the main content. G.Y. and H.G. designed and drew figures, and G.Y. summarized a table. G.Y. and T.Y. wrote the article. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (NSFC; Grant No. 31770199 and 31700215); Natural Science Foundation of Gansu province (21JR7RA487); the Natural Science Foundation of Gansu Province, Gansu Excellent Doctoral Program (No. 23JRRA1159 and 22JR5RA415); the Foundation of the Key Laboratory of Cell Activities and Stress Adaptations, Ministry of Education of China (lzujbky-2021-kb05); the Fundamental Research Funds for the Central Universities (lzujbky-2021-43).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
This is a review article. All data used in this research are included in this article and its supplementary information files.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Blanchoin, L.; Boujemaa-Paterski, R.; Sykes, C.; Plastino, J. Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 2014, 94, 235–263. [Google Scholar] [CrossRef]
- Pollard, T.D.; Cooper, J.A. Actin, a central player in cell shape and movement. Science 2009, 326, 1208–1212. [Google Scholar] [CrossRef] [PubMed]
- Peremyslov, V.V.; Cole, R.A.; Fowler, J.E.; Dolja, V.V. Myosin-powered membrane compartment drives cytoplasmic streaming, cell expansion and plant development. PLoS ONE 2015, 10, e0139331. [Google Scholar] [CrossRef] [PubMed]
- Szymanski, D.; Staiger, C.J. The actin cytoskeleton: Functional arrays for cytoplasmic organization and cell shape control. Plant Physiol. 2018, 176, 106–118. [Google Scholar] [CrossRef]
- Lian, N.; Wang, X.W.; Jing, Y.P.; Lin, J.X. Regulation of cytoskeleton-associated protein activities: Linking cellular signals to plant cytoskeletal function. J. Integr. Plant Biol. 2021, 63, 241–250. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, Y.Y.; Du, F.; Cao, L.J.; Dong, H.J.; Ren, H. Arabidopsis VILLIN4 is involved in root hair growth through regulating actin organization in a Ca2+-dependent manner. New Phytol. 2011, 190, 667–682. [Google Scholar] [CrossRef]
- Blanchoin, L.; Boujemaa-Paterski, R.; Henty, J.L.; Khurana, P.; Staiger, C.J. Actin dynamics in plant cells: A team effort from multiple proteins orchestrates this very fast-paced game. Curr. Opin. Plant Biol. 2010, 13, 714–723. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D.; Borisy, G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell 2003, 112, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Goodson, H.V.; Hawse, W.F. Molecular evolution of the actin family. J. Cell Sci. 2002, 115, 2619–2622. [Google Scholar] [CrossRef]
- Schafer, D.A.; Schroer, T.A. Actin-related proteins. Annu. Rev. Cell Dev. Biol. 1999, 15, 341–363. [Google Scholar] [CrossRef] [PubMed]
- McKinney, E.C.; Kandasamy, M.K.; Meagher, R.B. Arabidopsis contains ancient classes of differentially expressed actin-related protein genes. Plant Physiol. 2002, 128, 997–1007. [Google Scholar] [CrossRef]
- Pollard, T.D.; Beltzner, C.C. Structure and function of the Arp2/3 complex. Curr. Opin. Struct. Biol. 2002, 12, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Olave, I.A.; Reck-Peterson, S.L.; Crabtree, G.R. Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 2002, 71, 755–781. [Google Scholar] [CrossRef] [PubMed]
- Fyodorov, D.V.; Kadonaga, J.T. The many faces of chromatin remodeling: SWItching beyond transcription. Cell 2001, 106, 523–525. [Google Scholar] [CrossRef] [PubMed]
- Caillaud, M.C. Methods to visualize the actin cytoskeleton during plant cell division. Methods Mol. Biol. 2022, 2382, 1–16. [Google Scholar] [CrossRef]
- Nick, P. Microtubules, signalling and abiotic stress. Plant J. 2013, 75, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Wada, M.; Kong, S.G. Actin-mediated movement of chloroplasts. J. Cell Sci. 2018, 131, jcs210310. [Google Scholar] [CrossRef]
- Sedbrook, J.C. MAPs in plant cells: Delineating microtubule growth dynamics and organization. Curr. Opin. Plant Biol. 2004, 7, 632–640. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Meagher, R.B.; McKinney, E.C.; Vitale, A.V. The evolution of new structures: Clues from plant cytoskeletal genes. Trends Genet. 1999, 15, 278–284. [Google Scholar] [CrossRef]
- Nagao, R.T.; Shah, D.M.; Eckenrode, V.K.; Meagher, R.B. Multigene family of actin-related sequences isolated from a soybean genomic library. DNA 1981, 1, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Fukui, Y.; Katsumaru, H. Nuclear actin bundles in Amoeba, Dictyostelium and human HeLa cells induced by dimethyl sulfoxide. Exp. Cell Res. 1979, 120, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Clark, T.G.; Rosenbaum, J.L. An actin filament matrix in hand-isolated nuclei of X. laevis oocytes. Cell 1979, 18, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Bettinger, B.T.; Gilbert, D.M.; Amberg, D.C. Actin up in the nucleus. Nat. Rev. Mol. Cell Biol. 2004, 5, 410–415. [Google Scholar] [CrossRef]
- Scheer, U.; Hinssen, H.; Franke, W.W.; Jockusch, B.M. Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell 1984, 39, 111–122. [Google Scholar] [CrossRef]
- Wasser, M.; Chia, W. The EAST protein of drosophila controls an expandable nuclear endoskeleton. Nat. Cell Biol. 2000, 2, 268–275. [Google Scholar] [CrossRef]
- Rando, O.J.; Zhao, K.; Janmey, P.; Crabtree, G.R. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc. Natl. Acad. Sci. USA 2002, 99, 2824–2829. [Google Scholar] [CrossRef]
- Wei, M.; Fan, X.Y.; Ding, M.; Li, R.F.; Shao, S.P.; Hou, Y.P.; Meng, S.S.; Tang, F.C.; Li, C.; Sun, Y.J. Nuclear actin regulates inducible transcription by enhancing RNA polymerase II clustering. Sci. Adv. 2020, 6, eaay6515. [Google Scholar] [CrossRef]
- Kapoor, P.; Shen, X. Mechanisms of nuclear actin in chromatin-remodeling complexes. Trends Cell Biol. 2014, 24, 238–246. [Google Scholar] [CrossRef]
- Percipalle, P. Co-transcriptional nuclear actin dynamics. Nucleus 2013, 4, 43–52. [Google Scholar] [CrossRef]
- Hofmann, W.A.; Stojiljkovic, L.; Fuchsova, B.; Vargas, G.M.; Mavrommatis, E.; Philimonenko, V.; Kysela, K.; Goodrich, J.A.; Lessard, J.L.; Hope, T.J.; et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat. Cell Biol. 2004, 6, 1094–1101. [Google Scholar] [CrossRef] [PubMed]
- Philimonenko, V.V.; Zhao, J.; Iben, S.; Dingova, H.; Kysela, K.; Kahle, M.; Zentgraf, H.; Hofmann, W.A.; de Lanerolle, P.; Hozak, P.; et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat. Cell Biol. 2004, 6, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Vartiainen, M.K.; Guettler, S.; Larijani, B.; Treisman, R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 2007, 316, 1749–1752. [Google Scholar] [CrossRef] [PubMed]
- Dundr, M.; Ospina, J.K.; Sung, M.H.; John, S.; Upender, M.; Ried, T.; Hager, G.L.; Matera, A.G. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J. Cell Biol. 2007, 179, 1095–1103. [Google Scholar] [CrossRef]
- Baarlink, C.; Plessner, M.; Sherrard, A.; Morita, K.; Misu, S.; Virant, D.; Kleinschnitz, E.M.; Harniman, R.; Alibhai, D.; Baumeister, S.; et al. A transient pool of nuclear F-actin at mitotic exit controls chromatin organization. Nat. Cell Biol. 2017, 19, 1389–1399. [Google Scholar] [CrossRef]
- Schrank, B.R.; Aparicio, T.; Li, Y.; Chang, W.; Chait, B.T.; Gundersen, G.G.; Gottesman, M.E.; Gautier, J. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 2018, 559, 61–66. [Google Scholar] [CrossRef]
- Caridi, C.P.; D’Agostino, C.; Ryu, T.; Zapotoczny, G.; Delabaere, L.; Li, X.; Khodaverdian, V.Y.; Amaral, N.; Lin, E.; Rau, A.R.; et al. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 2018, 559, 54–60. [Google Scholar] [CrossRef]
- Percipalle, P.; Zhao, J.; Pope, B.; Weeds, A.; Lindberg, U.; Daneholt, B. Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J. Cell Biol. 2001, 153, 229–236. [Google Scholar] [CrossRef]
