Repression of GhTUBB1 Reduces Plant Height in Gossypium hirsutum
Abstract
:1. Introduction
2. Results
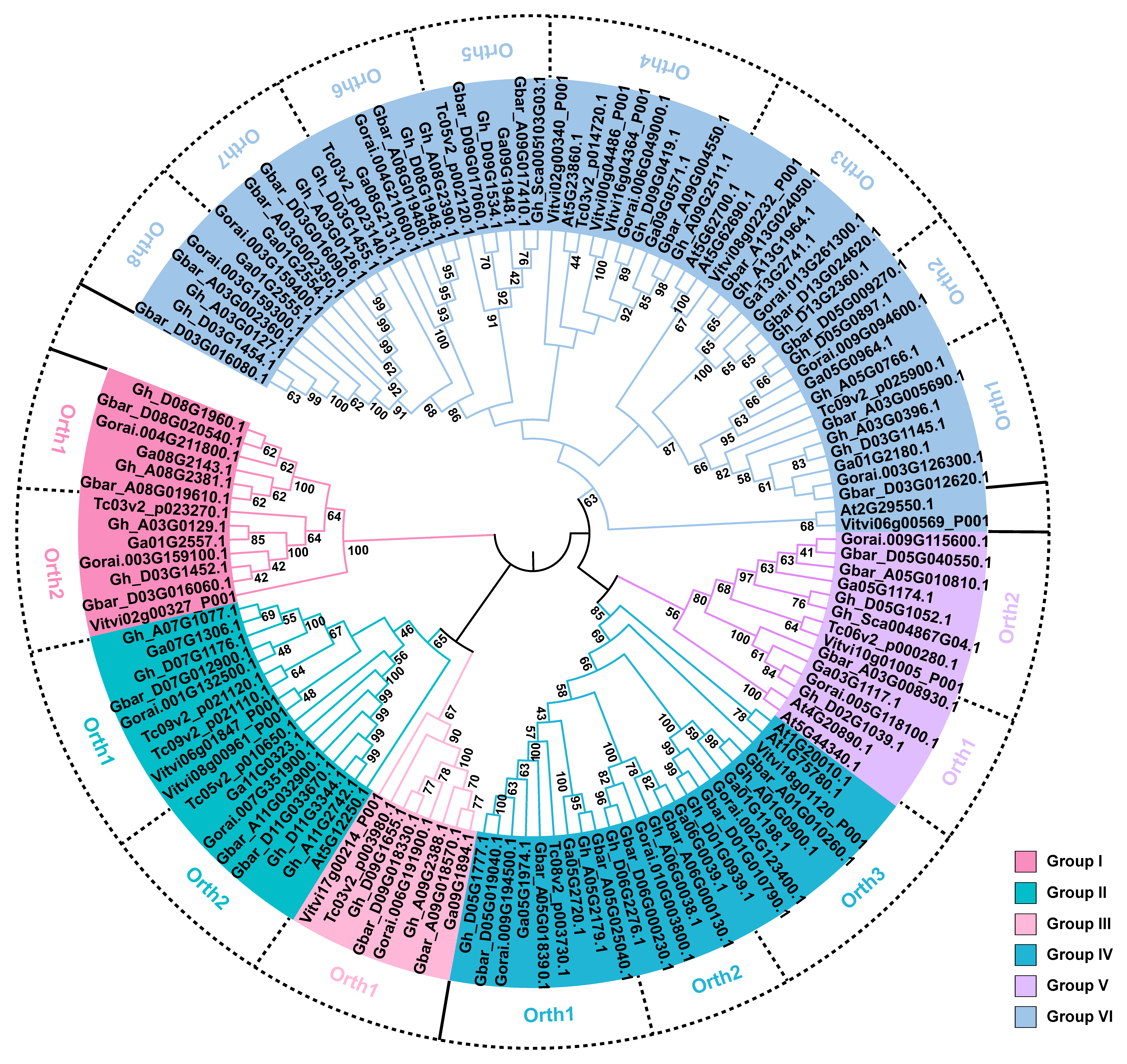
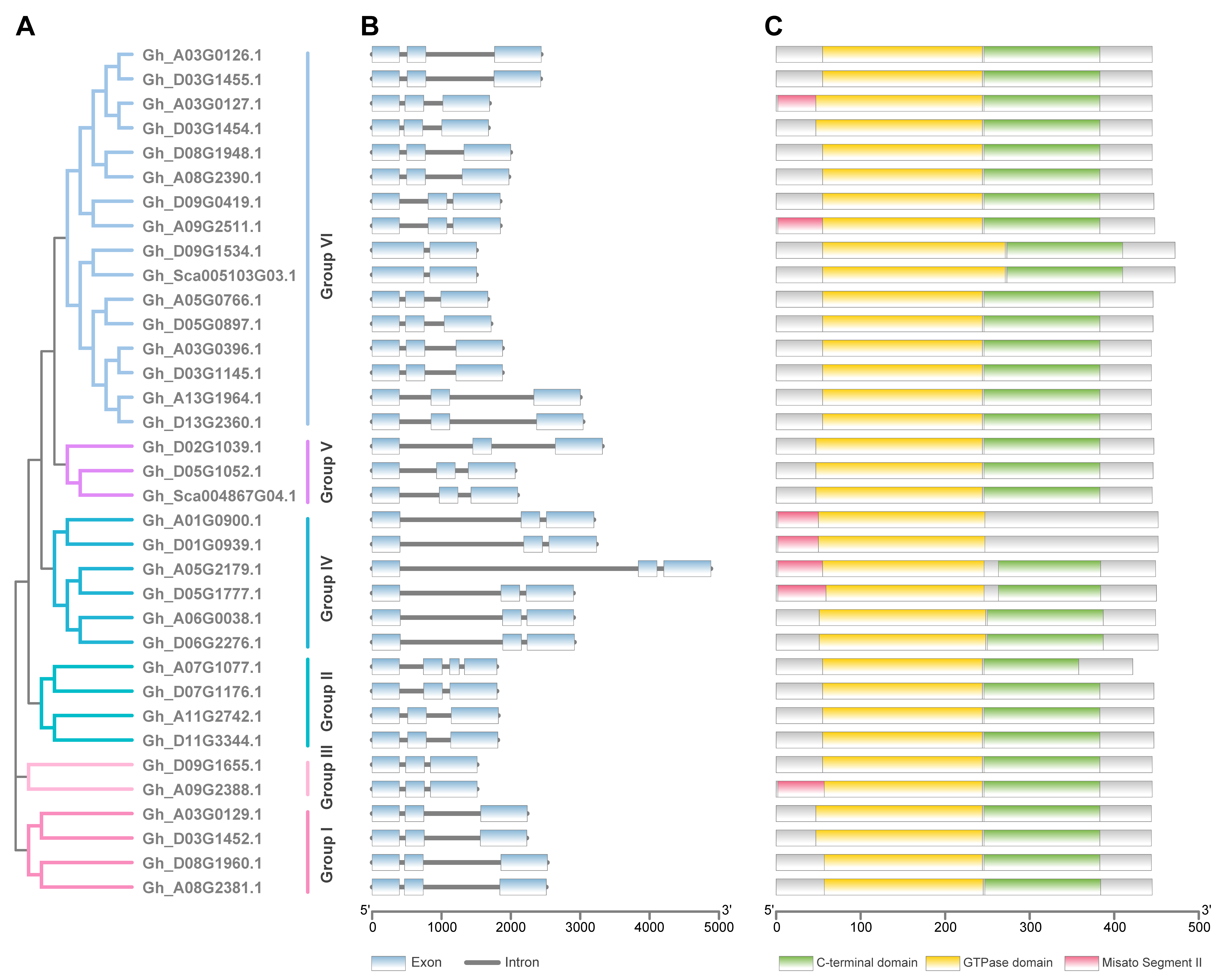
2.1. Identification of Cotton GhTUBB Proteins
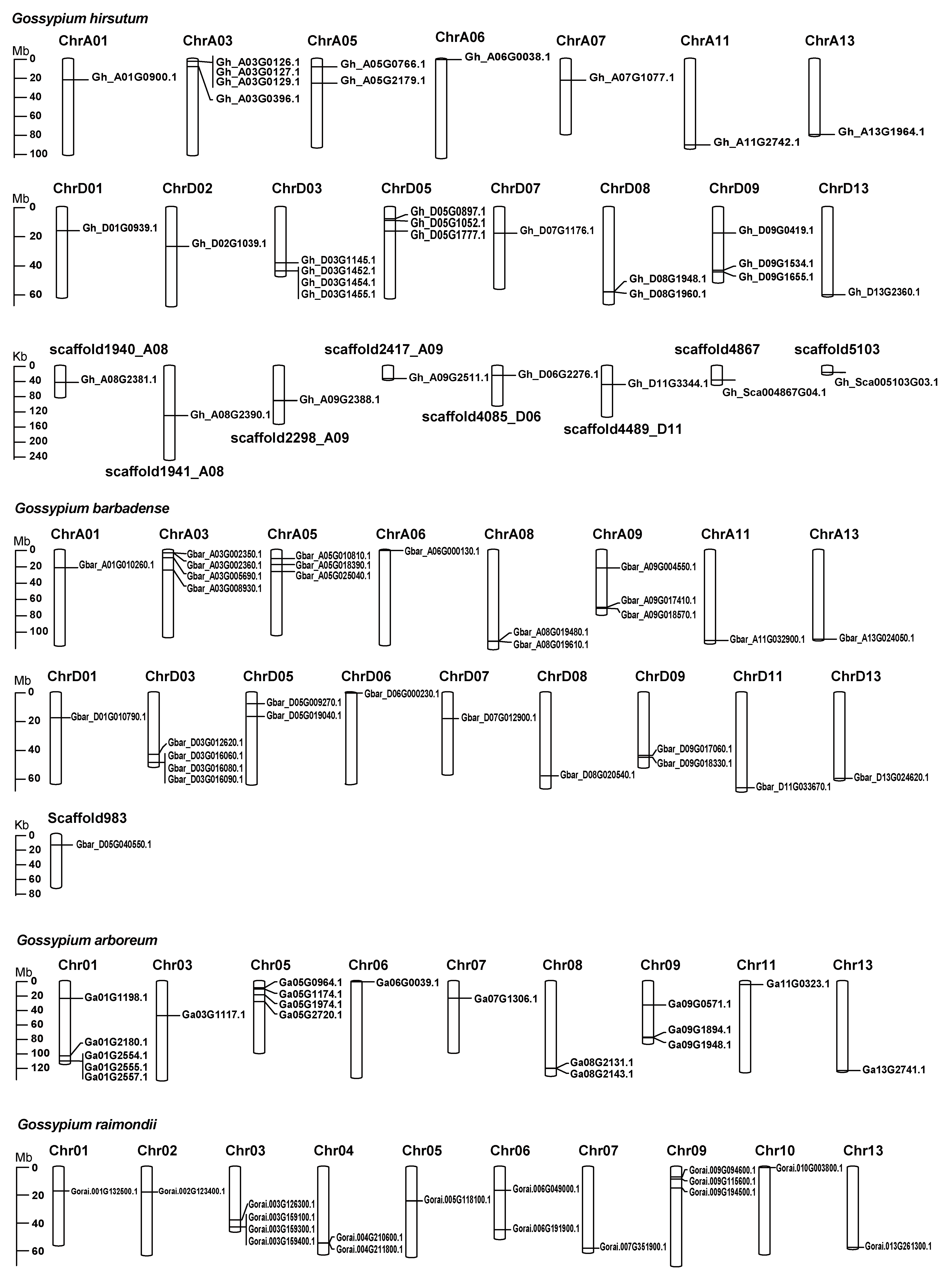
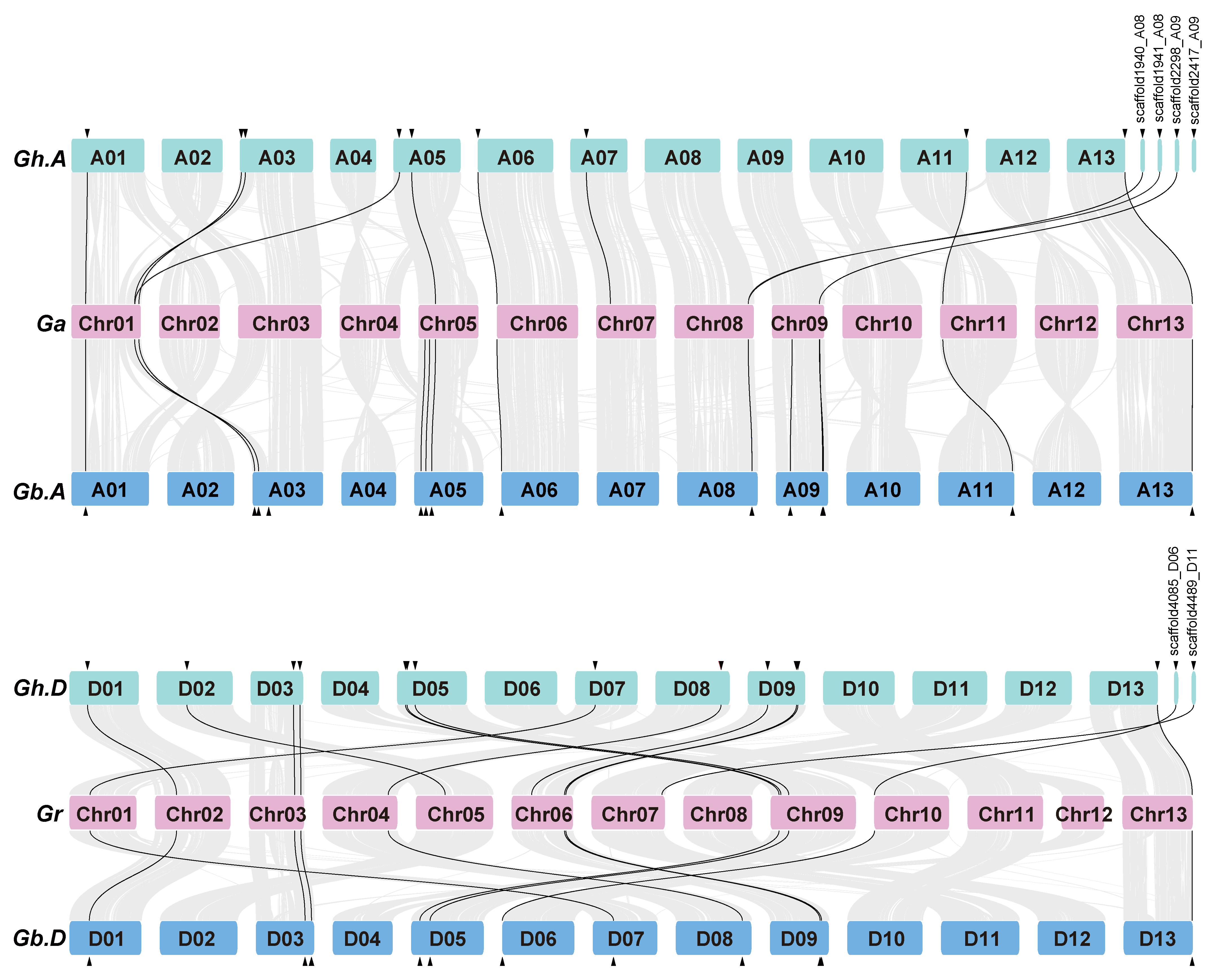
2.2. Chromosome Distribution and Synteny of G. hirsutum TUBB1 Family Members
2.3. Silencing of the GhTUBB1 Gene Reduces Plant Height
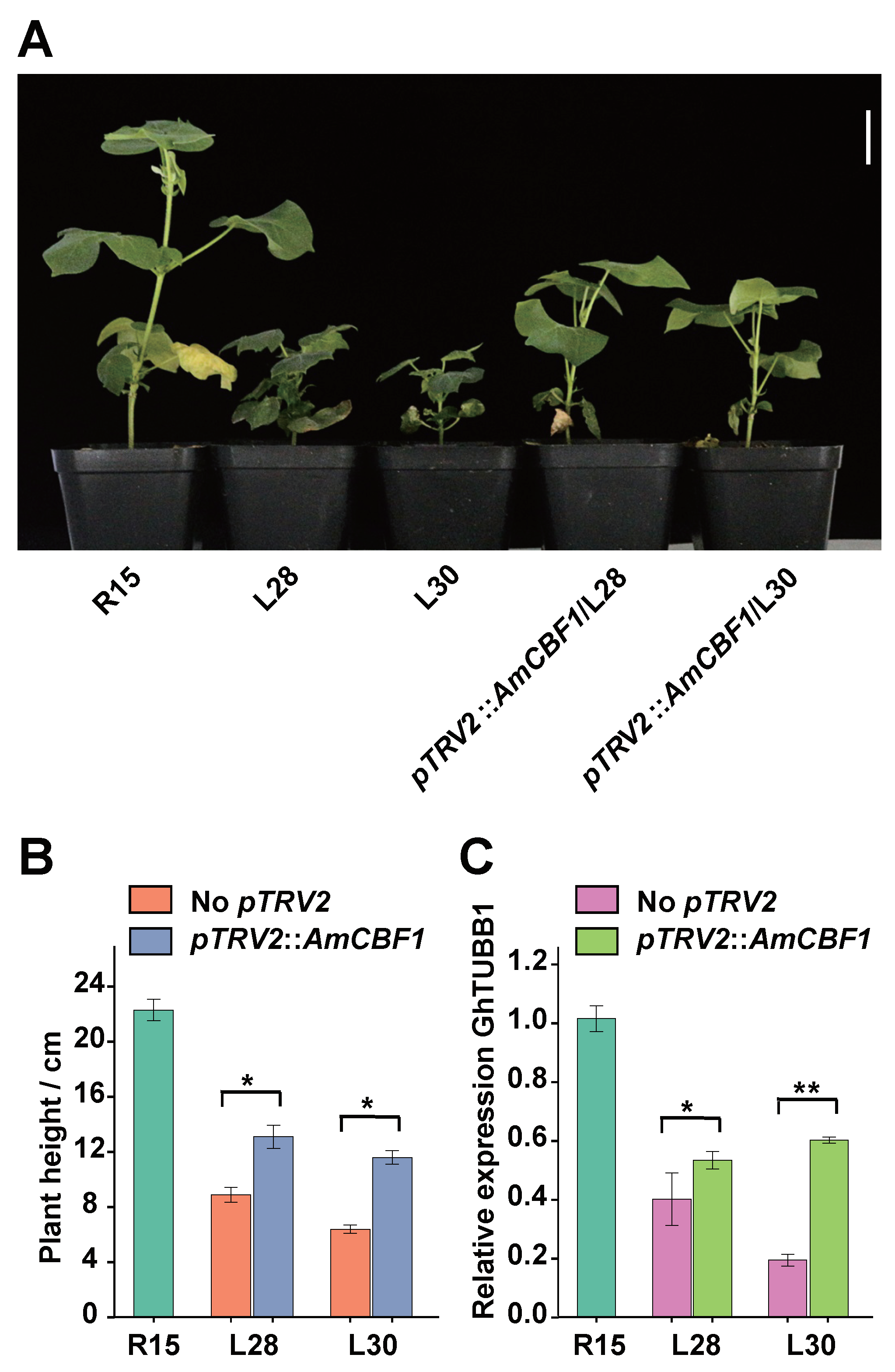
2.4. AmCBF1 Silencing Partially Restores Plant Height and GhTUBB1 Expression
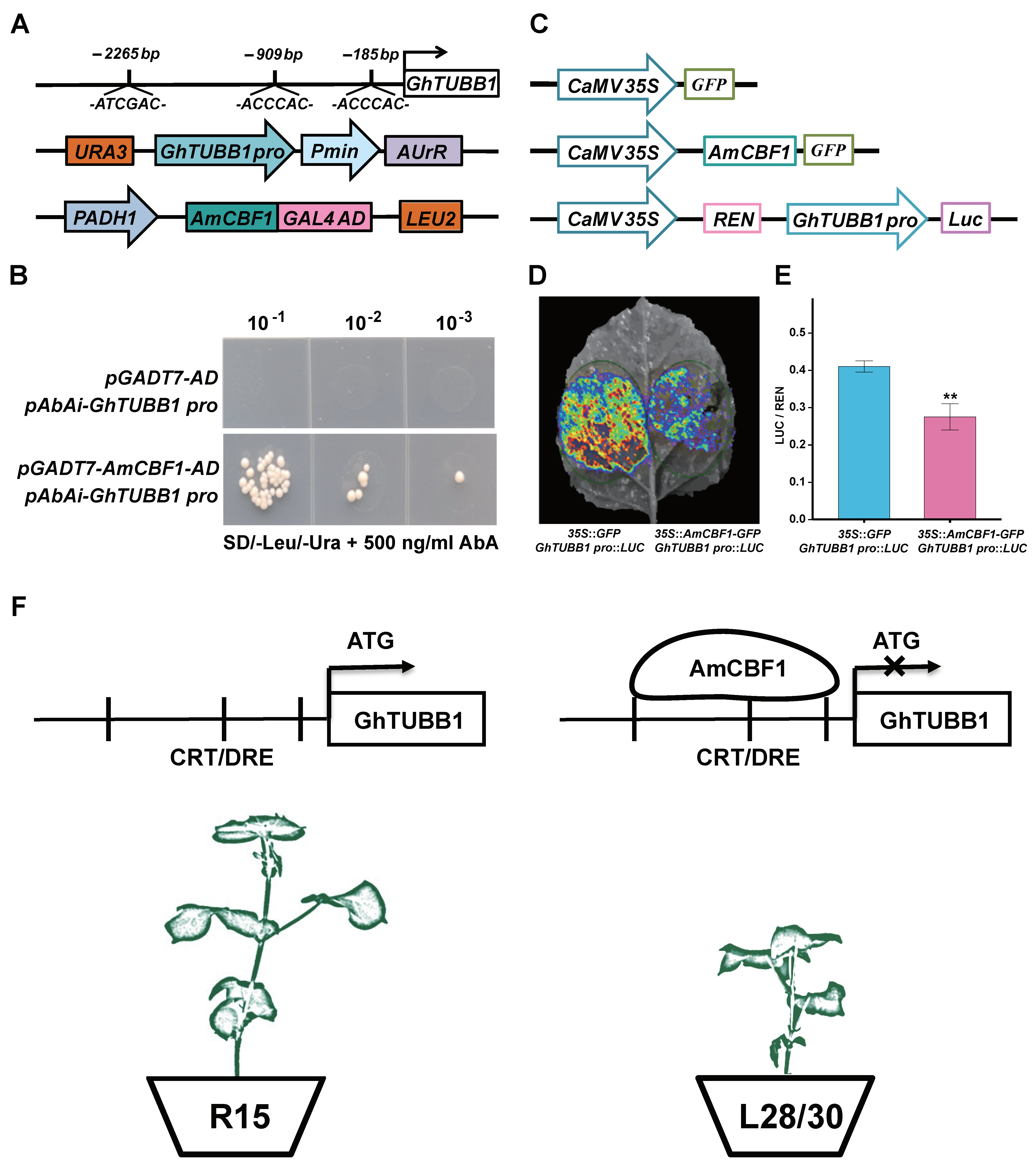
2.5. AmCBF1 Binds Directly to the GhTUBB1 Promoter
3. Discussion
4. Materials and Methods
4.1. Identification of TUB Family Members in Cotton
4.2. Phylogenetic Analysis
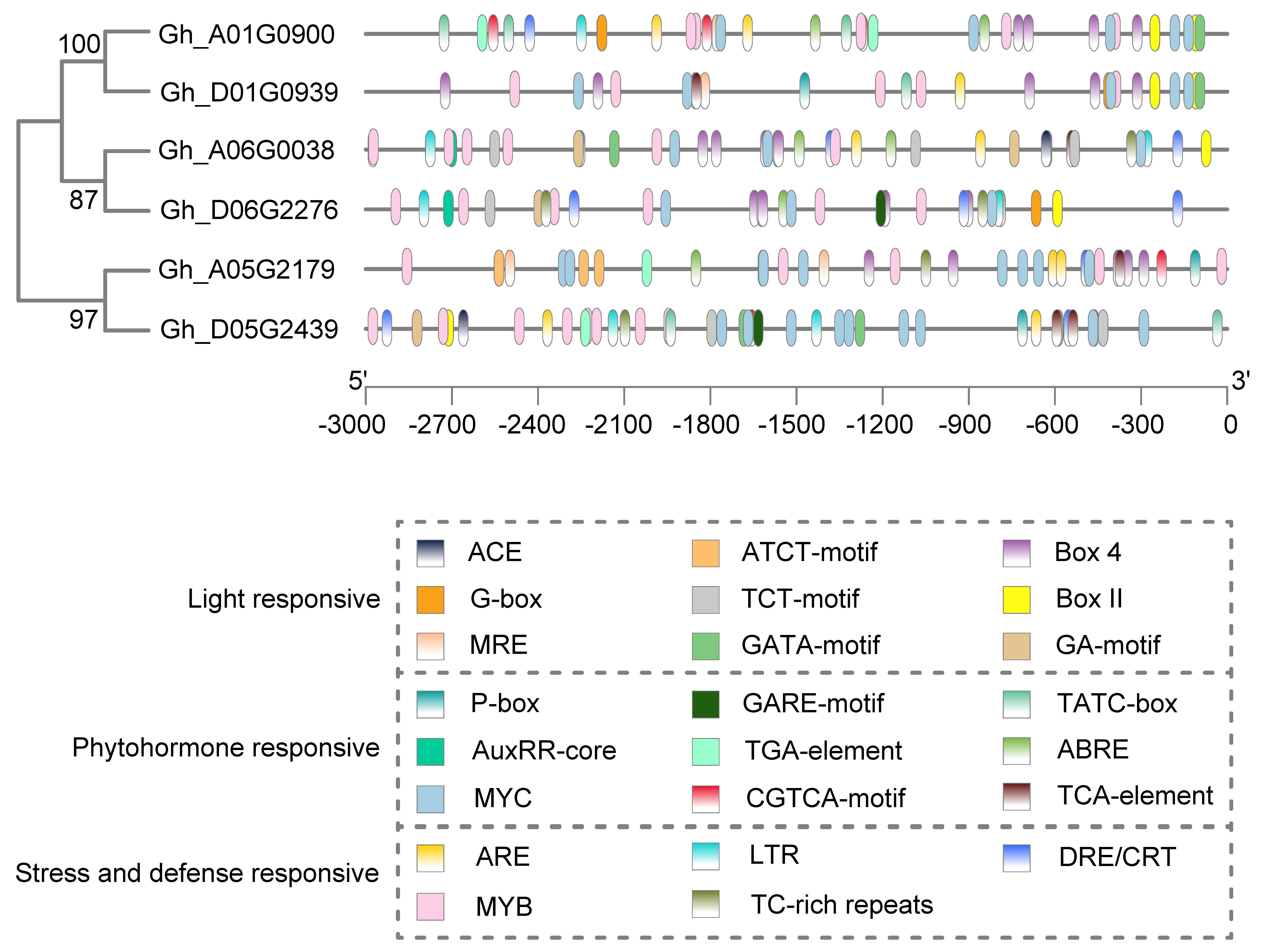
4.3. Gene Structure, Conserved Domain and Promoter Analysis
4.4. Chromosomal Mapping and Gene Syntenic Analysis
4.5. Plant Materials and Growing Conditions
4.6. Quantitative Real-Time PCR
4.7. Virus-Induced Gene Silencing
4.8. Yeast One-Hybrid Assay
4.9. Dual-Luciferase Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, J.; Hill, C.B.; Shabala, S.; Li, C.; Zhou, M. Manipulating GA-Related Genes for Cereal Crop Improvement. Int. J. Mol. Sci. 2022, 23, 14046. [Google Scholar] [CrossRef]
- Hedden, P. The genes of the Green Revolution. Trends Genet. 2003, 19, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.G. Novel Rht-1 dwarfing genes: Tools for wheat breeding and dissecting the function of DELLA proteins. J. Exp. Bot. 2017, 68, 354–358. [Google Scholar] [CrossRef]
- Bednarz, C.W.; Shurley, W.D.; Anthony, W.S.; Nichols, R.L. Yield, Quality, and Profitability of Cotton Produced at Varying Plant Densities. Agron. J. 2005, 97, 235–240. [Google Scholar] [CrossRef]
