Integrated Transcriptomic and Metabolomic Analysis Reveal the Underlying Mechanism of Anthocyanin Biosynthesis in Toona sinensis Leaves
Abstract
1. Introduction
2. Results
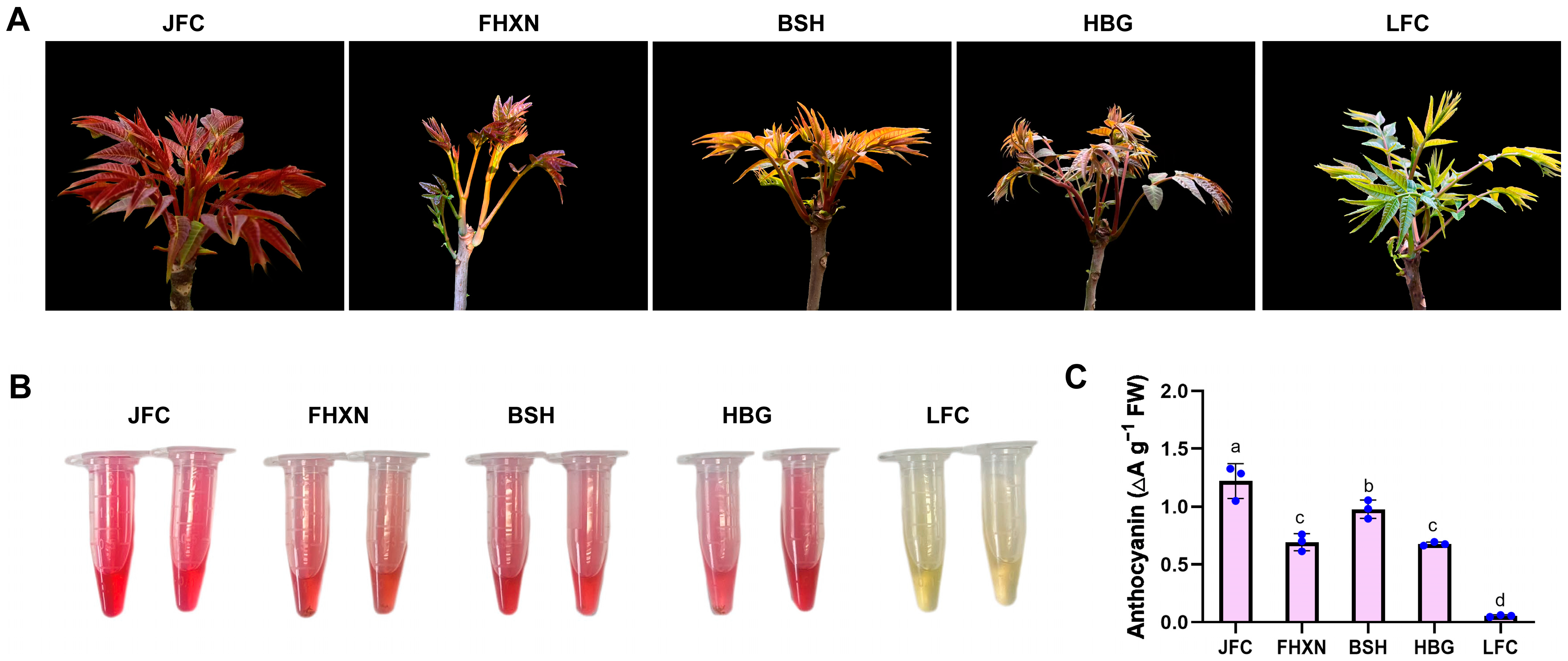
2.1. Morphological Observation and Anthocyanin Content of Leaves in Toona sinensis
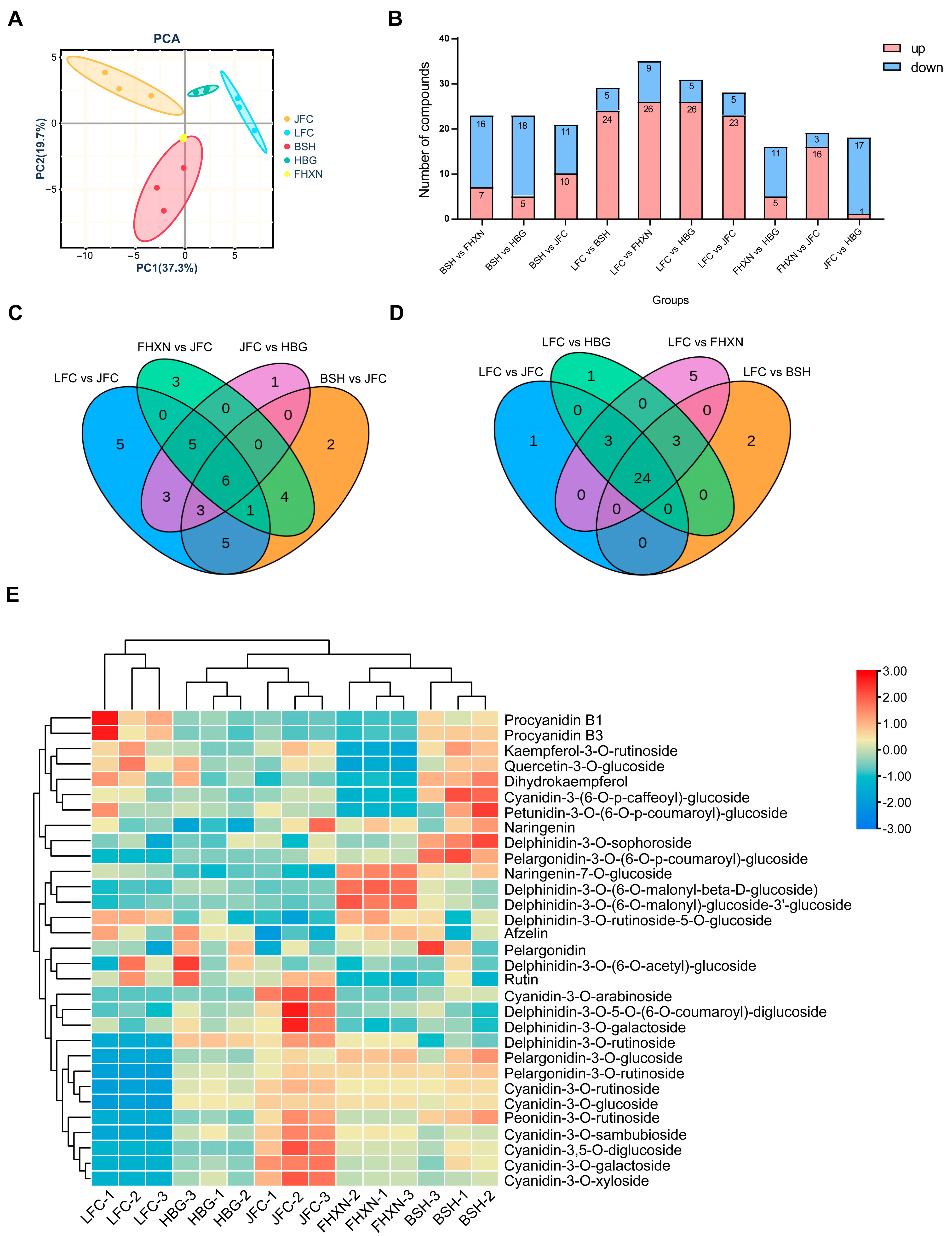
2.2. Metabolome Analysis of Anthocyanin in Five Toona Clones
2.3. Transcriptome Analysis of Five Toona sinensis Clones
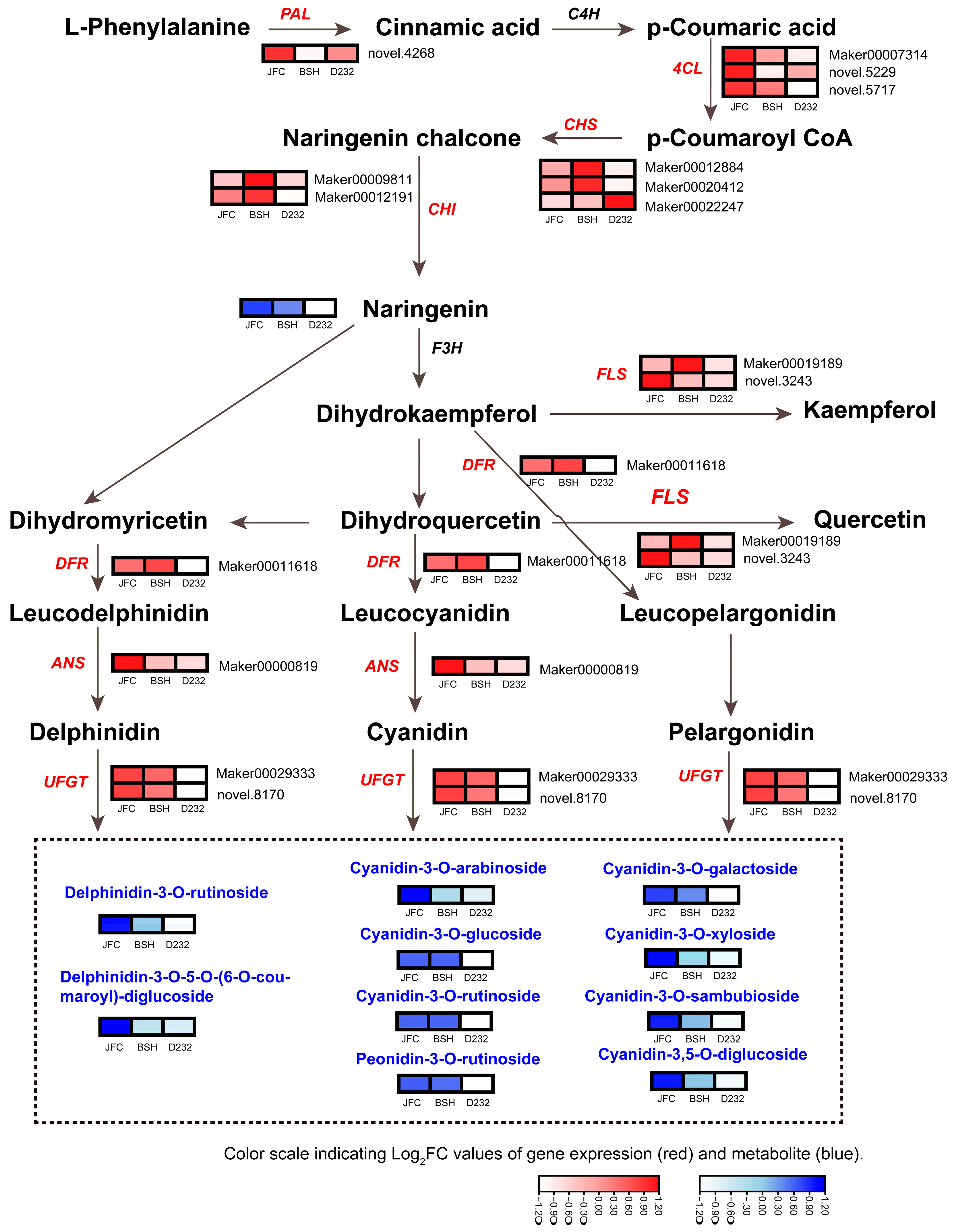
2.4. Analysis of Anthocyanidin Biosynthetic Pathway Genes and Anthocyanin Derivates in JFC, BSH, and LFC
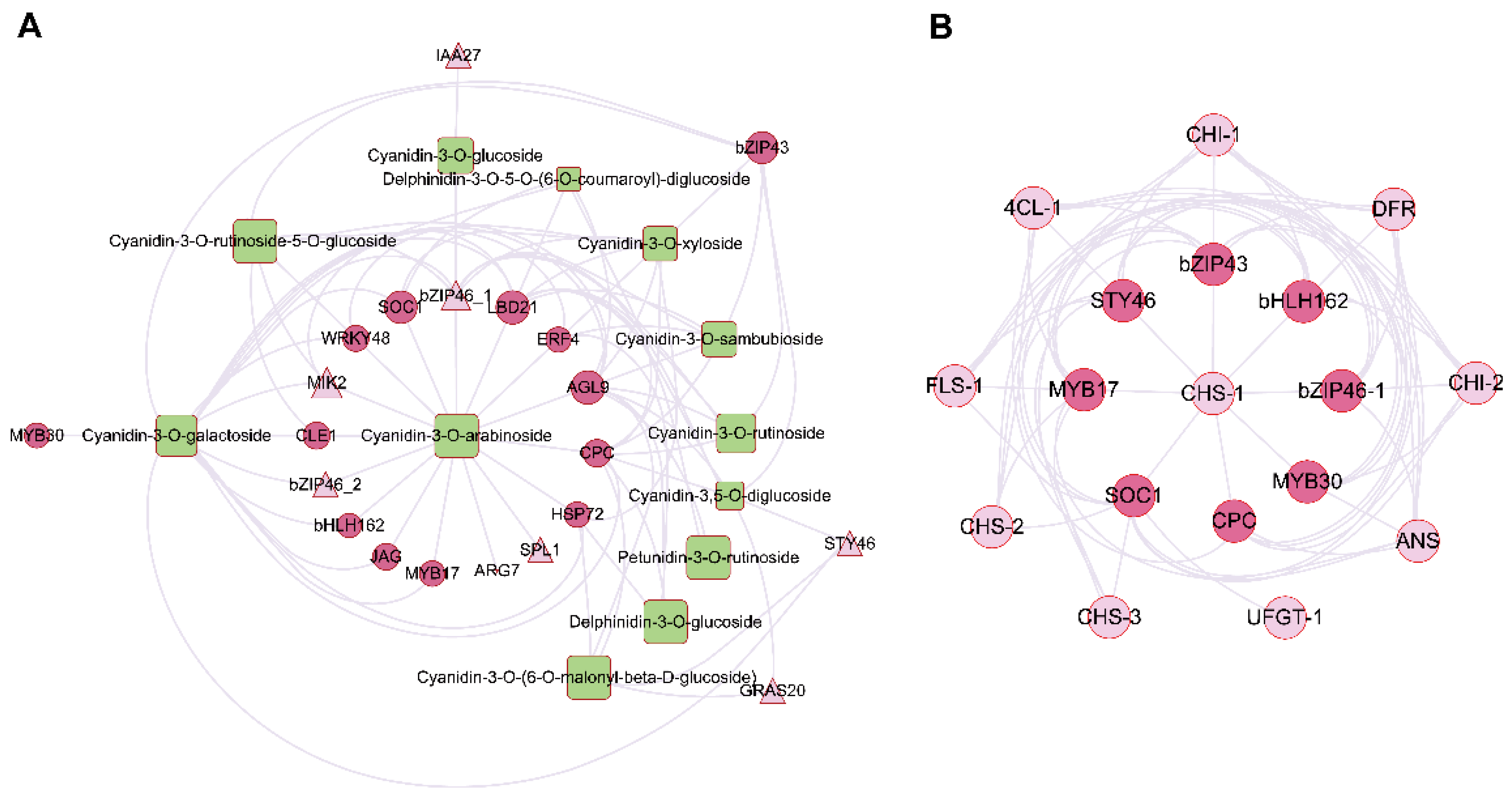
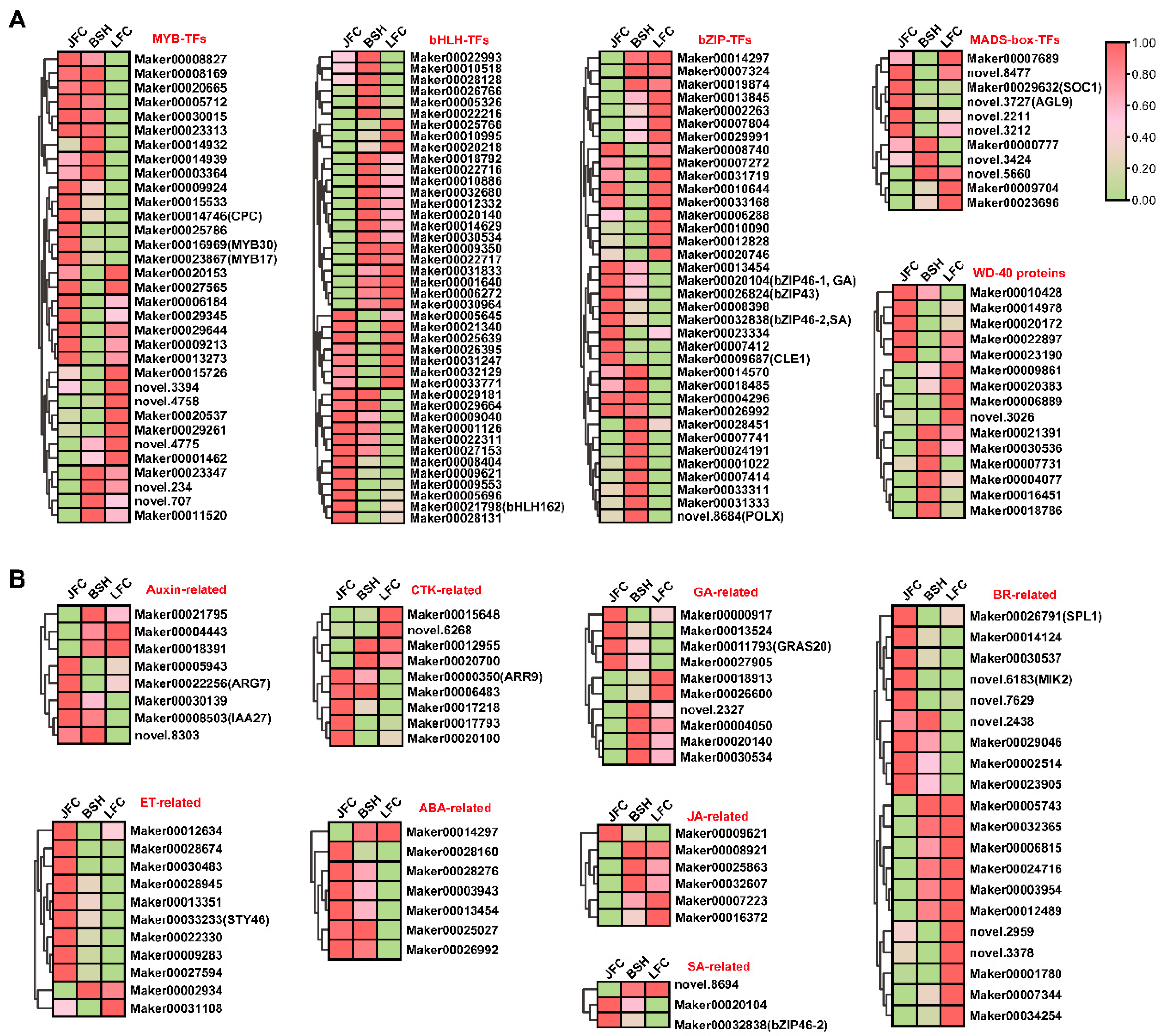
2.5. Correlation Analysis of Differentially Expressed Transcripts and Anthocyanin Compounds to Identify Key TFs and Plant Hormones Genes
2.6. Expression Profiles of TFs and Plant Hormone Genes Related to Anthocyanin Biosynthesis
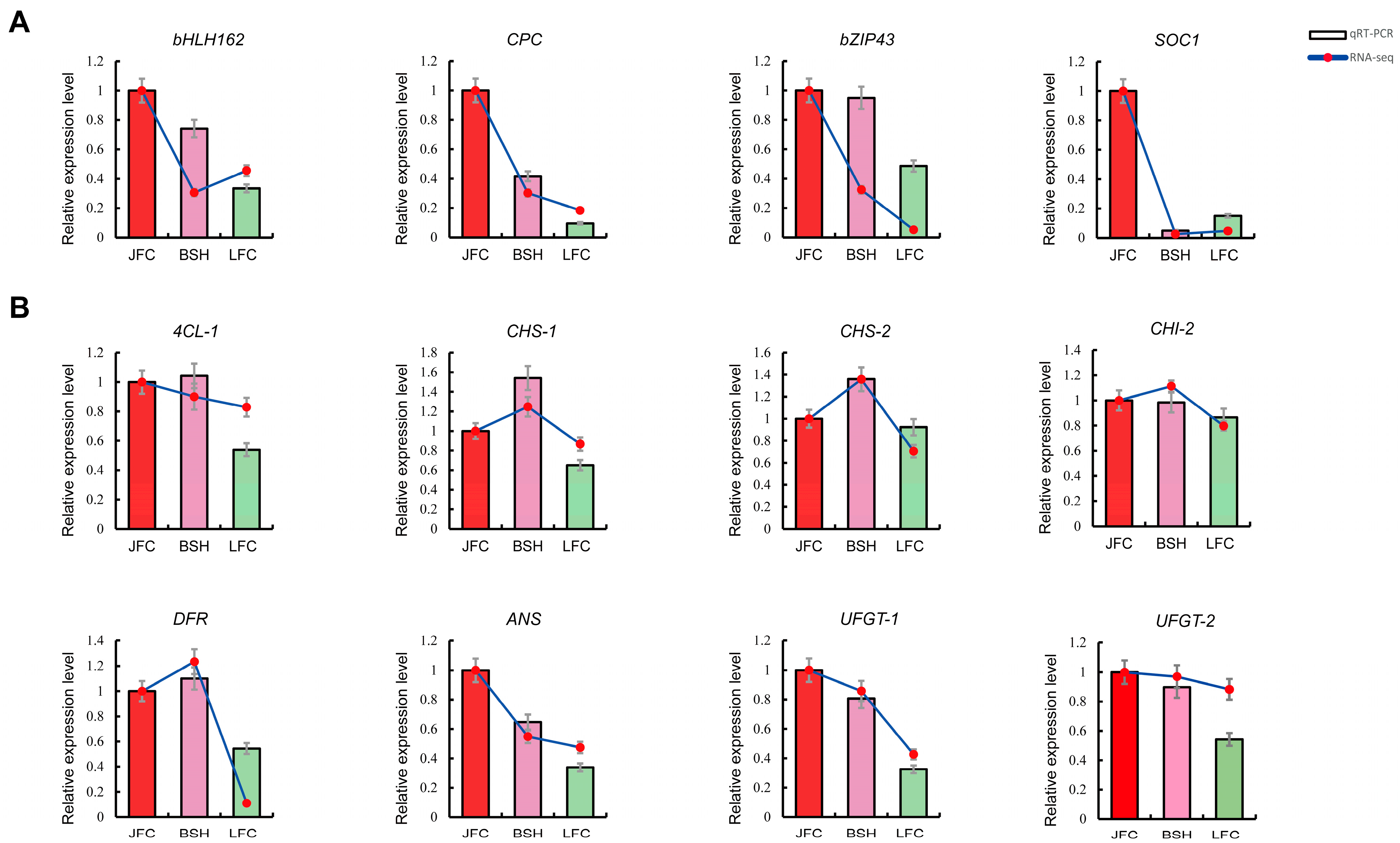
2.7. Expression Analysis of Genes through qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Sample Preparation and Extraction
4.3. UPLC Conditions
4.4. ESI-MS/MS Conditions
4.5. Identification of Differentially Accumulated Metabolites
4.6. RNA Sequencing
4.7. Transcript Profiles and Annotation
4.8. Correlation Analysis of Transcriptome and Metabolome
4.9. Verification of RNA-Seq Data by qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Parada, J.; Aguilera, J.M. Food microstructure affects the bioavailability of several nutrients. J. Food Sci. 2007, 72, R21–R32. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Xu, M.; Deng, P.; Gu, Q.; Yin, H.; Xia, G.; Jia, X.; Yang, H.; Tam, J. Establishment of a rapid method to quantify eight flavonol glycosides for quality assessment of red toon using UPLC. Acta Chromatogr. 2018, 30, 31–37. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, L.; Cao, Y.; Gu, Q.; Yang, H.; Tam, J.P. Quantitative Analysis and Comparison of Four Major Flavonol Glycosides in the Leaves of Toona sinensis (A. Juss.) Roemer (Chinese Toon) from Various Origins by High-Performance Liquid Chromatography-Diode Array Detector and Hierarchical Clustering Analysis. Pharmacogn. Mag. 2016, 12 (Suppl. S2), S270–S276. [Google Scholar] [PubMed]
- Wang, K.J.; Yang, C.R.; Zhang, Y.J. Phenolic antioxidants from Chinese toon (fresh young leaves and shoots of Toona sinensis). Food Chem. 2007, 101, 365–371. [Google Scholar] [CrossRef]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- He, J.; Giusti, M.M. Anthocyanins: Natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef]
- Yousuf, B.; Gul, K.; Wani, A.A.; Singh, P. Health Benefits of Anthocyanins and Their Encapsulation for Potential Use in Food Systems: A Review. Crit. Rev. Food Sci. Nutr. 2016, 56, 2223–2230. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef]