- Kukalev, A.; Nord, Y.; Palmberg, C.; Bergman, T.; Percipalle, P. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat. Struct. Mol. Biol. 2005, 12, 238–244. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; McKinney, E.C.; Meagher, R.B. Differential sublocalization of actin variants within the nucleus. Cytoskeleton 2010, 67, 729–743. [Google Scholar] [CrossRef]
- Kijima, S.T.; Staiger, C.J.; Katoh, K.; Nagasaki, A.; Ito, K.; Uyeda, T.Q.P. Arabidopsis vegetative actin isoforms, AtACT2 and AtACT7, generate distinct filament arrays in living plant cells. Sci. Rep. 2018, 8, 4381. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.F.; Fatema, U.; Peng, X.B.; Hacker, S.W.; Maruyama, D.; Sun, M.X.; Kawashima, T. ARP2/3-independent WAVE/SCAR pathway and class XI myosin control sperm nuclear migration in flowering plants. Proc. Natl. Acad. Sci. USA 2020, 117, 32757–32763. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.J.; Liu, J.Y.; Xue, X.H.; Tan, K.; Wang, C.B.; Su, H. The migration direction of hair cell nuclei is closely related to the perinuclear actin filaments in Arabidopsis. Biochem. Biophys. Res. Commun. 2019, 519, 783–789. [Google Scholar] [CrossRef]
- Pitzalis, N.; Heinlein, M. The roles of membranes and associated cytoskeleton in plant virus replication and cell-to-cell movement. J. Exp. Bot. 2018, 69, 117–132. [Google Scholar] [CrossRef] [PubMed]
- White, R.G.; Barton, D.A. The cytoskeleton in plasmodesmata: A role in intercellular transport? J. Exp. Bot. 2011, 62, 5249–5266. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Su, S.Z.; Liu, Z.H.; Chen, C.; Zhang, Y.; Wang, X.; Zhu, L.; Miao, L.; Wang, X.C.; Yuan, M. Cucumber mosaic virus movement protein severs actin filaments to increase the plasmodesmal size exclusion limit in tobacco. Plant Cell 2010, 22, 1373–1387. [Google Scholar] [CrossRef]
- Faulkner, C.R.; Blackman, L.M.; Collings, D.A.; Cordwell, S.J.; Overall, R.L. Anti-tropomyosin antibodies co-localise with actin microfilaments and label plasmodesmata. Eur. J. Cell Biol. 2009, 88, 357–369. [Google Scholar] [CrossRef]
- Deeks, M.J.; Calcutt, J.R.; Ingle, E.K.; Hawkins, T.J.; Chapman, S.; Richardson, A.C.; Mentlak, D.A.; Dixon, M.R.; Cartwright, F.; Smertenko, A.P.; et al. A superfamily of actin-binding proteins at the actin-membrane nexus of higher plants. Curr. Biol. 2012, 22, 1595–1600. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; McKinney, E.C.; Meagher, R.B. Functional nonequivalency of actin isovariants in Arabidopsis. Mol. Biol. Cell 2002, 13, 251–261. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; Burgos-Rivera, B.; McKinney, E.C.; Ruzicka, D.R.; Meagher, R.B. Class-specific interaction of profilin and ADF isovariants with actin in the regulation of plant development. Plant Cell 2007, 19, 3111–3126. [Google Scholar] [CrossRef]
- Kim, J.Y.; Ahn, J.; Bong, H.; Wada, M.; Kong, S.G. ACTIN2 functions in chloroplast photorelocation movement in Arabidopsis thaliana. J. Plant Biol. 2020, 63, 379–389. [Google Scholar] [CrossRef]
- Sheahan, M.B.; Collings, D.A.; Rose, R.J.; McCurdy, A.D.W. ACTIN7 is required for perinuclear clustering of chloroplasts during Arabidopsis protoplast culture. Plants 2020, 9, 225. [Google Scholar] [CrossRef] [PubMed]
- Mendrinna, A.; Persson, S. Root hair growth: It’s a one way street. F1000Prime Rep. 2015, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Mathur, J.; Mathur, N.; Kirik, V.; Kernebeck, B.; Srinivas, B.P.; Hulskamp, M. Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 2003, 130, 3137–3146. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Bi, S.T.; Wang, L.; Li, H.P.; Gao, B.A.; Huang, S.J.; Qu, X.L.; Cheng, J.N.; Wang, S.C.; Liu, C.Y.; et al. GLABRA2 regulates actin bundling protein VILLIN1 in root hair growth in response to osmotic stress. Plant Physiol. 2020, 184, 176–193. [Google Scholar] [CrossRef] [PubMed]
- Ringli, C.; Baumberger, N.; Diet, A.; Frey, B.; Keller, B. ACTIN2 is essential for bulge site selection and tip growth during root hair development of Arabidopsis. Plant Physiol. 2002, 129, 1464–1472. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; McKinney, E.C.; Meagher, R.B. A single vegetative actin isovariant overexpressed under the control of multiple regulatory sequences is sufficient for normal Arabidopsis development. Plant Cell 2009, 21, 701–718. [Google Scholar] [CrossRef]
- Nishimura, T.; Yokota, E.; Wada, T.; Shimmen, T.; Okada, K. An Arabidopsis ACT2 dominant-negative mutation, which disturbs F-actin polymerization, reveals its distinctive function in root development. Plant Cell Physiol. 2003, 44, 1131–1140. [Google Scholar] [CrossRef]
- Lanza, M.; Garcia-Ponce, B.; Castrillo, G.; Catarecha, P.; Sauer, M.; Rodriguez-Serrano, M.; Paez-Garcia, A.; Sanchez-Bermejo, E.; T C, M.; Leo del Puerto, Y.; et al. Role of actin cytoskeleton in brassinosteroid signaling and in its integration with the auxin response in plants. Dev. Cell 2012, 22, 1275–1285. [Google Scholar] [CrossRef]
- Gilliland, L.U.; Pawloski, L.C.; Kandasamy, M.K.; Meagher, R.B. Arabidopsis actin gene ACT7 plays an essential role in germination and root growth. Plant J. 2003, 33, 319–328. [Google Scholar] [CrossRef]
- Gilliland, L.U.; McKinney, E.C.; Asmussen, M.A.; Meagher, R.B. Detection of deleterious genotypes in multigenerational studies. I. Disruptions in individual Arabidopsis actin genes. Genetics 1998, 149, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Jasik, J.; Micieta, K.; Siao, W.; Voigt, B.; Stuchlik, S.; Schmelzer, E.; Turna, J.; Baluška, F. Actin3 promoter reveals undulating F-actin bundles at shanks and dynamic F-actin meshworks at tips of tip-growing pollen tubes. Plant Signal Behav. 2016, 11, e1146845. [Google Scholar] [CrossRef] [PubMed]
- Ketelaar, T.; de Ruijter, N.C.A.; Emons, A.M.C. Unstable F-actin specifies the area and microtubule direction of cell expansion in Arabidopsis root hairs. Plant Cell 2003, 15, 285–292. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Szymanski, D.B.; Cosgrove, D.J. Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr. Biol. 2009, 19, R800–R811. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, M.; Kojima, H.; Yokota, E.; Nakamori, R.; Anson, M.; Shimmen, T.; Oiwa, K. Calcium-induced mechanical change in the neck domain alters the activity of plant myosin XI. J. Biol. Chem. 2012, 287, 30711–30718. [Google Scholar] [CrossRef]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef]
- Takatsuka, H.; Umeda, M. Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 2014, 65, 2633–2643. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, S.S.; Mao, T.L.; Qu, X.L.; Cao, W.H.; Zhang, L.; Zhang, W.; He, L.; Li, S.D.; Ren, S.L.; et al. The plant-specific actin binding protein SCAB1 stabilizes actin filaments and regulates stomatal movement in Arabidopsis. Plant Cell 2011, 23, 2314–2330. [Google Scholar] [CrossRef]
- Takatsuka, H.; Higaki, T.; Umeda, M. Actin reorganization triggers rapid cell elongation in roots. Plant Physiol. 2018, 178, 1130–1141. [Google Scholar] [CrossRef]
- Mathur, J.; Mathur, N.; Kernebeck, B.; Hulskamp, M. Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 2003, 15, 1632–1645. [Google Scholar] [CrossRef]
- Li, X.B.; Fan, X.P.; Wang, X.L.; Cai, L.; Yang, W.C. The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 2005, 17, 859–875. [Google Scholar] [CrossRef]
- Sun, Y.W.; Liang, W.H.; Shen, W.J.; Feng, H.; Chen, J.D.; Si, Z.F.; Hu, Y.; Zhang, T.Z. G65V substitution in actin disturbs polymerization leading to inhibited cell elongation in Cotton. Front. Plant Sci. 2019, 10, 1486. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; Gilliland, L.U.; McKinney, E.C.; Meagher, R.B. One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell 2001, 13, 1541–1554. [Google Scholar] [CrossRef] [PubMed]