- Dai, J.; Li, W.; Tang, W.; Zhang, D.; Li, Z.; Lu, H.; Eneji, A.E.; Dong, H. Manipulation of dry matter accumulation and partitioning with plant density in relation to yield stability of cotton under intensive management. Field Crop. Res. 2015, 180, 207–215. [Google Scholar] [CrossRef]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ’Green revolution‘ genes encode mutant gibberellin response modulators. Nature. 1999, 400, 256–261. [Google Scholar] [CrossRef]
- Flintham, J.E.; Börner, A.; Worland, A.J.; Gale, M.D. Optimizing wheat grain yield: Effects of Rht (gibberellin-insensitive) dwarfing genes. J. Agric. Sci. 1997, 128, 11–25. [Google Scholar] [CrossRef]
- Pearce, S.; Saville, R.; Vaughan, S.P.; Chandler, P.M.; Wilhelm, E.P.; Sparks, C.A.; Al-Kaff, N.; Korolev, A.; Boulton, M.I.; Phillips, A.L.; et al. Molecular Characterization of Rht-1 Dwarfing Genes in Hexaploid Wheat. Plant Physiol. 2011, 157, 1820–1831. [Google Scholar] [CrossRef]
- Wu, J.; Kong, X.; Wan, J.; Liu, X.; Zhang, X.; Guo, X.; Zhou, R.; Zhao, G.; Jing, R.; Fu, X.; et al. Dominant and pleiotropic effects of a GAI gene in wheat results from a lack of interaction between DELLA and GID1. Plant Physiol. 2011, 157, 2120–2130. [Google Scholar] [CrossRef]
- Würschum, T.; Langer, S.M.; Longin, C.F.H.; Tucker, M.R.; Leiser, W.L. A modern Green Revolution gene for reduced height in wheat. Plant J. 2017, 92, 892–903. [Google Scholar] [CrossRef]
- Tian, X.; Xia, X.; Xu, D.; Liu, Y.; Xie, L.; Hassan, M.A.; Song, J.; Li, F.; Wang, D.; Zhang, Y.; et al. Rht24b, an ancient variation of TaGA2ox-A9, reduces plant height without yield penalty in wheat. New Phytol. 2021, 233, 738–750. [Google Scholar] [CrossRef] [PubMed]
