Comparison of the Molecular Motility of Tubulin Dimeric Isoforms: Molecular Dynamics Simulations and Diffracted X-ray Tracking Study
Abstract
:1. Introduction
2. Results
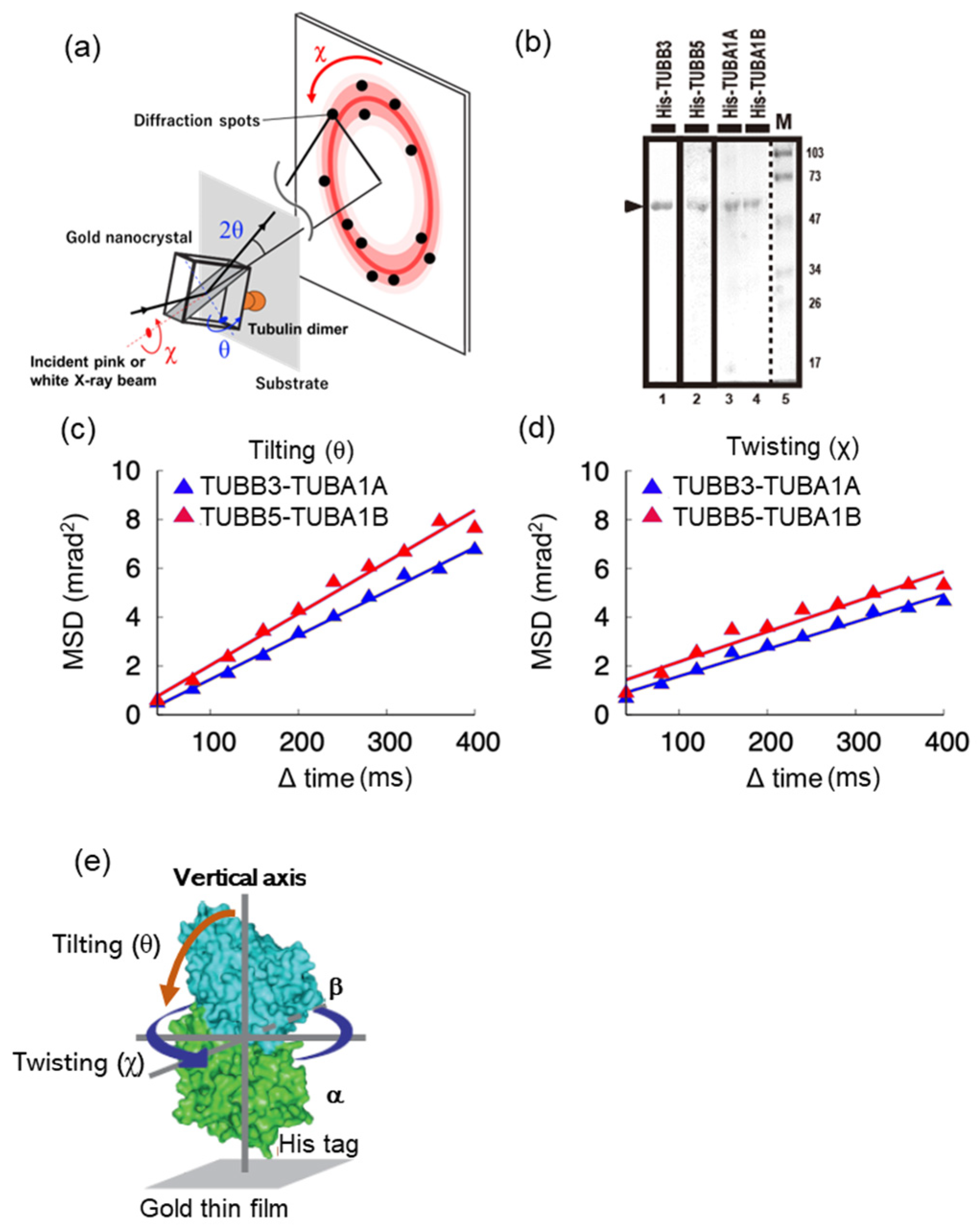
2.1. Molecular Motility around the Vertical axis of Neuronal Tubulin Dimers Is Lower Than That of Ubiquitous Tubulin Dimers
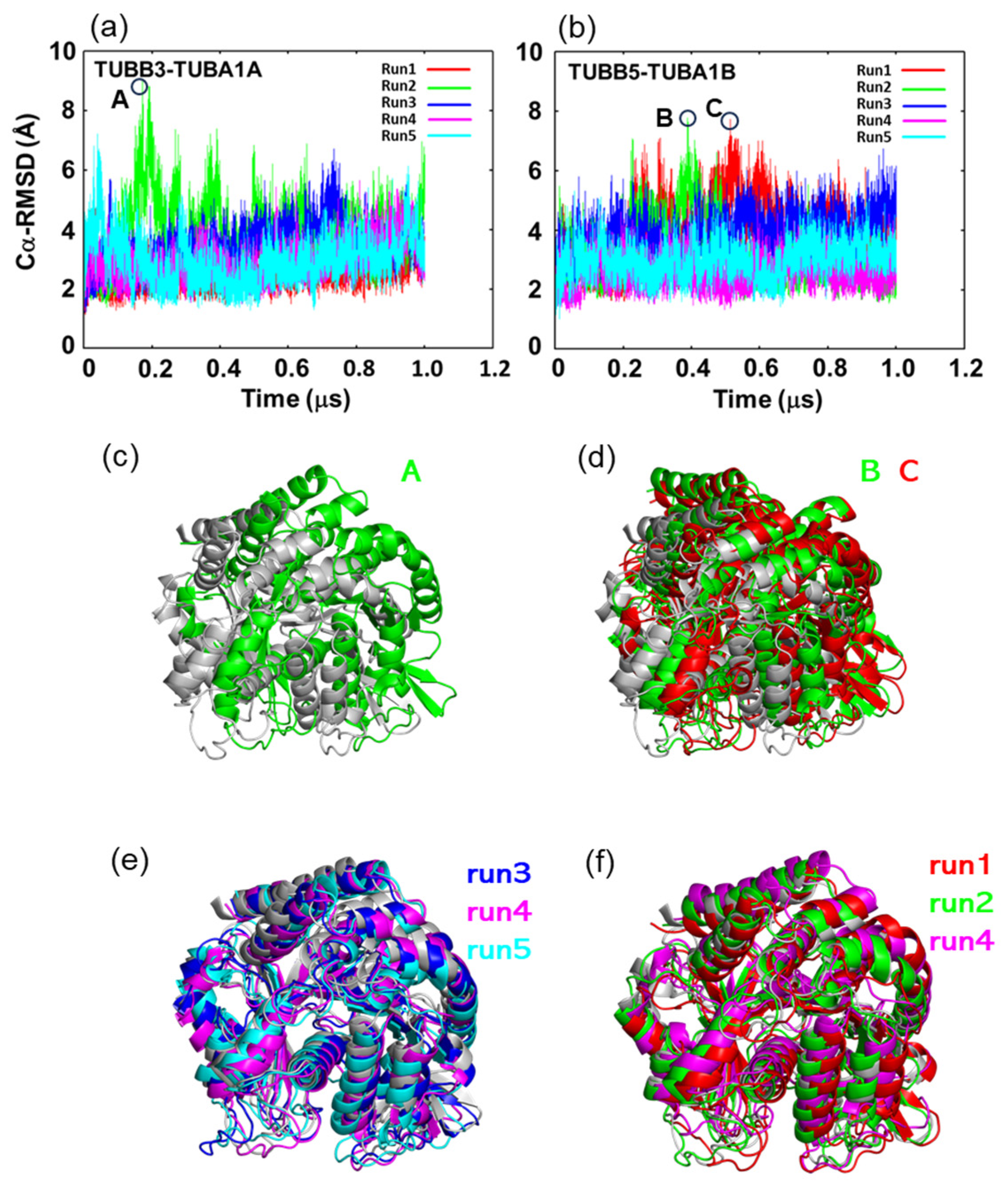
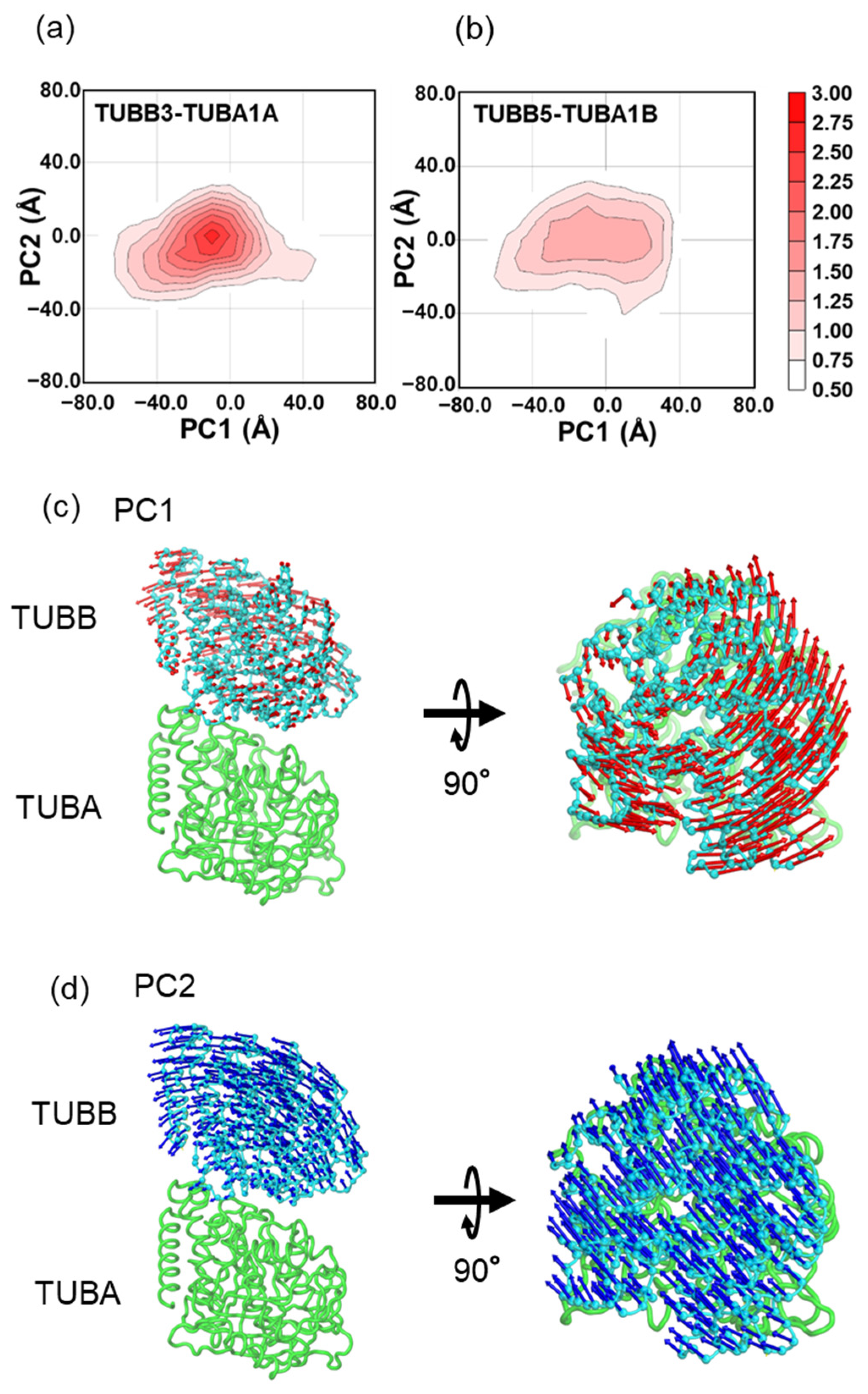
2.2. In MD Simulations, the Motility of Neurons and Ubiquitous Tubulin Dimers Was Consistent with the Results from DXT Analysis
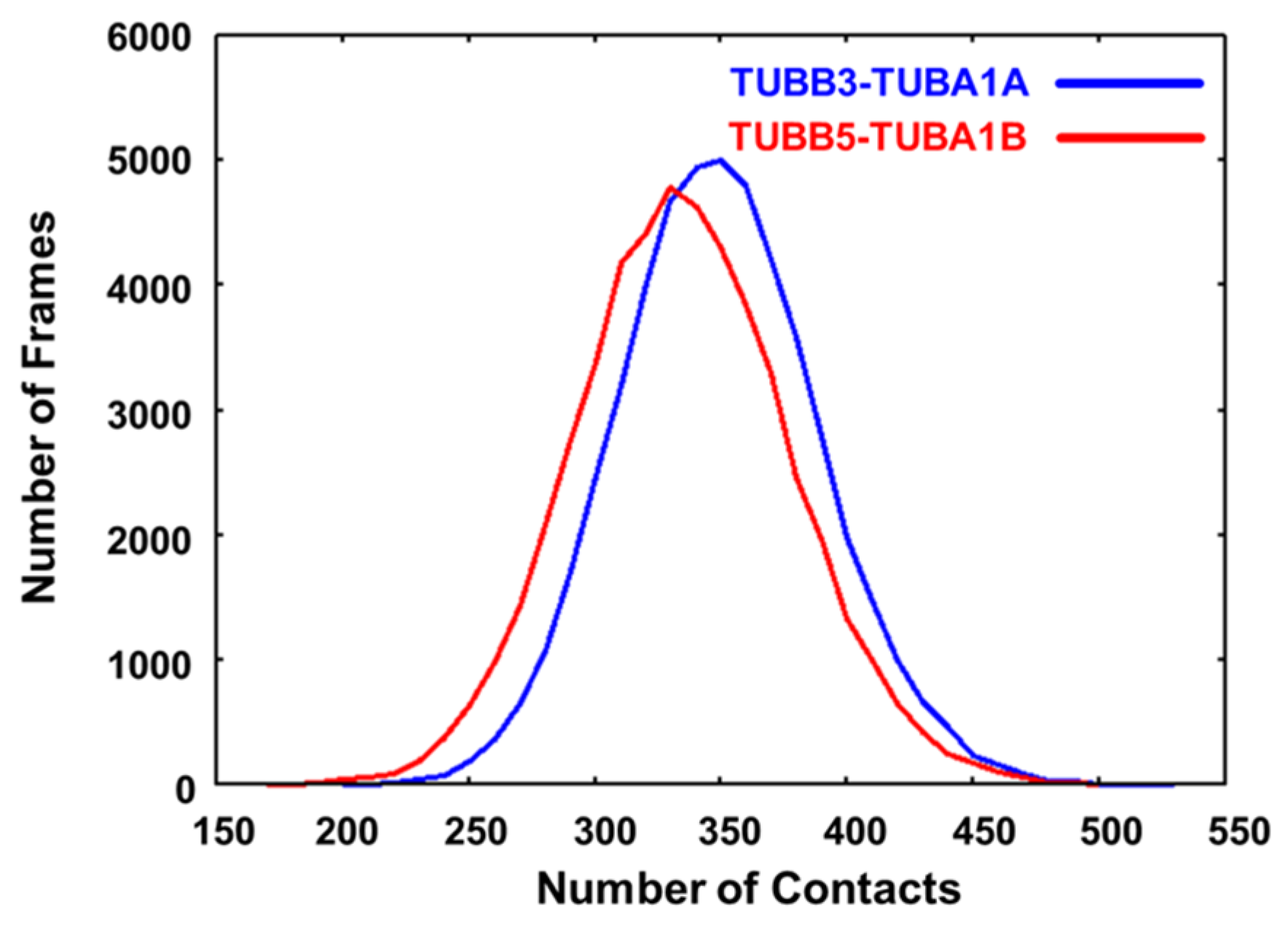
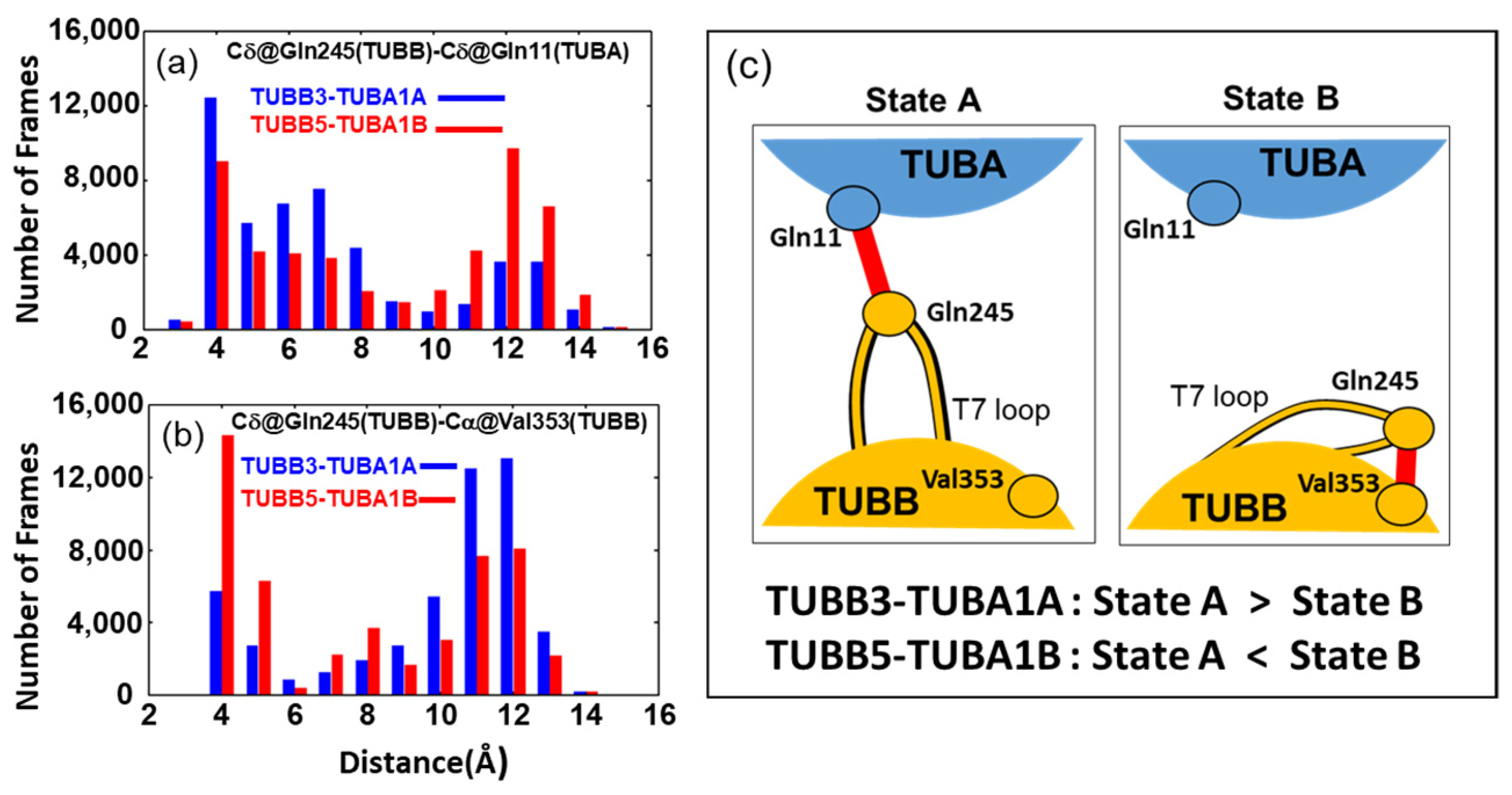
2.3. Neuronal Tubulin Dimers Have More TUBA-TUBB Interface Contacts Than Ubiquitous Ones
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Preparation of Tubulin Dimer
4.3. Sample Preparation for Diffracted X-ray Tracking
4.4. Diffracted X-ray Tracking (DXT)
4.5. Modeling of Tubulin Dimers
4.6. Molecular Dynamics Simulation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goodson, H.V.; Jonasson, E.M. Microtubules and Microtubule-Associated Proteins. Cold Spring Harb. Perspect. Biol. 2018, 10, a022608. [Google Scholar] [CrossRef] [PubMed]
- Nogales, E.; Whittaker, M.; Milligan, R.A.; Downing, K.H. High-resolution model of the microtubule. Cell 1999, 96, 79–88. [Google Scholar] [CrossRef]