- Numata, T.; Sugita, K.; Rahman, A.A.; Rahman, A. Actin isovariant ACT7 controls root meristem development in Arabidopsis through modulating auxin and ethylene responses. J. Exp. Bot. 2022, 73, 6255–6271. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, L.; Yuan, M.; Ge, Y.; Liu, Y.; Fan, J.; Ruan, Y.; Cui, Z.; Tong, S.; Zhang, S. The microfilament cytoskeleton plays a vital role in salt and osmotic stress tolerance in Arabidopsis. Plant Biol. 2010, 12, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, Z.J.; Guo, G.Q.; Guo, Y. Microfilament dynamics is required for root growth under alkaline stress in Arabidopsis. J. Integr. Plant Biol. 2010, 52, 952–958. [Google Scholar] [CrossRef]
- Parrotta, L.; Faleri, C.; Cresti, M.; Cai, G. Heat stress affects the cytoskeleton and the delivery of sucrose synthase in tobacco pollen tubes. Planta 2016, 243, 43–63. [Google Scholar] [CrossRef]
- Branco, R.; Pearsall, E.J.; Rundle, C.A.; White, R.G.; Bradby, J.E.; Hardham, A.R. Quantifying the plant actin cytoskeleton response to applied pressure using nanoindentation. Protoplasma 2017, 254, 1127–1137. [Google Scholar] [CrossRef]
- Li, J.J.; Staiger, C.J. Understanding cytoskeletal dynamics during the plant immune response. Annu. Rev. Psychol. 2018, 56, 513–533. [Google Scholar] [CrossRef]
- Cooper, J.A.; Schafer, D.A. Control of actin assembly and disassembly at filament ends. Curr. Opin. Cell Biol. 2000, 12, 97–103. [Google Scholar] [CrossRef]
- Hurley, J.H. The sugar kinase/heat shock protein 70/actin superfamily: Implications of conserved structure for mechanism. Annu. Rev. Biophys. Biomol. Struct. 1996, 25, 137–162. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Hendrickson, W.A. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 2007, 131, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Shida, M.; Arakawa, A.; Ishii, R.; Kishishita, S.; Takagi, T.; Kukimoto-Niino, M.; Sugano, S.; Tanaka, A.; Shirouzu, M.; Yokoyama, S. Direct inter-subdomain interactions switch between the closed and open forms of the Hsp70 nucleotide-binding domain in the nucleotide-free state. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 223–232. [Google Scholar] [CrossRef]
- Smith, T.M.; Kirley, T.L. Site-directed mutagenesis of a human brain ecto-apyrase: Evidence that the E-type ATPases are related to the actin/heat shock 70/sugar kinase superfamily. Biochemistry 1999, 38, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Pettigrew, D.W. Amino acid substitutions in the sugar kinase/hsp70/actin superfamily conserved ATPase core of E. coli glycerol kinase modulate allosteric ligand affinity but do not alter allosteric coupling. Arch. Biochem. Biophys. 2009, 481, 151–156. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kandasamy, M.K.; Deal, R.B.; McKinney, E.C.; Meagher, R.B. Silencing the nuclear actin-related protein AtARP4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J. 2005, 41, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, M.K.; McKinney, E.C.; Meagher, R.B. Cell cycle-dependent association of Arabidopsis actin-related proteins AtARP4 and AtARP7 with the nucleus. Plant J. 2003, 33, 939–948. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; McKinney, E.C.; Deal, R.B.; Smith, A.P.; Meagher, R.B. Arabidopsis actin-related protein ARP5 in multicellular development and DNA repair. Dev. Biol. 2009, 335, 22–32. [Google Scholar] [CrossRef]
- Kang, H.J.; Zhang, C.; An, Z.X.; Shen, W.H.; Zhu, Y. AtINO80 and AtARP5 physically interact and play common as well as distinct roles in regulating plant growth and development. New Phytol. 2019, 223, 336–353. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; McKinney, E.C.; Deal, R.B.; Meagher, R.B. Arabidopsis ARP7 is an essential actin-related protein required for normal embryogenesis, plant architecture, and floral organ abscission. Plant Physiol. 2005, 138, 2019–2032. [Google Scholar] [CrossRef]
- Meagher, R.B.; Kandasarny, M.K.; Deal, R.B.; McKinney, E.C. Actin-related proteins in chromatin-level control of the cell cycle and developmental transitions. Trends Cell Biol. 2007, 17, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, M.K.; Deal, R.B.; McKinney, E.C.; Meagher, R.B. Plant actin-related proteins. Trends Plant Sci. 2004, 9, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Vidali, L.; Hepler, P.K. Actin and pollen tube growth. Protoplasma 2001, 215, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Mallery, E.L.; Szymanski, D.B. ARP2/3 localization in Arabidopsis leaf pavement cells: A diversity of intracellular pools and cytoskeletal interactions. Front. Plant Sci. 2013, 4, 238. [Google Scholar] [CrossRef]
- Higgs, H.N.; Blanchoin, L.; Pollard, T.D. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry 1999, 38, 15212–15222. [Google Scholar] [CrossRef]
- Campellone, K.G.; Welch, M.D. A nucleator arms race: Cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 2010, 11, 237–251. [Google Scholar] [CrossRef]
- Zou, J.J.; Zheng, Z.Y.; Xue, S.; Li, H.H.; Wang, Y.R.; Le, J. The role of Arabidopsis Actin-Related Protein 3 in amyloplast sedimentation and polar auxin transport in root gravitropism. J. Exp. Bot. 2016, 67, 5325–5337. [Google Scholar] [CrossRef]
- Cifrova, P.; Oulehlova, D.; Kollarova, E.; Martinek, J.; Rosero, A.; Zarsky, V.; Schwarzerova, K.; Cvrckova, F. Division of labor between two actin nucleators-the formin FH1 and the ARP2/3 complex-in Arabidopsis epidermal cell morphogenesis. Front. Plant Sci. 2020, 11, 148. [Google Scholar] [CrossRef]
- Qin, L.; Liu, L.J.; Tu, J.Y.; Yang, G.G.; Wang, S.; Quilichini, T.D.; Gao, P.; Wang, H.; Peng, G.; Blancaflor, E.B.; et al. The ARP2/3 complex, acting cooperatively with Class I formins, modulates penetration resistance in Arabidopsis against powdery mildew invasion. Plant Cell 2021, 33, 3151–3175. [Google Scholar] [CrossRef]
- Jiang, K.; Sorefan, K.; Deeks, M.J.; Bevan, M.; Hussey, P.J.; Hetherington, A.M. The ARP2/3 complex mediates guard cell actin reorganization and stomatal movement in Arabidopsis. Plant Cell 2012, 24, 2031–2040. [Google Scholar] [CrossRef]
- Zhao, Y.; Pan, Z.; Zhang, Y.; Qu, X.; Zhang, Y.; Yang, Y.; Jiang, X.; Huang, S.; Yuan, M.; Schumaker, K.S.; et al. The actin-related Protein2/3 complex regulates mitochondrial-associated calcium signaling during salt stress in Arabidopsis. Plant Cell 2013, 25, 4544–4559. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, R.; Holmes, K.C. Actin structure and function. Annu. Rev. Biophys. 2011, 40, 169–186. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Deng, L.; Chang, D.; Chen, S.; Wang, X.; Kang, Z. TaADF3, an actin-depolymerizing factor, negatively modulates wheat resistance against Puccinia striiformis. Front. Plant Sci. 2015, 6, 1214. [Google Scholar] [CrossRef] [PubMed]
- Gunning, P.W.; Ghoshdastider, U.; Whitaker, S.; Popp, D.; Robinson, R.C. The evolution of compositionally and functionally distinct actin filaments. J. Cell Sci. 2015, 128, 2009–2019. [Google Scholar] [CrossRef] [PubMed]
- McCurdy, D.W.; Kovar, D.R.; Staiger, C.J. Actin and actin-binding proteins in higher plants. Protoplasma 2001, 215, 89–104. [Google Scholar] [CrossRef]
- Nan, Q.; Qian, D.; Niu, Y.; He, Y.X.; Tong, S.F.; Niu, Z.M.; Ma, J.C.; Yang, Y.; An, L.Z.; Wan, D.S.; et al. Plant actin-depolymerizing factors possess opposing biochemical properties arising from key amino acid changes throughout evolution. Plant Cell 2017, 29, 395–408. [Google Scholar] [CrossRef]
- Rozycka, M.; Khan, S.; Lopez, I.; Greenland, A.J.; Hussey, P.J. A Zea mays pollen cdna-encoding a putative actin-depolymerizing factor. Plant Physiol. 1995, 107, 1011–1012. [Google Scholar] [CrossRef]
- Lopez, I.; Anthony, R.G.; Maciver, S.K.; Jiang, C.J.; Khan, S.; Weeds, A.G.; Hussey, P.J. Pollen specific expression of maize genes encoding actin depolymerizing factor-like proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 7415–7420. [Google Scholar] [CrossRef]