- Herter, C.P.; Ebmeyer, E.; Kollers, S.; Korzun, V.; Leiser, W.L.; Wurschum, T.; Miedaner, T. Rht24 reduces height in the winter wheat population ’Solitar x Bussard‘ without adverse effects on Fusarium head blight infection. Theor. Appl. Genet. 2018, 131, 1263–1272. [Google Scholar] [CrossRef] [PubMed]
- Monna, L.; Kitazawa, N.; Yoshino, R.; Suzuki, J.; Masuda, H.; Maehara, Y.; Tanji, M.; Sato, M.; Nasu, S.; Minobe, Y. Positional Cloning of Rice Semidwarfing Gene, sd-1: Rice "Green Revolution Gene" Encodes a Mutant Enzyme Involved in Gibberellin Synthesis. DNA Res. 2002, 9, 11–17. [Google Scholar] [CrossRef]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature. 2002, 416, 701–702. [Google Scholar] [CrossRef]
- Spielmeyer, W.; Ellis, M.H.; Chandler, P.M. Semidwarf ( sd-1 ), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 2002, 99, 9043–9048. [Google Scholar] [CrossRef]
- Cho, Y.G.; Eun, M.Y.; McCouch, S.R.; Chae, Y.A. The semidwarf gene, sd-1, of rice (Oryza sativa L.). II. Molecular mapping and marker-assisted selection. Theor. Appl. Genet. 1994, 89, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Clouse, S.D.; Langford, M.; McMorris, T.C. A Brassinosteroid-Insensitive Mutant in Arabidopsis thaliana Exhibits Multiple Defects in Growth and Development. Plant Physiol. 1996, 111, 671–678. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Seto, H.; Fujioka, S.; Yoshida, S.; Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 2001, 410, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, L.; Qin, Y.; Chen, Y.; Liu, X.; Wang, M.; Mao, J.; Zhang, J.; He, Z.; Liu, L.; et al. A Temperature-Sensitive Misfolded bri1-301 Receptor Requires Its Kinase Activity to Promote Growth. Plant Physiol. 2018, 178, 1704–1719. [Google Scholar] [CrossRef]