- Okada, Y.; Hirokawa, N. Mechanism of the single-headed processivity: Diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc. Natl. Acad. Sci. USA 2000, 97, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Kamiguchi, H.; Akagawa, K. Syntaxin 1C, a soluble form of syntaxin, attenuates membrane recycling by destabilizing microtubules. J. Cell Sci. 2012, 125, 817–830. [Google Scholar] [CrossRef]
- Drechsler, H.; Xu, Y.; Geyer, V.F.; Zhang, Y.; Diez, S. Multivalent electrostatic microtubule interactions of synthetic peptides are sufficient to mimic advanced MAP-like behavior. Mol. Biol. Cell 2019, 30, 2953–2968. [Google Scholar] [CrossRef] [PubMed]
- Leandro-García, L.J.; Leskelä, S.; Landa, I.; Montero-Conde, C.; López-Jiménez, E.; Letón, R.; Cascón, A.; Robledo, M.; Rodríguez-Antona, C. Tumoral and tissue-specific expression of the major human β-tubulin isotypes. Cytoskeleton 2010, 67, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Roll-Mecak, A. The Tubulin Code in Microtubule Dynamics and Information Encoding. Dev. Cell 2020, 54, 7–20. [Google Scholar] [CrossRef]
- Ludueña, R.F. A hypothesis on the origin and evolution of tubulin. Int. Rev. Cell Mol. Biol. 2013, 302, 41–185. [Google Scholar] [CrossRef]