- Allwood, E.G.; Anthony, R.G.; Smertenko, A.P.; Reichelt, S.; Drøbak, B.K.; Doonan, J.H.; Weeds, A.G.; Hussey, P.J. Regulation of the pollen-specific actin-depolymerizing factor LlADF1. Plant Cell 2002, 14, 2915–2927. [Google Scholar] [CrossRef]
- Tholl, S.; Moreau, F.; Hoffmann, C.; Arumugam, K.; Dieterle, M.; Moes, D.; Neumann, K.; Steinmetz, A.; Thomas, C. Arabidopsis actin-depolymerizing factors (ADFs) 1 and 9 display antagonist activities. FEBS Lett. 2011, 585, 1821–1827. [Google Scholar] [CrossRef]
- Chen, C.Y.H.; Cheung, A.Y.; Wu, H.M. Actin-depolymerizing factor mediates Rac/Rop GTPase-regulated pollen tube growth. Plant Cell 2003, 15, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Nakayasu, T.; Yokota, E.; Shimmen, T. Purification of an actin-binding protein composed of 115-kDa polypeptide from pollen tubes of lily. Biochem. Biophys. Res. Commun. 1998, 249, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Klahre, U.; Friederich, E.; Kost, B.; Louvard, D.; Chua, N.H. Villin-like actin-binding proteins are expressed ubiquitously in Arabidopsis. Plant Physiol. 2000, 122, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Qu, X.L.; Bao, C.C.; Khurana, P.; Wang, Q.N.; Xie, Y.R.; Zheng, Y.Y.; Chen, N.Z.; Blanchoin, L.; Staiger, C.J.; et al. Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 2010, 22, 2749–2767. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Robinson, R.C.; Gao, L.Y.; Matsumoto, T.; Brunet, A.; Blanchoin, L.; Staiger, C.J. Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 2005, 17, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.Y.; Wu, H.M. Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 2004, 16, 257–269. [Google Scholar] [CrossRef]
- Ren, H.Y.; Xiang, Y. The function of actin-binding proteins in pollen tube growth. Protoplasma 2007, 230, 171–182. [Google Scholar] [CrossRef]
- Sun, T.T.; Li, S.W.; Ren, H.Y. Profilin as a regulator of the membrane-actin cytoskeleton interface in plant cells. Front. Plant Sci. 2013, 4, 512. [Google Scholar] [CrossRef]
- Sparkes, I. Recent advances in understanding plant myosin function: Life in the fast lane. Mol. Plant 2011, 4, 805–812. [Google Scholar] [CrossRef]
- Su, H.; Zhu, J.S.; Cai, C.; Pei, W.K.; Wang, J.J.; Dong, H.J.; Ren, H.Y. FIMBRIN1 is involved in lily pollen tube growth by stabilizing the actin fringe. Plant Cell 2012, 24, 4539–4554. [Google Scholar] [CrossRef]
- Kovar, D.R.; Staiger, C.J.; Weaver, E.A.; McCurdy, D.W. AtFim1 is an actin filament crosslinking protein from Arabidopsis thaliana. Plant J. 2000, 24, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Qu, X.; Zhang, M.; Jiang, Y.; Dai, A.; Zhao, W.; Cao, D.; Lan, Y.; Yu, R.; Wang, H.; et al. The balance between actin-bundling factors controls actin architecture in pollen tubes. iScience 2019, 16, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, F.; Guerin, C.; von Witsch, M.; Blanchoin, L.; Staiger, C.J. Identification of Arabidopsis cyclase-associated protein 1 as the first nucleotide exchange factor for plant actin. Mol. Biol. Cell 2007, 18, 3002–3014. [Google Scholar] [CrossRef] [PubMed]
- Mathur, J. The ARP2/3 complex: Giving plant cells a leading edge. Bioessays 2005, 27, 377–387. [Google Scholar] [CrossRef]
- Wang, H.J.; Wan, A.R.; Jauh, G.Y. An actin-binding protein, LlLIM1, mediates calcium and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol. 2008, 147, 1619–1636. [Google Scholar] [CrossRef]
- Thomas, C.; Hoffmann, C.; Dieterle, M.; Van Troys, M.; Ampe, C.; Steinmetz, A. Tobacco WLIM1 is a novel F-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell 2006, 18, 2194–2206. [Google Scholar] [CrossRef]
- Huang, S.J.; Blanchoin, L.; Kovar, D.R.; Staiger, C.J. Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J. Biol. Chem. 2003, 278, 44832–44842. [Google Scholar] [CrossRef]
- Harris, A.R.; Belardi, B.; Jreij, P.; Wei, K.; Shams, H.; Bausch, A.; Fletcher, D.A. Steric regulation of tandem calponin homology domain actin-binding affinity. Mol. Biol. Cell 2019, 30, 3112–3122. [Google Scholar] [CrossRef]
- Bretscher, A.; Weber, K. Fimbrin, a new microfilament-associated protein present in microvilli and other cell-surface structures. J. Cell Biol. 1980, 86, 335–340. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, R.H.; Qu, X.L.; Huang, S.J. Arabidopsis FIM5 decorates apical actin filaments and regulates their organization in the pollen tube. J. Exp. Bot. 2016, 67, 3407–3417. [Google Scholar] [CrossRef]
- Su, H.; Feng, H.L.; Chao, X.T.; Ding, X.; Nan, Q.; Wen, C.X.; Liu, H.D.; Xiang, Y.; Liu, W. Fimbrins 4 and 5 act synergistically during polarized pollen tube growth to ensure fertility in Arabidopsis. Plant Cell Physiol. 2017, 58, 2006–2016. [Google Scholar] [CrossRef] [PubMed]
- Kovar, D.R.; Gibbon, B.C.; McCurdy, D.W.; Staiger, C.J. Fluorescently-labeled fimbrin decorates a dynamic actin filament network in live plant cells. Planta 2001, 213, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.H.; Chang, M.; Zhang, M.; Wu, Y.J.; Qu, X.L.; Huang, S.J. The structurally plastic CH2 domain is linked to distinct functions of fimbrins/plastins. J. Biol. Chem. 2016, 291, 17881–17896. [Google Scholar] [CrossRef]
- McCurdy, D.W.; Staiger, C.J. Fimbrin. In Actin: A Dynamic Framework for Multiple Plant Cell Functions; Staiger, C.J., Baluska, F., Volkmann, D., Barlow, P., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2000; pp. 87–102. [Google Scholar]
- Blanchoin, L.; Staiger, C.J. Plant formins: Diverse isoforms and unique molecular mechanism. BBA-Mol. Cell Res. 2010, 1803, 201–206. [Google Scholar] [CrossRef]
- Lan, Y.X.; Liu, X.N.; Fu, Y.; Huang, S.J. Arabidopsis class I formins control membrane-originated actin polymerization at pollen tube tips. PLoS Genet. 2018, 14, e1007789. [Google Scholar] [CrossRef]
- Ingouff, M.; Gerald, J.N.F.; Guerin, C.; Robert, H.; Sorensen, M.B.; Van Damme, D.; Geelen, D.; Blanchoin, L.; Berger, F. Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 2005, 7, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Shen, Y.A.; Cai, C.; Zhong, C.C.; Zhu, L.; Yuan, M.; Ren, H.Y. The type II Arabidopsis formin14 interacts with microtubules and microfilaments to regulate cell division. Plant Cell 2010, 22, 2710–2726. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Y.; Tan, H.X.; Wang, Y.; Li, G.; Liang, W.Q.; Yuan, Z.; Hu, J.P.; Ren, H.Y.; Zhang, D.B. RICE MORPHOLOGY DETERMINANT encodes the type II formin FH5 and regulates rice morphogenesis. Plant Cell 2011, 23, 681–700. [Google Scholar] [CrossRef]
- Favery, B.; Chelysheva, L.A.; Lebris, M.; Jammes, F.; Marmagne, A.; de Almeida-Engler, J.; Lecomte, P.; Vaury, C.; Arkowitz, R.A.; Abad, P. Arabidopsis formin AtFH6 is a plasma membrane-associated protein upregulated in giant cells induced by parasitic nematodes. Plant Cell 2004, 16, 2529–2540. [Google Scholar] [CrossRef]
- Chang, S.W.; Ren, Z.H.; Liu, C.; Du, P.Z.; Li, J.B.; Liu, Z.Y.; Zhang, F.L.; Hou, H.L.; Shi, J.X.; Liang, W.Q.; et al. OsFH3 encodes a type II formin required for rice morphogenesis. Int. J. Mol. Sci. 2021, 22, 13250. [Google Scholar] [CrossRef]
- van Gisbergen, P.A.C.; Bezanilla, M. Plant formins: Membrane anchors for actin polymerization. Trends Cell Biol. 2013, 23, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Oulehlova, D.; Kollarova, E.; Cifrova, P.; Pejchar, P.; Zarsky, V.; Cvrckova, F. Arabidopsis class I formin FH1 relocates between membrane compartments during root cell ontogeny and associates with plasmodesmata. Plant Cell Physiol. 2019, 60, 1855–1870. [Google Scholar] [CrossRef] [PubMed]
- Diao, M.; Ren, S.L.; Wang, Q.N.; Qian, L.C.; Shen, J.F.; Liu, Y.L.; Huang, S.J. Arabidopsis formin 2 regulates cell-to-cell trafficking by capping and stabilizing actin filaments at plasmodesmata. eLife 2018, 7, e36316. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Dong, G.J.; Wu, L.M.; Wang, X.W.; Chen, F.; Xiong, E.H.; Xiong, G.S.; Zhou, Y.H.; Kong, Z.S.; Fu, Y.; et al. Formin protein DRT1 affects gross morphology and chloroplast relocation in rice. Plant Physiol. 2023, 191, 280–298. [Google Scholar] [CrossRef]