- Mussig, C. Brassinosteroid-promoted growth. Plant Biol. 2005, 7, 110–117. [Google Scholar] [CrossRef]
- Schrager-Lavelle, A.; Gath, N.N.; Devisetty, U.K.; Carrera, E.; López-Díaz, I.; Blázquez, M.A.; Maloof, J.N. The role of a class III gibberellin 2-oxidase in tomato internode elongation. Plant J. 2018, 97, 603–615. [Google Scholar] [CrossRef]
- Sauter, M.; Mekhedov, S.L.; Kende, H. Gibberellin promotes histone H1 kinase activity and the expression of cdc2 and cyclin genes during the induction of rapid growth in deepwater rice internodes. Plant J. 1995, 7, 623–632. [Google Scholar] [CrossRef]
- Dong, H.; Li, D.; Yang, R.; Zhang, L.; Zhang, Y.; Liu, X.; Kong, X.; Sun, J. GSK3 phosphorylates and regulates the Green Revolution protein Rht-B1b to reduce plant height in wheat. Plant Cell 2023, 35, 1970–1983. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef]
- Achard, P.; Gong, F.; Cheminant, S.; Alioua, M.; Hedden, P.; Genschik, P. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell. 2008, 20, 2117–2129. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.G.; Yang, M.; Liu, P.; Yang, G.D.; Wu, C.A.; Zheng, C.C. GhDREB1 enhances abiotic stress tolerance, delays GA-mediated development and represses cytokinin signalling in transgenic Arabidopsis. Plant Cell Environ. 2009, 32, 1132–1145. [Google Scholar] [CrossRef]
- Li, W.-X.; Chen, M.; Chen, W.-T.; Qiao, C.-K.; Li, M.-H.; Han, L.-J. Determination of mepiquat chloride in cotton crops and soil and its dissipation rates. Ecotoxicol. Environ. Saf. 2012, 85, 137–143. [Google Scholar] [CrossRef]