- Jaakola, L.; Hohtola, A. Effect of latitude on flavonoid biosynthesis in plants. Plant Cell Environ. 2010, 33, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Li, M.; Ma, F.; Cheng, L. Phenylpropanoid metabolites and expression of key genes involved in anthocyanin biosynthesis in the shaded peel of apple fruit in response to sun exposure. Plant Physiol. Bioch. 2013, 69, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, Z.; Wu, Y.; Zheng, L.; Zhang, G. Regulatory Mechanisms of Anthocyanin Biosynthesis in Apple and Pear. Int. J. Mol. Sci. 2021, 22, 8441. [Google Scholar] [CrossRef]
- Mattoo, A.K.; Dwivedi, S.L.; Dutt, S.; Singh, B.; Garg, M.; Ortiz, R. Anthocyanin-Rich Vegetables for Human Consumption-Focus on Potato, Sweetpotato and Tomato. Int. J. Mol. Sci. 2022, 23, 2634. [Google Scholar] [CrossRef]
- Sun, C.; Deng, L.; Du, M.; Zhao, J.; Chen, Q.; Huang, T.; Jiang, H.; Li, C.B.; Li, C. A Transcriptional Network Promotes Anthocyanin Biosynthesis in Tomato Flesh. Mol. Plant. 2020, 13, 42–58. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Song, Z.; Zhang, H. Repression of MYBL2 by Both microRNA858a and HY5 Leads to the Activation of Anthocyanin Biosynthetic Pathway in Arabidopsis. Mol. Plant. 2016, 9, 1395–1405. [Google Scholar] [CrossRef]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef]
- Li, L.; Ban, Z.J.; Li, X.H.; Wu, M.Y.; Wang, A.L.; Jiang, Y.Q.; Jiang, Y.H. Differential Expression of Anthocyanin Biosynthetic Genes and Transcription Factor PcMYB10 in Pears (Pyrus communis L.). PLoS ONE 2012, 7, e46070. [Google Scholar] [CrossRef]
- An, X.H.; Tian, Y.; Chen, K.Q.; Wang, X.F.; Hao, Y.J. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 2012, 169, 710–717. [Google Scholar] [CrossRef]
- Chen, L.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Bioch. 2019, 136, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Das, P.K.; Shin, D.H.; Choi, S.-B.; Park, Y.-I. Sugar-hormone cross-talk in anthocyanin biosynthesis. Mol. Cells 2012, 34, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Lei, Z.; Zhou, C.; Ji, X.; Wei, G.; Huang, Y.; Yu, W.; Luo, Y.; Qiu, Y. Transcriptome Analysis Reveals genes involved in flavonoid biosynthesis and accumulation in Dendrobium catenatum from different locations. Sci. Rep. 2018, 8, 6373. [Google Scholar] [CrossRef]
- Li, H.; Yang, Z.; Zeng, Q.; Wang, S.; Luo, Y.; Huang, Y.; Xin, Y.; He, N. Abnormal expression of bHLH3 disrupts a flavonoid homeostasis network, causing differences in pigment composition among mulberry fruits. Hortic. Res. 2020, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef]
- Lau, O.S.; Deng, X.W. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012, 17, 584–593. [Google Scholar] [CrossRef]