- Breuss, M.W.; Leca, I.; Gstrein, T.; Hansen, A.H.; Keays, D.A. Tubulins and brain development—The origins of functional specification. Mol. Cell. Neurosci. 2017, 84, 58–67. [Google Scholar] [CrossRef]
- Yang, L.Q.; Sang, P.; Tao, Y.; Fu, Y.X.; Zhang, K.Q.; Xie, Y.H.; Liu, S.Q. Protein dynamics and motions in relation to their functions: Several case studies and the underlying mechanisms. J. Biomol. Struct. Dyn. 2014, 32, 372–393. [Google Scholar] [CrossRef]
- Rai, K.; Kumbhar, B.V.; Panda, D.; Kunwar, A. Computational study of interactions of anti-cancer drug eribulin with human tubulin isotypes. Phys. Chem. Chem. Phys. 2022, 24, 16694–16700. [Google Scholar] [CrossRef] [PubMed]
- Zúñiga-Bustos, M.; Vásquez, P.A.; Jaña, G.A.; Guzmán, J.L.; Alderete, J.B.; Jiménez, V.A. Mechanism-Based Rational Discovery and In Vitro Evaluation of Novel Microtubule Stabilizing Agents with Non-Taxol-Competitive Activity. J. Chem. Inf. Model. 2020, 60, 3204–3213. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Srivastava, G.; Singh, A.; Prakasham, A.P.; Negi, A.S.; Sharma, A. Insight into microtubule destabilization mechanism of 3,4,5-trimethoxyphenyl indanone derivatives using molecular dynamics simulation and