- Siebert, J.D.; Stewart, A.M. Influence of Plant Density on Cotton Response to Mepiquat Chloride Application. Agron. J. 2006, 98, 1634–1639. [Google Scholar] [CrossRef]
- Wang, L.; Mu, C.; Du, M.; Chen, Y.; Tian, X.; Zhang, M.; Li, Z. The effect of mepiquat chloride on elongation of cotton (Gossypium hirsutum L.) internode is associated with low concentration of gibberellic acid. Plant Sci. 2014, 225, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Sun, J.-L.; Jia, Y.-H.; Wang, J.; Xu, Z.-J.; Du, X.-M. Morphological characters, inheritance and response to exogenous hormones of a cotton super-dwarf mutant of Gossypium hirsutum. Plant Breed. 2011, 130, 67–72. [Google Scholar] [CrossRef]
- Jia, Y.; Ding, Y.; Shi, Y.; Zhang, X.; Gong, Z.; Yang, S. The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 2016, 212, 345–353. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, C.; Yang, X.; Liu, K.; Wu, Z.; Zhang, X.; Zheng, W.; Xun, Q.; Liu, C.; Lu, L.; et al. PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol. 2014, 203, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Zhou, B.; Zhang, T. Isolation and characterization of a sterile-dwarf mutant in Asian cotton (Gossypium arboreum L.). J. Genet. Genom. 2009, 36, 343–353. [Google Scholar] [CrossRef]
- Ma, C.; Rehman, A.; Li, H.G.; Zhao, Z.B.; Sun, G.; Du, X.M. Mapping of dwarfing QTL of Ari1327, a semi-dwarf mutant of upland cotton. BMC Plant Biol. 2022, 22, 1–14. [Google Scholar] [CrossRef]