- Stracke, R.; Favory, J.J.; Gruber, H.; Bartelniewoehner, L.; Bartels, S.; Binkert, M.; Funk, M.; Weisshaar, B.; Ulm, R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 2010, 33, 88–103. [Google Scholar]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef]
- Zhao, Y.; Min, T.; Chen, M.; Wang, H.; Zhu, C.; Jin, R.; Allan, A.C.; Lin-Wang, K.; Xu, C. The Photomorphogenic Transcription Factor PpHY5 Regulates Anthocyanin Accumulation in Response to UVA and UVB Irradiation. Front. Plant Sci. 2020, 11, 603178. [Google Scholar] [CrossRef]
- Gao, H.-N.; Jiang, H.; Cui, J.-Y.; You, C.-X.; Li, Y.-Y. Review: The effects of hormones and environmental factors on anthocyanin biosynthesis in apple. Plant Sci. 2021, 312, 111024. [Google Scholar] [CrossRef] [PubMed]
- An, J.-P.; Yao, J.-F.; Xu, R.-R.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 2018, 41, 2678–2692. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Zhang, M.; Wen, C.; Xie, X.; Tian, W.; Wen, S.; Lu, R.; Liu, L. Integrated metabolomic and transcriptomic analysis of the anthocyanin regulatory networks in Salvia miltiorrhiza Bge. flowers. BMC Plant Biol. 2020, 20, 349. [Google Scholar] [CrossRef]
- Xie, Z.; Wang, J.; Wang, W.; Wang, Y.; Xu, J.; Li, Z.; Zhao, X.; Fu, B. Integrated Analysis of the Transcriptome and Metabolome Revealed the Molecular Mechanisms Underlying the Enhanced Salt Tolerance of Rice Due to the Application of Exogenous Melatonin. Front. Plant Sci. 2020, 11, 618680. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.T.; Xiu, Z.; Chen, C.H.; Wang, Y.; Yang, J.X.; Sui, J.J.; Jiang, S.J.; Wang, P.; Yue, S.Y.; Zhang, Q.Q.; et al. Long read sequencing of Toona sinensis (A. Juss) Roem: A chromosome-level reference genome for the family Meliaceae. Mol. Ecol. Resour. 2021, 21, 1243–1255. [Google Scholar] [CrossRef]
- Xue, L.; Wang, J.; Zhao, J.; Zheng, Y.; Wang, H.F.; Wu, X.; Xian, C.; Lei, J.J.; Zhong, C.F.; Zhang, Y.T. Study on cyanidin metabolism in petals of pink-flowered strawberry based on transcriptome sequencing and metabolite analysis. BMC Plant Biol. 2019, 19, 423. [Google Scholar] [CrossRef]
- Bogs, J.; Jaffé, F.W.; Takos, A.M.; Walker, A.R.; Robinson, S.P. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 2007, 143, 1347–1361. [Google Scholar] [CrossRef]
- Clark, S.T.; Verwoerd, W.S. A systems approach to identifying correlated gene targets for the loss of colour pigmentation in plants. BMC Bioinform. 2011, 12, 343. [Google Scholar] [CrossRef]
- Terrier, N.; Torregrosa, L.; Ageorges, A.; Vialet, S.; Verries, C.; Cheynier, V.; Romieu, C. Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 2009, 149, 1028–1041. [Google Scholar] [CrossRef]