conformational modes analysis. J. Comput. Aided Mol. Des. 2018, 32, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Manandhar, A.; Kang, M.; Chakraborty, K.; Loverde, S.M. Effect of Nucleotide State on the Protofilament Conformation of Tubulin Octamers. J. Phys. Chem. B 2018, 122, 6164–6178. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, K.; Mohan, J.; Senapati, S. Relating nucleotide-dependent conformational changes in free tubulin dimers to tubulin assembly. Biopolymers 2013, 99, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.C.; Okumura, Y.; Adachi, S.; Suda, H.; Taniguchi, Y.; Yagi, N. Picometer-scale dynamical X-ray imaging of single DNA molecules. Phys. Rev. Lett. 2001, 87, 248102. [Google Scholar] [CrossRef]
- Sekiguchi, H.; Nakagawa, A.; Moriya, K.; Makabe, K.; Ichiyanagi, K.; Nozawa, S.; Sato, T.; Adachi, S.; Kuwajima, K.; Yohda, M.; et al. ATP dependent rotational motion of group II chaperonin observed by X-ray single molecule tracking. PLoS ONE 2013, 8, e64176. [Google Scholar] [CrossRef]
- Sekiguchi, H.; Suzuki, Y.; Nishino, Y.; Kobayashi, S.; Shimoyama, Y.; Cai, W.; Nagata, K.; Okada, M.; Ichiyanagi, K.; Ohta, N.; et al. Real time ligand-induced motion mappings of AChBP and nAChR using X-ray single molecule tracking. Sci. Rep. 2014, 4, 6384. [Google Scholar] [CrossRef]