- Mishra, P.K.; Sharma, A.; Prakash, A. Current research and development in cotton harvesters: A review with application to Indian cotton production systems. Heliyon 2023, 9, e16124. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wang, L.; Zhang, Q.; Ma, C.; Su, X.; Cheng, H.; Guo, H. AmCBF1 Transcription Factor Regulates Plant Architecture by Repressing GhPP2C1 or GhPP2C2 in Gossypium hirsutum. Front. Plant Sci. 2022, 13, 914206. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, C.; Hussey, P. Microtubule-associated proteins in plants — why we need a map. Nat. Rev. Mol. Cell Biol. 2001, 2, 40–47. [Google Scholar] [CrossRef]
- Hamada, T. Microtubule organization and microtubule-associated proteins in plant cells. Int. Rev. Cell Mol. Biol. 2014, 312, 1–52. [Google Scholar]
- Chen, Y.; Hou, M.; Liu, L.; Wu, S.; Shen, Y.; Ishiyama, K.; Kobayashi, M.; McCarty, D.R.; Tan, B.-C. The Maize DWARF1 Encodes a Gibberellin 3-Oxidase and Is Dual Localized to the Nucleus and Cytosol. Plant Physiol. 2014, 166, 2028–2039. [Google Scholar] [CrossRef]
- Lawit, S.J.; Wych, H.M.; Xu, D.; Kundu, S.; Tomes, D.T. Maize DELLA Proteins dwarf plant8 and dwarf plant9 as Modulators of Plant Development. Plant Cell Physiol. 2010, 51, 1854–1868. [Google Scholar] [CrossRef]
- Li, W.; Ge, F.; Qiang, Z.; Zhu, L.; Zhang, S.; Chen, L.; Wang, X.; Li, J.; Fu, Y. Maize ZmRPH1 encodes a microtubule-associated protein that controls plant and ear height. Plant Biotechnol. J. 2020, 18, 1345–1347. [Google Scholar] [CrossRef]
- Song, Q.; Gao, W.; Du, C.; Wang, J.; Zuo, K. Cotton microtubule-associated protein GhMAP20L5 mediates fiber elongation through the interaction with the tubulin GhTUB13. Plant Sci. 2023, 327, 111545. [Google Scholar] [CrossRef]