- Debes, M.A.; Arias, M.E.; Grellet-Bournonville, C.F.; Wulff, A.F.; Martínez-Zamora, M.G.; Castagnaro, A.P.; Díaz-Ricci, J.C. White-fruited Duchesnea indica (Rosaceae) is impaired in ANS gene expression. Am. J. Bot. 2011, 98, 2077–2083. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, J.; Pei, T.; Bai, Z.; Han, R.; Liang, Z. Overexpression of SmANS Enhances Anthocyanin Accumulation and Alters Phenolic Acids Content in Salvia miltiorrhiza and Salvia miltiorrhiza Bge f. alba Plantlets. Int. J. Mol. Sci. 2019, 20, 2225. [Google Scholar] [CrossRef]
- Liu, H.; Su, J.; Zhu, Y.; Yao, G.; Allan, A.C.; Ampomah-Dwamena, C.; Shu, Q.; Lin-Wang, K.; Zhang, S.; Wu, J. The involvement of PybZIPa in light-induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G-boxes in its promoter. Hortic. Res. 2019, 6, 134. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Jiang, S.; Zhang, T.; Xu, H.; Fang, H.; Zhang, J.; Su, M.; Wang, Y.; Zhang, Z.; Wang, N.; et al. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 2019, 289, 110286. [Google Scholar] [CrossRef] [PubMed]
- Alabd, A.; Ahmad, M.; Zhang, X.; Gao, Y.; Peng, L.; Zhang, L.; Ni, J.; Bai, S.; Teng, Y. Light-responsive transcription factor PpWRKY44 induces anthocyanin accumulation by regulating PpMYB10 expression in pear. Hortic. Res. 2022, 9, uhac199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Z.; Zhao, Y.; Guo, D.; Zhao, X.; Gao, W.; Zhang, J.; Song, B.; Cahill, D. StWRKY13 promotes anthocyanin biosynthesis in potato (Solanum tuberosum) tubers. Funct. Plant Biol. 2021, 49, 102–114. [Google Scholar] [CrossRef]
- Qi, F.; Liu, Y.; Luo, Y.; Cui, Y.; Lu, C.; Li, H.; Huang, H.; Dai, S. Functional analysis of the ScAG and ScAGL11 MADS-box transcription factors for anthocyanin biosynthesis and bicolour pattern formation in Senecio cruentus ray florets. Hortic. Res. 2022, 9, uhac071. [Google Scholar] [CrossRef]
- Qi, X.; Liu, C.; Song, L.; Li, M. PaMADS7, a MADS-box transcription factor, regulates sweet cherry fruit ripening and softening. Plant Sci. 2020, 301, 110634. [Google Scholar] [CrossRef]
- Shi, H.; Liu, G.; Wei, Y.; Chan, Z. The zinc-finger transcription factor ZAT6 is essential for hydrogen peroxide induction of anthocyanin synthesis in Arabidopsis. Plant Mol. Biol. 2018, 97, 165–176. [Google Scholar] [CrossRef]
- Wang, D.R.; Yang, K.; Wang, X.; Lin, X.L.; Rui, L.; Liu, H.F.; Liu, D.D.; You, C.X. Overexpression of MdZAT5, an C2H2-Type Zinc Finger Protein, Regulates Anthocyanin Accumulation and Salt Stress Response in Apple Calli and Arabidopsis. Int. J. Mol. Sci. 2022, 23, 1897. [Google Scholar] [CrossRef]
- Kimura, Y.; Aoki, S.; Ando, E.; Kitatsuji, A.; Watanabe, A.; Ohnishi, M.; Takahashi, K.; Inoue, S.I.; Nakamichi, N.; Tamada, Y.; et al. A flowering integrator, SOC1, affects stomatal opening in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Ryu, C.H.; Lee, S.; Cho, L.H.; Kim, S.L.; Lee, Y.S.; Choi, S.C.; Jeong, H.J.; Yi, J.; Park, S.J.; Han, C.D.; et al. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 2009, 32, 1412–1427. [Google Scholar] [CrossRef]