- Kozono, H.; Matsushita, Y.; Ogawa, N.; Kozono, Y.; Miyabe, T.; Sekiguchi, H.; Ichiyanagi, K.; Okimoto, N.; Taiji, M.; Kanagawa, O.; et al. Single-molecule motions of MHC class II rely on bound peptides. Biophys. J. 2015, 108, 350–359. [Google Scholar] [CrossRef]
- Sasaki, Y.C.; Suzuki, Y.; Yagi, N.; Adachi, S.; Ishibashi, M.; Suda, H.; Toyota, K.; Yanagihara, M. Tracking of individual nanocrystals using diffracted x rays. Phys. Rev. E 2000, 62, 3843–3847. [Google Scholar] [CrossRef]
- Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 2.0; Schrodinger, LLC: New York, NY, USA, 2018. [Google Scholar]
- Ravelli, R.B.; Gigant, B.; Curmi, P.A.; Jourdain, I.; Lachkar, S.; Sobel, A.; Knossow, M. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 2004, 428, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Dorléans, A.; Gigant, B.; Ravelli, R.B.G.; Mailliet, P.; Mikol, V.; Knossow, M. Variations in the colchicine-binding domain provide insight into the structural switch of tubulin. Proc. Natl. Acad. Sci. USA 2009, 106, 13775–13779. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyaya, S.; Chakravorty, D.; Basu, G. A collective motion description of tubulin βT7 loop dynamics. Biophys. Physicobiol. 2019, 16, 264–273. [Google Scholar] [CrossRef]