- Ma, Z.M.; Liu, X.L.; Nath, S.; Sun, H.; Tran, T.M.; Yang, L.; Mayor, S.; Miao, Y.S. Formin nanoclustering-mediated actin assembly during plant flagellin and DSF signaling. Cell Rep. 2021, 34, 108884. [Google Scholar] [CrossRef]
- Ma, Z.M.; Sun, Y.B.; Zhu, X.L.; Yang, L.; Chen, X.; Miao, Y.S. Membrane nanodomains modulate formin condensation for actin remodeling in Arabidopsis innate immune responses. Plant Cell 2022, 34, 374–394. [Google Scholar] [CrossRef]
- Li, J.J.; Henty-Ridilla, J.L.; Huang, S.J.; Wang, X.; Blanchoin, L.; Staiger, C.J. Capping protein modulates the dynamic behavior of actin filaments in response to phosphatidic acid in Arabidopsis. Plant Cell 2012, 24, 3742–3754. [Google Scholar] [CrossRef]
- Li, J.J.; Henty-Ridilla, J.L.; Staiger, B.H.; Day, B.; Staiger, C.J. Capping protein integrates multiple MAMP signalling pathways to modulate actin dynamics during plant innate immunity. Nat. Commun. 2015, 6, 7206. [Google Scholar] [CrossRef]
- Li, J.J.; Cao, L.Y.; Staiger, C.J. Capping protein modulates actin remodeling in response to reactive oxygen species during plant innate immunity. Plant Physiol. 2017, 173, 1125–1136. [Google Scholar] [CrossRef]
- Wang, J.; Qian, D.; Fan, T.T.; Jia, H.L.; An, L.Z.; Xiang, Y. Arabidopsis actin capping protein (AtCP) subunits have different expression patterns, and downregulation of AtCPB confers increased thermotolerance of Arabidopsis after heat shock stress. Plant Sci. 2012, 193, 110–119. [Google Scholar] [CrossRef]
- Huang, S.J.; Qu, X.L.; Zhang, R.H. Plant villins: Versatile actin regulatory proteins. J. Integr. Plant Biol. 2015, 57, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Tholl, S.; Moes, D.; Dieterle, M.; Papuga, J.; Moreau, F.; Steinmetz, A. Actin bundling in plants. Cell Motil. Cytoskel 2009, 66, 940–957. [Google Scholar] [CrossRef] [PubMed]
- Khurana, P.; Henty, J.L.; Huang, S.J.; Staiger, A.M.; Blanchoin, L.; Staiger, C.J. Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 2010, 22, 2727–2748. [Google Scholar] [CrossRef]
- Bi, S.; Li, M.; Liu, C.; Liu, X.; Cheng, J.; Wang, L.; Wang, J.; Lv, Y.; He, M.; Cheng, X.; et al. Actin depolymerizing factor ADF7 inhibits actin bundling protein VILLIN1 to regulate root hair formation in response to osmotic stress in Arabidopsis. PLoS Genet. 2022, 18, e1010338. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.X.; Guo, M.M.; Zhou, Z.Y.; Wang, B.X.; Pan, Q.; Li, J.J.; Zhou, J.M.; Li, J.J. MPK3-and MPK6-mediated VLN3 phosphorylation regulates actin dynamics during stomatal immunity in Arabidopsis. Nat. Commun. 2021, 12, 6474. [Google Scholar] [CrossRef]
- Zou, M.; Ren, H.; Li, J. An auxin transport inhibitor targets villin-mediated actin dynamics to regulate polar auxin transport. Plant Physiol. 2019, 181, 161–178. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.L.; Zhang, H.; Xie, Y.R.; Wang, J.; Chen, N.Z.; Huang, S.J. Arabidopsis villins promote actin turnover at pollen tube tips and facilitate the construction of actin collars. Plant Cell 2013, 25, 1803–1817. [Google Scholar] [CrossRef]
- van der Honing, H.S.; Kieft, H.; Emons, A.M.C.; Ketelaar, T. Arabidopsis VILLIN2 and VILLIN3 are required for the generation of thick actin filament bundles and for directional organ growth. Plant Physiol. 2012, 158, 1426–1438. [Google Scholar] [CrossRef]
- Papuga, J.; Hoffmann, C.; Dieterle, M.; Moes, D.; Moreau, F.; Tholl, S.; Steinmetz, A.; Thomas, C. Arabidopsis LIM proteins: A family of actin bundlers with distinct expression patterns and modes of regulation. Plant Cell 2010, 22, 3034–3052. [Google Scholar] [CrossRef]
- Peremyslov, V.V.; Prokhnevsky, A.I.; Dolja, V.V. Class XI myosins are required for development, cell expansion, and F-Actin organization in Arabidopsis. Plant Cell 2010, 22, 1883–1897. [Google Scholar] [CrossRef]
- Reichelt, S.; Knight, A.E.; Hodge, T.P.; Baluška, F.; Šamaj, J.; Volkmann, D.; Kendrick-Jones, J. Characterization of the unconventional myosin VIII in plant cells and its localization at the post-cytokinetic cell wall. Plant J. 1999, 19, 555–567. [Google Scholar] [CrossRef]
- Haraguchi, T.; Tominaga, M.; Matsumoto, R.; Sato, K.; Nakano, A.; Yamamoto, K.; Ito, K. Molecular characterization and subcellular localization of Arabidopsis class VIII myosin, ATM1. J. Biol. Chem. 2014, 289, 12343–12355. [Google Scholar] [CrossRef] [PubMed]
- Henn, A.; Sadot, E. The unique enzymatic and mechanistic properties of plant myosins. Curr. Opin. Plant Biol. 2014, 22, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Henty-Ridilla, J.L.; Szymanski, D.B.; Staiger, C.J. Arabidopsis myosin XI: A motor rules the tracks. Plant Physiol. 2014, 166, 1359–1370. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Cai, C.; Staiger, C.J. Myosins XI are involved in exocytosis of cellulose synthase complexes. Plant Physiol. 2019, 179, 1537–1555. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.W.; Huang, L.; Zhang, C.H.; Staiger, C.J. Arabidopsis myosin XIK interacts with the exocyst complex to facilitate vesicle tethering during exocytosis. Plant Cell 2021, 33, 2454–2478. [Google Scholar] [CrossRef] [PubMed]
- Hepler, P.K.; Winship, L.J. The pollen tube clear zone: Clues to the mechanism of polarized growth. J. Integr. Plant Biol. 2015, 57, 79–92. [Google Scholar] [CrossRef]
- Tominaga, M.; Kimura, A.; Yokota, E.; Haraguchi, T.; Shimmen, T.; Yamamoto, K.; Nakano, A.; Ito, K. Cytoplasmic streaming velocity as a plant size determinant. Dev. Cell 2013, 27, 345–352. [Google Scholar] [CrossRef]
- Tian, X.; Wang, X.; Li, Y. Myosin XI-B is involved in the transport of vesicles and organelles in pollen tubes of Arabidopsis thaliana. Plant J. 2021, 108, 1145–1161. [Google Scholar] [CrossRef]
- Abu-Abied, M.; Belausov, E.; Hagay, S.; Peremyslov, V.; Dolja, V.; Sadot, E. Myosin XI-K is involved in root organogenesis, polar auxin transport, and cell division. J. Exp. Bot. 2018, 69, 2869–2881. [Google Scholar] [CrossRef]
- Sohn, R.H.; Goldschmidt-Clermont, P.J. Profilin-at the crossroads of signal-transduction and the actin cytoskeleton. Bioessays 1994, 16, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Pandey, D.K.; Chaudhary, B. Evolutionary expansion and structural functionalism of the ancient family of profilin proteins. Gene 2017, 626, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Baluška, F.; von Witsch, M.; Peters, M.; Hlavacka, A.; Volkmann, D. Mastoparan alters subcellular distribution of profilin and remodels F-actin cytoskeleton in cells of maize root apices. Plant Cell Physiol. 2001, 42, 912–922. [Google Scholar] [CrossRef] [PubMed]
- Pandey, D.K.; Chaudhary, B. Evolution of functional diversity among actin-binding profilin genes in land plants. Front. Cell Dev. Biol. 2020, 8, 588689. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Christensen, H.E.M.; Ishimaru, Y.; Dong, C.H.; Chao-Ming, W.; Cleary, A.L.; Chua, N.H. Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol. 2000, 124, 1637–1647. [Google Scholar] [CrossRef] [PubMed]
- Zweifel, M.E.; Courtemanche, N. Competition for delivery of profilin-actin to barbed ends limits the rate of formin-mediated actin filament elongation. J. Biol. Chem. 2020, 295, 4513–4525. [Google Scholar] [CrossRef]
- Horan, B.G.; Zerze, G.H.; Kim, Y.C.; Vavylonis, D.; Mittal, J. Computational modeling highlights the role of the disordered Formin Homology 1 domain in profilin-actin transfer. FEBS Lett. 2018, 592, 1804–1816. [Google Scholar] [CrossRef]
- Cao, L.Y.; Henty-Ridilla, J.L.; Blanchoin, L.; Staiger, C.J. Profilin-dependent nucleation and assembly of actin filaments controls cell elongation in Arabidopsis. Plant Physiol. 2016, 170, 220–233. [Google Scholar] [CrossRef]