- Ma, L.; Yan, Y. GhSOC1s Evolve to Respond Differently to the Environmental Cues and Promote Flowering in Partially Independent Ways. Front. Plant Sci. 2022, 13, 882946. [Google Scholar] [CrossRef]
- Wang, C.K.; Han, P.L.; Zhao, Y.W.; Ji, X.L.; Yu, J.Q.; You, C.X.; Hu, D.G.; Hao, Y.J. Auxin regulates anthocyanin biosynthesis through the auxin repressor protein MdIAA26. Biochem. Biophys. Res. Commun. 2020, 533, 717–722. [Google Scholar] [CrossRef]
- Wang, P.; Ge, M.; Yu, A.; Song, W.; Fang, J.; Leng, X. Effects of ethylene on berry ripening and anthocyanin accumulation of “Fujiminori” grape in protected cultivation. J. Sci. Food Agric. 2022, 102, 1124–1136. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | Log2FC_gene (JFC: LFC) | Gene Name | AGI | Gene Annotation | Compounds | Log2FC_meta (JFC: LFC) | Coefficient | p Value |
|---|---|---|---|---|---|---|---|---|
| Maker00012886 | 1.5011 | ERF4 | At5g44210 | AP2 domain | Cyanidin-3-O-galactoside | 7.9568 | 0.8479 | 6.52 × 10−5 |
| Cyanidin-3-O-sambubioside | 5.4217 | 0.8049 | 0.0002953 | |||||
| Cyanidin-3-O-arabinoside | 9.025 | 0.8034 | 0.000309044 | |||||
| Cyanidin-3-O-xyloside | 5.2648 | 0.8327 | 0.000116859 | |||||
| Delphinidin-3-O-glucoside | 9.025 | 0.8197 | 0.00018361 | |||||
| Maker00009687 | 2.2469 | CLE1 | At2g22850 | bZIP transcription factor | Cyanidin-3-O-rutinoside-5-O-glucoside | 9.025 | 0.8066 | 0.000279669 |
| Cyanidin-3-O-galactoside | 7.9568 | 0.8819 | 1.37 × 10−5 | |||||
| Cyanidin-3-O-arabinoside | 9.025 | 0.8696 | 2.54 × 10−5 | |||||
| Maker00020104 | 2.8632 | bZIP46_1 | At1g68640 | bZIP transcription factor | Cyanidin-3-O-rutinoside-5-O-glucoside | 9.025 | 0.8042 | 0.000301857 |
| Cyanidin-3-O-rutinoside | 7.2774 | 0.8418 | 8.30 × 10−5 | |||||
| Cyanidin-3-O-galactoside | 7.9568 | 0.8692 | 2.59 × 10−5 | |||||
| Cyanidin-3-O-sambubioside | 5.4217 | 0.8169 | 0.000201339 | |||||
| Cyanidin-3-O-glucoside | 5.7951 | 0.8047 | 0.000296932 | |||||
| Cyanidin-3-O-arabinoside | 9.025 | 0.8256 | 0.000149917 | |||||
| Cyanidin-3,5-O-diglucoside | 2.6736 | 0.8462 | 6.98 × 10−5 | |||||
| Cyanidin-3-O-xyloside | 5.2648 | 0.8548 | 4.91 × 10−5 | |||||
| Maker00026824 | 4.2921 | bZIP43 | At2g22850 | bZIP transcription factor | Cyanidin-3-O-rutinoside-5-O-glucoside | 9.025 | 0.844 | 7.62 × 10−5 |
| Cyanidin-3-O-rutinoside | 7.2774 | 0.8201 | 0.00018127 | |||||
| Cyanidin-3-O-galactoside | 7.9568 | 0.8575 | 4.39 × 10−5 | |||||
| Cyanidin-3-O-sambubioside | 5.4217 | 0.8221 | 0.000169159 | |||||
| Cyanidin-3,5-O-diglucoside | 2.6736 | 0.8355 | 0.000105219 | |||||
| Cyanidin-3-O-xyloside | 5.2648 | 0.8607 | 3.81 × 10−5 | |||||
| Maker00032838 | 1.3156 | bZIP46_2 | At1g68640 | bZIP transcription factor | Cyanidin-3-O-galactoside | 7.9568 | 0.8368 | 0.00010049 |