- Kawashima, Y.; Sasaki, Y.C.; Sugita, Y.; Yoda, T.; Okamoto, Y. Replica-exchange molecular dynamics simulation of diffracted X-ray tracking. Mol. Simul. 2007, 33, 97–102. [Google Scholar] [CrossRef]
- Shao, Q.; Hall, C.K. Binding Preferences of Amino Acids for Gold Nanoparticles: A Molecular Simulation Study. Langmuir 2016, 32, 7888–7896. [Google Scholar] [CrossRef]
- Buglak, A.A.; Kononov, A.I. Comparative study of gold and silver interactions with amino acids and nucleobases. RSC Adv. 2020, 10, 34149–34160. [Google Scholar] [CrossRef] [PubMed]
- Pamula, M.C.; Ti, S.-C.; Kapoor, T.M. The structured core of human β tubulin confers isotype-specific polymerization properties. J. Cell Biol. 2016, 213, 425–433. [Google Scholar] [CrossRef]
- Narvi, E.; Jaakkola, K.; Winsel, S.; Oetken-Lindholm, C.; Halonen, P.; Kallio, L.; Kallio, M.J. Altered TUBB3 expression contributes to the epothilone response of mitotic cells. Br. J. Cancer 2013, 108, 82–90. [Google Scholar] [CrossRef]
- Shelanski, M.L.; Gaskin, F.; Cantor, C.R. Microtubule assembly in the absence of added nucleotides. Proc. Natl. Acad. Sci. USA 1973, 70, 765–768. [Google Scholar] [CrossRef]