- Sun, H.; Qiao, Z.; Chua, K.P.; Tursic, A.; Liu, X.; Gao, Y.G.; Mu, Y.G.; Hou, X.L.; Miao, Y.S. Profilin negatively regulates formin-mediated actin assembly to modulate PAMP-triggered plant immunity. Curr. Biol. 2018, 28, 1882–1895. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Y.; Ren, H.Y. Profilin promotes formin-mediated actin filament assembly and vesicle transport during polarity formation in pollen. Plant Cell 2021, 33, 1252–1267. [Google Scholar] [CrossRef]
- Gerst, J.E.; Ferguson, K.; Vojtek, A.; Wigler, M.; Field, J. Cap is a bifunctional component of the Saccharomyces-cerevisiae adenylyl cyclase complex. Mol. Cell Biol. 1991, 11, 1248–1257. [Google Scholar] [CrossRef] [PubMed]
- Perelroizen, I.; Didry, D.; Christensen, H.; Chua, N.H.; Carlier, M.F. Role of nucleotide exchange and hydrolysis in the function of profilin in actin assembly. J. Biol. Chem. 1996, 271, 12302–12309. [Google Scholar] [CrossRef] [PubMed]
- Barrero, R.A.; Umeda, M.; Yamamura, S.; Uchimiya, H. Arabidopsis CAP regulates the actin cytoskeleton necessary for plant cell elongation and division. Plant Cell 2002, 14, 149–163. [Google Scholar] [CrossRef][Green Version]
- Jiang, Y.X.; Chang, M.; Lan, Y.X.; Huang, S.J. Mechanism of CAP1-mediated apical actin polymerization in pollen tubes. Proc. Natl. Acad. Sci. USA 2019, 116, 12084–12093. [Google Scholar] [CrossRef] [PubMed]
- Deeks, M.J.; Rodrigues, C.; Dimmock, S.; Ketelaar, T.; Maciver, S.K.; Malho, R.; Hussey, P.J. Arabidopsis CAP1-a key regulator of actin organisation and development. J. Cell Sci. 2007, 120, 2609–2618. [Google Scholar] [CrossRef]
- Suarez, C.; Roland, J.; Boujemaa-Paterski, R.; Kang, H.; McCullough, B.R.; Reymann, A.C.; Guerin, C.; Martiel, J.L.; De La Cruz, E.M.; Blanchoin, L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries (vol 21, pg 862, 2011). Curr. Biol. 2011, 21, 904. [Google Scholar] [CrossRef][Green Version]
- Galkin, V.E.; Orlova, A.; Kudryashov, D.S.; Solodukhin, A.; Reisler, E.; Schroder, G.F.; Egelman, E.H. Remodeling of actin filaments by ADF/cofilin proteins. Proc. Natl. Acad. Sci. USA 2011, 108, 20568–20572. [Google Scholar] [CrossRef]
- Ruzicka, D.R.; Kandasamy, M.K.; McKinney, E.C.; Burgos-Rivera, B.; Meagher, R.B. The ancient subclasses of Arabidopsis ACTIN DEPOLYMERIZING FACTOR genes exhibit novel and differential expression. Plant J. 2007, 52, 460–472. [Google Scholar] [CrossRef]
- Zheng, Y.Y.; Xie, Y.R.; Jiang, Y.X.; Qu, X.L.; Huang, S.J. Arabidopsis ACTIN-DEPOLYMERIZING FACTOR7 severs actin filaments and regulates actin cable turnover to promote normal pollen tube growth. Plant Cell 2013, 25, 3405–3423. [Google Scholar] [CrossRef]
- Daher, F.B.; Geitmann, A. Actin depolymerizing factors ADF7 and ADF10 play distinct roles during pollen development and pollen tube growth. Plant Signal Behav. 2012, 7, 879–881. [Google Scholar] [CrossRef]
- Daher, F.B.; van Oostende, C.; Geitmann, A. Spatial and temporal expression of actin depolymerizing factors ADF7 and ADF10 during male gametophyte development in Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 1177–1192. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.X.; Wang, J.; Xie, Y.R.; Chen, N.Z.; Huang, S.J. ADF10 shapes the overall organization of apical actin filaments by promoting their turnover and ordering in pollen tubes. J. Cell Sci. 2017, 130, 3988–4001. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Wong, E.I.; Vidali, L.; Estavillo, A.; Hepler, P.K.; Wu, H.M.; Cheung, A.Y. The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell 2002, 14, 2175–2190. [Google Scholar] [CrossRef] [PubMed]
- Augustine, R.C.; Vidali, L.; Kleinman, K.P.; Bezanilla, M. Actin depolymerizing factor is essential for viability in plants, and its phosphoregulation is important for tip growth. Plant J. 2008, 54, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.H.; Xia, G.X.; Hong, Y.; Ramachandran, S.; Kost, B.; Chua, N.H. ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 2001, 13, 1333–1346. [Google Scholar] [CrossRef]
- Burgos-Rivera, B.; Ruzicka, D.R.; Deal, R.B.; McKinney, E.C.; King-Reid, L.; Meagher, R.B. ACTIN DEPOLYMERIZING FACTOR9 controls development and gene expression in Arabidopsis. Plant Mol. Biol. 2008, 68, 619–632. [Google Scholar] [CrossRef]
- Henty, J.L.; Bledsoe, S.W.; Khurana, P.; Meagher, R.B.; Day, B.; Blanchoin, L.; Staiger, C.J. Arabidopsis actin depolymerizing factor4 modulates the stochastic dynamic behavior of actin filaments in the cortical array of epidermal cells. Plant Cell 2011, 23, 3711–3726. [Google Scholar] [CrossRef]
- Tian, M.Y.; Chaudhry, F.; Ruzicka, D.R.; Meagher, R.B.; Staiger, C.J.; Day, B. Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB. Plant Physiol. 2009, 150, 815–824. [Google Scholar] [CrossRef]
- Henty-Ridilla, J.L.; Li, J.J.; Day, B.; Staiger, C.J. ACTIN DEPOLYMERIZING FACTOR4 regulates actin dynamics during innate immune signaling in Arabidopsis. Plant Cell 2014, 26, 340–352. [Google Scholar] [CrossRef]
- Lu, Y.J.; Li, P.; Shimono, M.; Corrion, A.; Higaki, T.; He, S.Y.; Day, B. Arabidopsis calcium-dependent protein kinase 3 regulates actin cytoskeleton organization and immunity. Nat. Commun. 2020, 11, 6234. [Google Scholar] [CrossRef]
- Qian, D.; Zhang, Z.; He, J.X.; Zhang, P.; Ou, X.B.; Li, T.; Niu, L.P.; Nan, Q.; Niu, Y.; He, W.L.; et al. Arabidopsis ADF5 promotes stomatal closure by regulating actin cytoskeleton remodeling in response to ABA and drought stress. J. Exp. Bot. 2019, 70, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Qian, D.; Luo, C.; Niu, Y.; Li, T.; Li, C.; Xiang, Y.; Wang, X.; Niu, Y. Arabidopsis ADF5 acts as a downstream target gene of CBFs in response to low-temperature stress. Front. Cell Dev. Biol. 2021, 9, 635533. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.B. Small GTPases: Versatile signaling switches in plants. Plant Cell 2002, 14, S375–S388. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.L.; Yang, Z. The Rop GTPase: An emerging signaling switch in plants. Plant Mol. Biol. 2000, 44, 1–9. [Google Scholar] [CrossRef]
- Fu, Y.; Li, H.; Yang, Z.B. The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 2002, 14, 777–794. [Google Scholar] [CrossRef]
- Fu, Y.; Wu, G.; Yang, Z.B. Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 2001, 152, 1019–1032. [Google Scholar] [CrossRef]
- Allwood, E.G.; Smertenko, A.P.; Hussey, P.J. Phosphorylation of plant actin-depolymerising factor by calmodulin-like domain protein kinase. FEBS Lett. 2001, 499, 97–100. [Google Scholar] [CrossRef]
- Smertenko, A.P.; Jiang, C.J.; Simmons, N.J.; Weeds, A.G.; Davies, D.R.; Hussey, P.J. Ser6 in the maize actin-depolymerizing factor, ZmADF3, is phosphorylated by a calcium-stimulated protein kinase and is essential for the control of functional activity. Plant J. 1998, 14, 187–193. [Google Scholar] [CrossRef]
- Goldy, C.; Caillaud, M.C. Connecting the plant cytoskeleton to the cell surface via the phosphoinositides. Curr. Opin. Plant Biol. 2023, 73, 102365. [Google Scholar] [CrossRef]
- Szymanski, D.B. Breaking the WAVE complex: The point of Arabidopsis trichomes. Curr. Opin. Plant Biol. 2005, 8, 103–112. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Stephan, O.O.H. Effects of environmental stress factors on the actin cytoskeleton of fungi and plants: Ionizing radiation and ROS. Cytoskeleton 2023, 80, 330–355. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Lee, Y.; Jeon, B.W.; Staiger, C.J.; Lee, Y. Phosphatidylinositol 3- and 4-phosphate modulate actin filament reorganization in guard cells of day flower. Plant Cell Environ. 2008, 31, 366–377. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, J.H.; Wang, W.; Chen, N.Z.; Ma, T.S.; Xi, Y.N.; Zhang, X.L.; Lin, H.F.; Bai, Y.; Huang, S.J.; et al. ARP2/3 complex-mediated actin dynamics is required for hydrogen peroxide-induced stomatal closure in Arabidopsis. Plant Cell Environ. 2014, 37, 1548–1560. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, H.; Zhang, Q.; Li, M.; Yan, M.; Wang, R.; Wang, L.; Welti, R.; Zhang, W.; Wang, X. Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 2009, 21, 2357–2377. [Google Scholar] [CrossRef]