- Li, L.; Wang, X.-L.; Huang, G.-Q.; Li, X.-B. Molecular characterization of cotton GhTUA9 gene specifically expressed in fibre and involved in cell elongation. J. Exp. Bot. 2007, 58, 3227–3238. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-B.; Cai, L.; Cheng, N.-H.; Liu, J.-W. Molecular Characterization of the Cotton GhTUB1 Gene That Is Preferentially Expressed in Fiber. Plant Physiol. 2002, 130, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Bishop, D.L.; Bugbee, B.G. Photosynthetic capacity and dry mass partitioning in dwarf and semi-dwarf wheat (Triticum aestivum L.). J. Plant Physiol. 1998, 153, 558–565. [Google Scholar] [CrossRef]
- Yaish, M.W.; El-Kereamy, A.; Zhu, T.; Beatty, P.H.; Good, A.G.; Bi, Y.-M.; Rothstein, S.J. The APETALA-2-Like Transcription Factor OsAP2-39 Controls Key Interactions between Abscisic Acid and Gibberellin in Rice. PLOS Genet. 2010, 6, e1001098. [Google Scholar] [CrossRef]
- Shu, K.; Zhou, W.; Yang, W. APETALA 2-domain-containing transcription factors: Focusing on abscisic acid and gibberellins antagonism. New Phytol. 2017, 217, 977–983. [Google Scholar] [CrossRef]
- Shu, K.; Chen, Q.; Wu, Y.; Liu, R.; Zhang, H.; Wang, P.; Li, Y.; Wang, S.; Tang, S.; Liu, C.; et al. ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 2015, 85, 348–361. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007, 449, 463–467. [Google Scholar] [PubMed]
- Argout, X.; Martin, G.; Droc, G.; Fouet, O.; Labadie, K.; Rivals, E.; Aury, J.; Lanaud, C. The cacao Criollo genome v2.0: An improved version of the genome for genetic and functional genomic studies. BMC Genom. 2017, 18, 730. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Tang, Z.; Wang, M.; Gao, W.; Tu, L.; Jin, X.; Chen, L.; He, Y.; Zhang, L.; Zhu, L.; et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 2015, 5, 17662. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Fan, G.; Wang, K.; Sun, F.; Yuan, Y.; Song, G.; Li, Q.; Ma, Z.; Lu, C.; Zou, C.; et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014, 46, 567–572. [Google Scholar] [CrossRef]