- Herzog, W.; Weber, K. In vitro assembly of pure tubulin into microtubules in the absence of microtubule-associated proteins and glycerol. Proc. Natl. Acad. Sci. USA 1977, 74, 1860–1864. [Google Scholar] [CrossRef]
- Löwe, J.; Li, H.; Downing, K.H.; Nogales, E. Refined structure of αβ-tubulin at 3.5 Å resolution11Edited by I. A. Wilson. J. Mol. Biol. 2001, 313, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Sali, A. Comparative protein modeling by satisfaction of spatial restraints. Mol. Med. Today 1995, 1, 270–277. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Lindahl, E.; Hess, B.; van der Spoel, D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. Mol. Model. Annu. 2001, 7, 306–317. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Crystal Structure and Pair Potentials: A Molecular-Dynamics Study. Phys. Rev. Lett. 1980, 45, 1196–1199. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Søndergaard, C.R.; Olsson, M.H.M.; Rostkowski, M.; Jensen, J.H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamane, T.; Nakayama, T.; Ekimoto, T.; Inoue, M.; Ikezaki, K.; Sekiguchi, H.; Kuramochi, M.; Terao, Y.; Judai, K.; Saito, M.; et al. Comparison of the Molecular Motility of Tubulin Dimeric Isoforms: Molecular Dynamics Simulations and Diffracted X-ray Tracking Study. Int. J. Mol. Sci. 2023, 24, 15423. https://doi.org/10.3390/ijms242015423