- Cao, L.Y.; Wang, W.Y.; Zhang, W.W.; Staiger, C.J. Lipid signaling requires ROS production to elicit actin cytoskeleton remodeling during plant innate immunity. Int. J. Mol. Sci. 2022, 23, 2447. [Google Scholar] [CrossRef]
- Kushwah, S.; Jones, A.M.; Laxmi, A. Cytokinin-induced root growth involves actin filament reorganization. Plant Signal Behav. 2011, 6, 1848–1850. [Google Scholar] [CrossRef]
- Pizarro, L.; Munoz, D.; Marash, I.; Gupta, R.; Anand, G.; Leibman-Markus, M.; Bar, M. Cytokinin modulates cellular trafficking and the cytoskeleton, enhancing defense responses. Cells 2021, 10, 1634. [Google Scholar] [CrossRef]
- Iwabuchi, K.; Takagi, S. Actin-based mechanisms for light-dependent intracellular positioning of nuclei and chloroplasts in Arabidopsis. Plant Signal Behav. 2010, 5, 1010–1013. [Google Scholar] [CrossRef]
- Iwabuchi, K.; Minamino, R.; Takagi, S. Actin reorganization underlies phototropin-dependent positioning of nuclei in Arabidopsis leaf cells. Plant Physiol. 2010, 152, 1309–1319. [Google Scholar] [CrossRef]
- Kadota, A.; Yamada, N.; Suetsugu, N.; Hirose, M.; Saito, C.; Shoda, K.; Ichikawa, S.; Kagawa, T.; Nakano, A.; Wada, M. Short actin-based mechanism for light-directed chloroplast movement in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 13106–13111. [Google Scholar] [CrossRef]
- Whippo, C.W.; Khurana, P.; Davis, P.A.; DeBlasio, S.L.; DeSloover, D.; Staiger, C.J.; Hangarter, R.P. THRUMIN1 is a light-regulated actin-bundling protein involved in chloroplast motility. Curr. Biol. 2011, 21, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.Q.; Xing, J.J.; Wan, Y.L.; Lv, X.Q.; Fan, L.S.; Zhang, Y.D.; Song, K.; Wang, L.; Wang, X.H.; Deng, X.; et al. Arabidopsis blue light receptor phototropin 1 undergoes blue light-induced activation in membrane microdomains. Mol. Plant 2018, 11, 846–859. [Google Scholar] [CrossRef] [PubMed]
- Krzeszowiec, W.; Rajwa, B.; Dobrucki, J.; Gabrys, H. Actin cytoskeleton in Arabidopsis thaliana under blue and red light. Biol. Cell 2007, 99, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.Y.; Liu, Y.C.; Bibeau, J.P.; Lemoi, K.P.; Tuzel, E.; Vidali, L. The kinesin-like proteins, KAC1/2, regulate actin dynamics underlying chloroplast light-avoidance in Physcomitrella patens. J. Integr. Plant Biol. 2015, 57, 106–119. [Google Scholar] [CrossRef]
- Šamaj, J.; Ovecka, M.; Hlavacka, A.; Lecourieux, F.; Meskiene, I.; Lichtscheidl, I.; Lenart, P.; Salaj, J.; Volkmann, D.; Bogre, L.; et al. Involvement of the mitogen-activated protein kinase SIMK in regulation of root hair tip growth. Embo J. 2002, 21, 3296–3306. [Google Scholar] [CrossRef]
- Sangwan, V.; Orvar, B.L.; Beyerly, J.; Hirt, H.; Dhindsa, R.S. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 2002, 31, 629–638. [Google Scholar] [CrossRef]
- Šamaj, J.; Baluška, F.; Hirt, H. From signal to cell polarity: Mitogen-activated protein kinases as sensors and effectors of cytoskeleton dynamicity. J. Exp. Bot. 2004, 55, 189–198. [Google Scholar] [CrossRef]
- Staiger, C.J.; Franklin-Tong, V.E. The actin cytoskeleton is a target of the self-incompatibility response in Papaver rhoeas. J. Exp. Bot. 2003, 54, 103–113. [Google Scholar] [CrossRef]
- Zhao, W.; Qu, X.; Zhuang, Y.; Wang, L.; Bosch, M.; Franklin-Tong, V.E.; Xue, Y.; Huang, S. Villin controls the formation and enlargement of punctate actin foci in pollen tubes. J. Cell Sci. 2020, 133, jcs237404. [Google Scholar] [CrossRef]
- Wang, Y.F.; Fan, L.M.; Zhang, W.Z.; Zhang, W.; Wu, W.H. Ca2+-permeable channels in the plasma membrane of Arabidopsis pollen are regulated by actin microfilaments. Plant Physiol. 2004, 136, 3892–3904. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.S.; Han, X.M.; Zheng, L.Z.; Xie, Y.; Mu, Y.G.; Yates, J.R.; Drubin, D.G. Fimbrin phosphorylation by metaphase Cdk1 regulates actin cable dynamics in budding yeast. Nat. Commun. 2016, 7, 11265. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Holehouse, A.S.; Pappu, R.V. Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 2020, 49, 107–133. [Google Scholar] [CrossRef]
- Xie, Y.; Sun, J.L.; Han, X.; Tursic-Wunder, A.; Toh, J.D.W.; Hong, W.J.; Gao, Y.G.; Miao, Y.S. Polarisome scaffolder Spa2-mediated macromolecular condensation of Aip5 for actin polymerization. Nat. Commun. 2019, 10, 5078. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.M.; Miao, Y.S. Review: F-Actin remodelling during plant signal transduction via biomolecular assembly. Plant Sci. 2020, 301, 110663. [Google Scholar] [CrossRef]
- Pydiura, N.; Pirko, Y.; Galinousky, D.; Postovoitova, A.; Yemets, A.; Kilchevsky, A.; Blume, Y. Genome-wide identification, phylogenetic classification, and exon-intron structure characterization of the tubulin and actin genes in flax (Linum usitatissimum). Cell Biol. Int. 2019, 43, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Goldspink, D.A.; Matthews, Z.J.; Lund, E.K.; Wileman, T.; Mogensen, M.M. Immuno-fluorescent labeling of microtubules and centrosomal proteins in ex vivo intestinal tissue and 3D in vitro intestinal organoids. J. Vis. Exp. 2017, 13, 56662. [Google Scholar] [CrossRef]
- Lombardo, A.T.; Nelson, S.R.; Kennedy, G.G.; Trybus, K.M.; Walcott, S.; Warshaw, D.M. Myosin Va transport of liposomes in three-dimensional actin networks is modulated by actin filament density, position, and polarity. Proc. Natl. Acad. Sci. USA 2019, 116, 8326–8335. [Google Scholar] [CrossRef]
- Schneider, J.; Jasnin, M. Capturing actin assemblies in cells using in situ cryo-electron tomography. Eur. J. Cell Biol. 2022, 101, 151224. [Google Scholar] [CrossRef]
- Ren, Z.H.; Zhang, Y.; Zhang, Y.; He, Y.Q.; Du, P.Z.; Wang, Z.X.; Sun, F.; Ren, H.Y. Cryo-EM structure of actin filaments from Zea mays pollen. Plant Cell 2019, 31, 2855–2867. [Google Scholar] [CrossRef]
- Hussey, P.J.; Ketelaar, T.; Deeks, M.J. Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 2006, 57, 109–125. [Google Scholar] [CrossRef] [PubMed]
- Sampathkumar, A.; Gutierrez, R.; McFarlane, H.E.; Bringmann, M.; Lindeboom, J.; Emons, A.M.; Samuels, L.; Ketelaar, T.; Ehrhardt, D.W.; Persson, S. Patterning and lifetime of plasma membrane-localized cellulose synthase is dependent on actin organization in Arabidopsis interphase cells. Plant Physiol. 2013, 162, 675–688. [Google Scholar] [CrossRef]
- Appenzeller-Herzog, C.; Hauri, H.P. The ER-Golgi intermediate compartment (ERGIC): In search of its identity and function. J. Cell Sci. 2006, 119, 2173–2183. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.W.; Hussey, P.J. Interactions between plant endomembrane systems and the actin cytoskeleton. Front. Plant Sci. 2015, 6, 422. [Google Scholar] [CrossRef] [PubMed]
- Monastyrska, I.; He, C.; Geng, J.F.; Hoppe, A.D.; Li, Z.J.; Klionsky, D.J. Arp2 links autophagic machinery with the actin cytoskeleton. Mol. Biol. Cell 2008, 19, 1962–1975. [Google Scholar] [CrossRef]
- Monastyrska, I.; Reggiori, F.; Klionsky, D.J. Harpooning the Cvt complex to the phagophore assembly site. Autophagy 2008, 4, 914–916. [Google Scholar] [CrossRef]
- Kast, D.J.; Zajac, A.L.; Holzbaur, E.L.F.; Ostap, E.M.; Dominguez, R. WHAMM directs the Arp2/3 complex to the ER for autophagosome biogenesis through an actin comet tail mechanism. Curr. Biol. 2015, 25, 1791–1797. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Condeelis, J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. BBA-Mol. Cell Res. 2007, 1773, 642–652. [Google Scholar] [CrossRef]
- Zhang, C.H.; Mallery, E.; Reagan, S.; Boyko, V.P.; Kotchoni, S.O.; Szymanski, D.B. The endoplasmic reticulum is a reservoir for WAVE/SCAR regulatory complex signaling in the Arabidopsis leaf. Plant Physiol. 2013, 162, 689–706. [Google Scholar] [CrossRef][Green Version]