| Cyanidin-3-O-arabinoside | 9.025 | 0.8284 | 0.000136002 | |||||
| novel.8684 | 2.8522 | POLX | At3g45520 | bZIP transcription factor | Pelargonidin-3-O-(6-O-p-coumaroyl)-glucoside | 4.1336 | 0.8159 | 0.000208096 |
| Maker00014746 | 2.4849 | CPC | At2g46410 | Myb-like DNA-binding domain | Cyanidin-3-O-(6-O-malonyl-beta-D-glucoside) | 9.025 | 0.801 | 0.000332643 |
| Cyanidin-3-O-rutinoside | 7.2774 | 0.8151 | 0.000213466 | |||||
| Cyanidin-3-O-galactoside | 7.9568 | 0.8915 | 8.09 × 10−6 | |||||
| Cyanidin-3-O-sambubioside | 5.4217 | 0.811 | 0.000243476 | |||||
| Cyanidin-3-O-arabinoside | 9.025 | 0.9149 | 1.78 × 10−6 | |||||
| Cyanidin-3,5-O-diglucoside | 2.6736 | 0.8358 | 0.000104028 | |||||
| Cyanidin-3-O-xyloside | 5.2648 | 0.8778 | 1.69 × 10−5 | |||||
| Delphinidin-3-O-glucoside | 9.025 | 0.8114 | 0.000240963 | |||||
| Delphinidin-3-O-5-O-(6-O-coumaroyl)-diglucoside | 1.0838 | 0.8249 | 0.000153878 | |||||
| Maker00016969 | 1.3602 | MYB30 | At4g09460 | Myb-like DNA-binding domain | Cyanidin-3-O-galactoside | 7.9568 | 0.8189 | 0.000188654 |
| Maker00023867 | 1.6363 | MYB17 | At3g61250 | Myb-like DNA-binding domain | Cyanidin-3-O-galactoside | 7.9568 | 0.8493 | 6.17 × 10−5 |
| Cyanidin-3-O-arabinoside | 9.025 | 0.8159 | 0.000207999 | |||||
| Maker00029632 | 4.4878 | SOC1 | At2g45660 | MADS-box transcription factor | Cyanidin-3-O-galactoside | 7.9568 | 0.9012 | 4.53 × 10−6 |
| Cyanidin-3-O-arabinoside | 9.025 | 0.9525 | 4.40 × 10−8 | |||||
| Cyanidin-3-O-xyloside | 5.2648 | 0.8064 | 0.000281454 | |||||
| Delphinidin-3-O-5-O-(6-O-coumaroyl)-diglucoside | 1.0838 | 0.8166 | 0.000203571 | |||||
| novel.3727 | 4.9012 | AGL9 | At1g24260 | MADS-box transcription factor | Cyanidin-3-O-rutinoside-5-O-glucoside | 9.025 | 0.8712 | 2.35 × 10−5 |
| Cyanidin-3-O-(6-O-malonyl-beta-D-glucoside) | 9.025 | 0.8 | 0.000341781 | |||||
| Cyanidin-3-O-rutinoside | 7.2774 | 0.8165 | 0.000204 | |||||
| Cyanidin-3-O-galactoside | 7.9568 | 0.9652 | 5.96 × 10−9 | |||||
| Cyanidin-3-O-sambubioside | 5.4217 | 0.8078 | 0.000269708 | |||||
| Cyanidin-3-O-arabinoside | 9.025 | 0.9734 | 1.07 × 10−9 | |||||
| Cyanidin-3,5-O-diglucoside | 2.6736 | 0.8416 | 8.35 × 10−5 | |||||
| Cyanidin-3-O-xyloside | 5.2648 | 0.8968 | 5.93 × 10−6 | |||||
| Delphinidin-3-O-glucoside | 9.025 | 0.8552 | 4.83 × 10−5 | |||||
| Delphinidin-3-O-5-O-(6-O-coumaroyl)-diglucoside | 1.0838 | 0.8735 | 2.10 × 10−5 | |||||
| Petunidin-3-O-rutinoside | 9.025 | 0.8018 | 0.000324031 | |||||
| Maker00006536 | 1.8404 | WRKY48 | At5g49520 | WRKY DNA-binding protein 48 | Cyanidin-3-O-rutinoside-5-O-glucoside | 9.025 | 0.832 | 0.000119819 |
| Cyanidin-3-O-galactoside | 7.9568 | 0.8644 | 3.22 × 10−5 | |||||
| Cyanidin-3-O-arabinoside | 9.025 | 0.8435 | 7.77 × 10−5 | |||||
| Cyanidin-3-O-xyloside | 5.2648 | 0.8093 | 0.000257445 | |||||