Yamane T, Nakayama T, Ekimoto T, Inoue M, Ikezaki K, Sekiguchi H, Kuramochi M, Terao Y, Judai K, Saito M, et al. Comparison of the Molecular Motility of Tubulin Dimeric Isoforms: Molecular Dynamics Simulations and Diffracted X-ray Tracking Study. International Journal of Molecular Sciences. 2023; 24(20):15423. https://doi.org/10.3390/ijms242015423
Chicago/Turabian StyleYamane, Tsutomu, Takahiro Nakayama, Toru Ekimoto, Masao Inoue, Keigo Ikezaki, Hiroshi Sekiguchi, Masahiro Kuramochi, Yasuo Terao, Ken Judai, Minoru Saito, and et al. 2023. "Comparison of the Molecular Motility of Tubulin Dimeric Isoforms: Molecular Dynamics Simulations and Diffracted X-ray Tracking Study" International Journal of Molecular Sciences 24, no. 20: 15423. https://doi.org/10.3390/ijms242015423
APA StyleYamane, T., Nakayama, T., Ekimoto, T., Inoue, M., Ikezaki, K., Sekiguchi, H., Kuramochi, M., Terao, Y., Judai, K., Saito, M., Ikeguchi, M., & Sasaki, Y. C. (2023). Comparison of the Molecular Motility of Tubulin Dimeric Isoforms: Molecular Dynamics Simulations and Diffracted X-ray Tracking Study. International Journal of Molecular Sciences, 24(20), 15423. https://doi.org/10.3390/ijms242015423