- Wang, P.W.; Richardson, C.; Hawes, C.; Hussey, P.J. Arabidopsis NAP1 regulates the formation of autophagosomes. Curr. Biol. 2016, 26, 2060–2069. [Google Scholar] [CrossRef]
- Hohfeld, J. Autophagy: Press and push for destruction. Curr. Biol. 2016, 26, R703–R705. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.W.; Gao, E.L.; Hussey, P.J. Autophagosome biogenesis in plants: An actin cytoskeleton perspective. Trends Plant Sci. 2020, 25, 850–858. [Google Scholar] [CrossRef]
- Franklin-Tong, V.E.; Gourlay, C.W. A role for actin in regulating apoptosis/programmed cell death: Evidence spanning yeast, plants and animals. Biochem. J. 2008, 413, 389–404. [Google Scholar] [CrossRef] [PubMed]
- Komis, G.; Novak, D.; Ovecka, M.; Samajova, O.; Šamaj, J. Advances in imaging plant cell dynamics. Plant Physiol. 2018, 176, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Conduit, P.T.; Wainman, A.; Novak, Z.A.; Weil, T.T.; Raff, J.W. Re-examining the role of Drosophila Sas-4 in centrosome assembly using two-colour-3D-SIM FRAP. eLife 2015, 4, e08483. [Google Scholar] [CrossRef]
- Lanzano, L.; Scipioni, L.; Di Bona, M.; Bianchini, P.; Bizzarri, R.; Cardarelli, F.; Diaspro, A.; Vicidomini, G. Measurement of nanoscale three-dimensional diffusion in the interior of living cells by STED-FCS. Nat. Commun. 2017, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Diao, M.; Li, X.; Huang, S.J. Arabidopsis AIP1-1 regulates the organization of apical actin filaments by promoting their turnover in pollen tubes. Sci. China Life Sci. 2020, 63, 239–250. [Google Scholar] [CrossRef]
- Higaki, T.; Kutsuna, N.; Sano, T.; Kondo, N.; Hasezawa, S. Quantification and cluster analysis of actin cytoskeletal structures in plant cells: Role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J. 2010, 61, 156–165. [Google Scholar] [CrossRef]
- Zhu, J.; Nan, Q.; Qin, T.; Qian, D.; Mao, T.; Yuan, S.; Wu, X.; Niu, Y.; Bai, Q.; An, L.; et al. Higher-ordered actin structures remodeled by Arabidopsis ACTIN-DEPOLYMERIZING FACTOR5 are important for pollen germination and pollen tube growth. Mol. Plant 2017, 10, 1065–1081. [Google Scholar] [CrossRef]
- Baluška, F.; Mancuso, S. Actin cytoskeleton and action potentials: Forgotten connections. In The Cytoskeleton Diverse Roles in a Plant’s Life; Sahi, V.P., Baluška, F., Eds.; Plant Cell Monographs; Springer: Cham, Switzerland, 2019; Volume 24, pp. 63–83. [Google Scholar]
- Cvrčková, F. Formins and membranes: Anchoring cortical actin to the cell wall and beyond. Front. Plant Sci. 2013, 4, 436. [Google Scholar] [CrossRef]
- Cheung, A.Y.; Niroomand, S.; Zou, Y.J.; Wu, H.M. A transmembrane formin nucleates subapical actin assembly and controls tip-focused growth in pollen tubes. Proc. Natl. Acad. Sci. USA 2010, 107, 16390–16395. [Google Scholar] [CrossRef]
- Baluška, F.; Hlavačka, A. Plant formins come of age: Something special about cross-walls. New Phytol. 2005, 168, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Deeks, M.J.; Cvrcková, F.; Machesky, L.M.; Mikitová, V.; Ketelaar, T.; Zársky, V.; Davies, B.; Hussey, P.J. Arabidopsis group Ie formins localize to specific cell membrane domains, interact with actin-binding proteins and cause defects in cell expansion upon aberrant expression. New Phytol. 2005, 168, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Baluška, F.; Cvrcková, F.; Kendrick-Jones, J.; Volkmann, D. Sink plasmodesmata as gateways for phloem unloading. Myosin VIII and calreticulin as molecular determinants of sink strength? Plant Physiol. 2001, 126, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Amari, K.; Di Donato, M.; Dolja, V.V.; Heinlein, M. Myosins VIII and XI play distinct roles in reproduction and transport of Tobacco mosaic virus. PLoS Path. 2014, 10, e1004448. [Google Scholar] [CrossRef]
- Volkmann, D.; Mori, T.; Tirlapur, U.K.; König, K.; Fujiwara, T.; Kendrick-Jones, J.; Baluška, F. Unconventional myosins of the plant-specific class VIII: Endocytosis, cytokinesis, plasmodesmata/pit-fields, and cell-to-cell coupling. Cell Biol. Int. 2003, 27, 289–291. [Google Scholar] [CrossRef]
- Baluška, F.; Mancuso, S. Root apex transition zone as oscillatory zone. Front. Plant Sci. 2013, 4, 354. [Google Scholar] [CrossRef]
- Baluška, F.; Mancuso, S.; Volkmann, D.; Barlow, P.W. Root apex transition zone: A signalling-response nexus in the root. Trends Plant Sci. 2010, 15, 402–408. [Google Scholar] [CrossRef]
- Goode, B.L.; Drubin, D.G.; Barnes, G. Functional cooperation between the microtubule and actin cytoskeletons. Curr. Opin. Cell Biol. 2000, 12, 63–71. [Google Scholar] [CrossRef]
- Collings, D.A.; Wasteneys, G.O. Actin microfilament and microtubule distribution patterns in the expanding root of Arabidopsis thaliana. Can. J. Bot. 2005, 83, 579–590. [Google Scholar] [CrossRef]
- Sampathkumar, A.; Lindeboom, J.J.; Debolt, S.; Gutierrez, R.; Ehrhardt, D.W.; Ketelaar, T.; Persson, S. Live cell imaging reveals structural associations between the actin and microtubule cytoskeleton in Arabidopsis. Plant Cell 2011, 23, 2302–2313. [Google Scholar] [CrossRef] [PubMed]
- Zang, J.Z.; Klemm, S.; Pain, C.; Duckney, P.; Bao, Z.R.; Stamm, G.; Kriechbaumer, V.; Burstenbinder, K.; Hussey, P.J.; Wang, P.W. A novel plant actin-microtubule bridging complex regulates cytoskeletal and ER structure at ER-PM contact sites. Curr. Biol. 2021, 31, 1251–1260. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.C.; Zhang, H.; Xia, Q.; Yu, J.J.; Zhu, D.Y.; Zhao, Q. ZmGLR, a cell membrane localized microtubule-associated protein, mediated leaf morphogenesis in maize. Plant Sci. 2019, 289, 110248. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Liu, X.M.; Li, J.J.; Sun, J.B.; Song, L.N.; Mao, T.L. Arabidopsis microtubule-destabilizing protein 25 functions in pollen tube growth by severing actin filaments. Plant Cell 2014, 26, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Wang, X.L.; Qin, T.; Zhang, Y.; Liu, X.M.; Sun, J.B.; Zhou, Y.; Zhu, L.; Zhang, Z.D.; Yuan, M.; et al. MDP25, a novel calcium regulatory protein, mediates hypocotyl cell elongation by destabilizing cortical microtubules in Arabidopsis. Plant Cell 2011, 23, 4411–4427. [Google Scholar] [CrossRef]
- Yang, P.; Jin, J.; Zhang, J.; Wang, D.; Bai, X.; Xie, W.; Hu, T.; Zhao, X.; Mao, T.; Qin, T. MDP25 mediates the fine-tuning of microtubule organization in response to salt stress. J. Integr. Plant Biol. 2022, 64, 1181–1195. [Google Scholar] [CrossRef]
- Smertenko, A.; Hewitt, S.L.; Jacques, C.N.; Kacprzyk, R.; Liu, Y.; Marcec, M.J.; Moyo, L.; Ogden, A.; Oung, H.M.; Schmidt, S.; et al. Phragmoplast microtubule dynamics-a game of zones. J. Cell Sci. 2018, 131, jcs203331. [Google Scholar] [CrossRef]
- Muller, S.; Jürgens, G. Plant cytokinesis-No ring, no constriction but centrifugal construction of the partitioning membrane. Semin. Cell Dev. Biol. 2016, 53, 10–18. [Google Scholar] [CrossRef]
- Maeda, K.; Higaki, T. Disruption of actin filaments delays accumulation of cell plate membranes after chromosome separation. Plant Signal. Behav. 2021, 16, 1873586. [Google Scholar] [CrossRef]
- Wu, S.Z.; Bezanilla, M. Actin and microtubule cross talk mediates persistent polarized growth. J. Cell Biol. 2018, 217, 3531–3544. [Google Scholar] [CrossRef]
- Welf, E.S.; Driscoll, M.K.; Dean, K.M.; Schafer, C.; Chu, J.; Davidson, M.W.; Lin, M.Z.; Danuser, G.; Fiolka, R. Quantitative multiscale cell imaging in controlled 3D microenvironments. Dev. Cell 2016, 36, 462–475. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Ehrhardt, D.W.; Hashimoto, T. Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 2010, 12, 1064–1070. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Ren, H.Y. Development and application of probes for labeling the actin cytoskeleton in living plant cells. Protoplasma 2011, 248, 239–250. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).