| novel.6183 | 3.5074 | MIK2 | At4g08850 | Leucine-rich repeat receptor-like protein kinase | Cyanidin-3-O-rutinoside-5-O-glucoside | 9.025 | 0.8304 | 0.000126884 |
| Cyanidin-3-O-galactoside | 7.9568 | 0.9136 | 1.95 × 10−6 | |||||
| Cyanidin-3-O-arabinoside | 9.025 | 0.9219 | 1.03 × 10−6 | |||||
| Delphinidin-3-O-5-O-(6-O-coumaroyl)-diglucoside | 1.0838 | 0.8338 | 0.000112199 | |||||
| Maker00021798 | 1.1527 | bHLH162 | At4g20970 | Helix-loop-helix DNA-binding domain | Cyanidin-3-O-galactoside | 7.9568 | 0.8684 | 2.68 × 10−5 |
| Cyanidin-3-O-arabinoside | 9.025 | 0.8782 | 1.66 × 10−5 | |||||
| Maker00003242 | 2.1058 | JAG | At1g13400 | C2H2 and C2HC zinc fingers superfamily protein | Cyanidin-3-O-galactoside | 7.9568 | 0.8668 | 2.90 × 10−5 |
| Cyanidin-3-O-arabinoside | 9.025 | 0.8657 | 3.04 × 10−5 | |||||
| Maker00011793 | 1.1426 | GRAS20 | At3g54220 | GRAS domain family | Cyanidin-3-O-(6-O-malonyl-beta-D-glucoside) | 9.025 | 0.8327 | 0.000116807 |
| Cyanidin-3,5-O-diglucoside | 2.6736 | 0.8072 | 0.000275174 | |||||
| novel.4188 | 1.6057 | HSP72 | At1g56410 | heat shock protein 70 family protein | Cyanidin-3-O-(6-O-malonyl-beta-D-glucoside) | 9.025 | 0.8302 | 0.000127565 |
| Cyanidin-3-O-galactoside | 7.9568 | 0.9167 | 1.55 × 10−6 | |||||
| Cyanidin-3-O-arabinoside | 9.025 | 0.9144 | 1.84 × 10−6 | |||||
| Cyanidin-3-O-xyloside | 5.2648 | 0.807 | 0.00027619 | |||||
| Delphinidin-3-O-glucoside | 9.025 | 0.8147 | 0.000216435 | |||||
| novel.4220 | 4.8378 | LBD21 | At3g11090 | Lateral organ boundaries domain | Cyanidin-3-O-rutinoside-5-O-glucoside | 9.025 | 0.8909 | 8.38 × 10−6 |
| Cyanidin-3-O-rutinoside | 7.2774 | 0.8734 | 2.11 × 10−5 | |||||
| Cyanidin-3-O-galactoside | 7.9568 | 0.8584 | 4.21 × 10−5 | |||||
| Cyanidin-3-O-sambubioside | 5.4217 | 0.8307 | 0.00012547 | |||||
| Cyanidin-3-O-arabinoside | 9.025 | 0.8884 | 9.68 × 10−6 | |||||
| Cyanidin-3,5-O-diglucoside | 2.6736 | 0.8797 | 1.54 × 10−5 | |||||
| Cyanidin-3-O-xyloside | 5.2648 | 0.8999 | 4.89 × 10−6 | |||||
| Delphinidin-3-O-5-O-(6-O-coumaroyl)-diglucoside | 1.0838 | 0.9109 | 2.36 × 10−6 | |||||
| Petunidin-3-O-rutinoside | 9.025 | 0.8911 | 8.31 × 10−6 | |||||
| Maker00033233 | 1.0573 | STY46 | At4g35780 | Protein tyrosine kinase | Cyanidin-3-O-(6-O-malonyl-beta-D-glucoside) | 9.025 | 0.8235 | 0.000161377 |
| Cyanidin-3-O-galactoside | 7.9568 | 0.8026 | 0.00031603 | |||||
| Cyanidin-3,5-O-diglucoside | 2.6736 | 0.8017 | 0.000324943 | |||||
| Maker00000350 | 1.9217 | ARR9 | At3g57040 | Response regulator receiver domain | Cyanidin-3-O-rutinoside-5-O-glucoside | 9.025 | 0.8952 | 6.53 × 10−6 |
| Cyanidin-3-O-rutinoside | 7.2774 | 0.8032 | 0.000310451 | |||||
| Cyanidin-3-O-galactoside | 7.9568 | 0.842 | 8.23 × 10−5 | |||||
| Cyanidin-3-O-arabinoside | 9.025 | 0.8663 | 2.95 × 10−5 | |||||