- Paterson, A.H.; Wendel, J.F.; Gundlach, H.; Guo, H.; Jenkins, J.; Jin, D.; Llewellyn, D.; Showmaker, K.C.; Shu, S.; Udall, J.; et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 2012, 492, 423–427. [Google Scholar] [CrossRef]
- Zhu, T.; Liang, C.; Meng, Z.; Sun, G.; Meng, Z.; Guo, S.; Zhang, R. CottonFGD: An integrated functional genomics database for cotton. BMC Plant Biol. 2017, 17, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Becker, A.; Lange, M. VIGS--genomics goes functional. Trends Plant Sci. 2010, 15, 1–4. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Ma, C.; Wang, L.; Su, X.; Huang, J.; Cheng, H.; Guo, H. Repression of GhTUBB1 Reduces Plant Height in Gossypium hirsutum. Int. J. Mol. Sci. 2023, 24, 15424. https://doi.org/10.3390/ijms242015424
Zhang L, Ma C, Wang L, Su X, Huang J, Cheng H, Guo H. Repression of GhTUBB1 Reduces Plant Height in Gossypium hirsutum. International Journal of Molecular Sciences. 2023; 24(20):15424. https://doi.org/10.3390/ijms242015424
Chicago/Turabian StyleZhang, Lihua, Caixia Ma, Lihua Wang, Xiaofeng Su, Jinling Huang, Hongmei Cheng, and Huiming Guo. 2023. "Repression of GhTUBB1 Reduces Plant Height in Gossypium hirsutum" International Journal of Molecular Sciences 24, no. 20: 15424. https://doi.org/10.3390/ijms242015424
APA StyleZhang, L., Ma, C., Wang, L., Su, X., Huang, J., Cheng, H., & Guo, H. (2023). Repression of GhTUBB1 Reduces Plant Height in Gossypium hirsutum. International Journal of Molecular Sciences, 24(20), 15424. https://doi.org/10.3390/ijms242015424