| Cyanidin-3,5-O-diglucoside | 2.6736 | 0.833 | 0.000115529 | |||||
| Maker00026791 | 1.2059 | SPL1 | At3g57920 | SBP domain | Cyanidin-3-O-arabinoside | 9.025 | 0.8222 | 0.000168825 |
| Maker00008503 | 1.3275 | IAA27 | At4g29080 | AUX/IAA family | Cyanidin-3-O-glucoside | 5.7951 | 0.8337 | 0.000112681 |
| Maker00022256 | 1.0026 | ARG7 | At1g75590 | Auxin responsive protein | Cyanidin-3-O-arabinoside | 9.025 | 0.8388 | 9.31 × 10−5 |
| Primer Name | Primer Sequence F (5′-3′) | Primer Sequence R (5′-3′) |
|---|---|---|
| RT-bHLH162 (Maker00021798) | CAGGTTAATGGGAATCGAAAAG | CCAACCCAGTTACCAAAACAAT |
| RT-CPC (Maker00014746) | AAACTCGTTGGAGACAGGTG | GCAAACCCTTCTCCATGTTT |
| RT-bZIP43 (Maker00026824) | TCGGAGACTATGATCGGAAATC | ACTTGAAACGACAGGTTTTGGT |
| RT-SOC1 (Maker00029632) | CAATTGAAGCACGAAATAGCAA | TGTACCGTTTAGAGGGAAGGAA |
| RT-4CL-1 (Maker00007314) | AGGTTTGAGGCTTCGCAGTA | GTCCCCAGTGACAACAATCC |
| RT-CHS-1 (Maker00012884) | TGCAATTCTTGACCAAGTCG | GATGAACAAAACGCATGCAC |
| RT-CHS-2 (Maker00020412) | TGGTGCAATTGATGGACACT | TGCTTCGACCAAGCTTTTCT |
| RT-CHI-2 (Maker00012191) | GGGTGTTGGAAGAAAATCCA | GGTCGACTCCATCACCCTTA |
| RT-DFR (Maker00011618) | CCATTCCTGGCTTCAAGTGT | AAGCAATTGACCCCATTCTG |
| RT-ANS (Maker00000819) | GCTTTCAGGCAACTGGTCAT | TGCTCGGGAATATTTCTGCT |
| RT-UFGT-1 (Maker00029333) | GTGGTCGAGACTGGGAGAAG | AGGACCTGCAGTCCAAAGTG |
| RT-UFGT-2 (novel.8170) | GTGACAAAACCCTGGAAGGA | CAATTTTGGGAAAGCTGCAT |
| RT-Actin | TGATTGGGATGGAAGCAGCA | GAACATGGTTGAACCGCCAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Fan, Y.; Han, X.; Pan, H.; Dai, J.; Wei, Y.; Zhuo, R.; Liu, J. Integrated Transcriptomic and Metabolomic Analysis Reveal the Underlying Mechanism of Anthocyanin Biosynthesis in Toona sinensis Leaves. Int. J. Mol. Sci. 2023, 24, 15459. https://doi.org/10.3390/ijms242015459
Xu J, Fan Y, Han X, Pan H, Dai J, Wei Y, Zhuo R, Liu J. Integrated Transcriptomic and Metabolomic Analysis Reveal the Underlying Mechanism of Anthocyanin Biosynthesis in Toona sinensis Leaves. International Journal of Molecular Sciences. 2023; 24(20):15459. https://doi.org/10.3390/ijms242015459
Chicago/Turabian StyleXu, Jing, Yanru Fan, Xiaojiao Han, Huanhuan Pan, Jianhua Dai, Yi Wei, Renying Zhuo, and Jun Liu. 2023. "Integrated Transcriptomic and Metabolomic Analysis Reveal the Underlying Mechanism of Anthocyanin Biosynthesis in Toona sinensis Leaves" International Journal of Molecular Sciences 24, no. 20: 15459. https://doi.org/10.3390/ijms242015459
APA StyleXu, J., Fan, Y., Han, X., Pan, H., Dai, J., Wei, Y., Zhuo, R., & Liu, J. (2023). Integrated Transcriptomic and Metabolomic Analysis Reveal the Underlying Mechanism of Anthocyanin Biosynthesis in Toona sinensis Leaves. International Journal of Molecular Sciences, 24(20), 15459. https://doi.org/10.3390/ijms242015459







