Genome-Wide Analysis of Glycerol-3-Phosphate Acyltransferase (GPAT) Family in Perilla frutescens and Functional Characterization of PfGPAT9 Crucial for Biosynthesis of Storage Oils Rich in High-Value Lipids
Abstract
:1. Introduction
2. Results
2.1. Twenty-Four GPAT Family Members Were Identified in the P. frutescens Genome
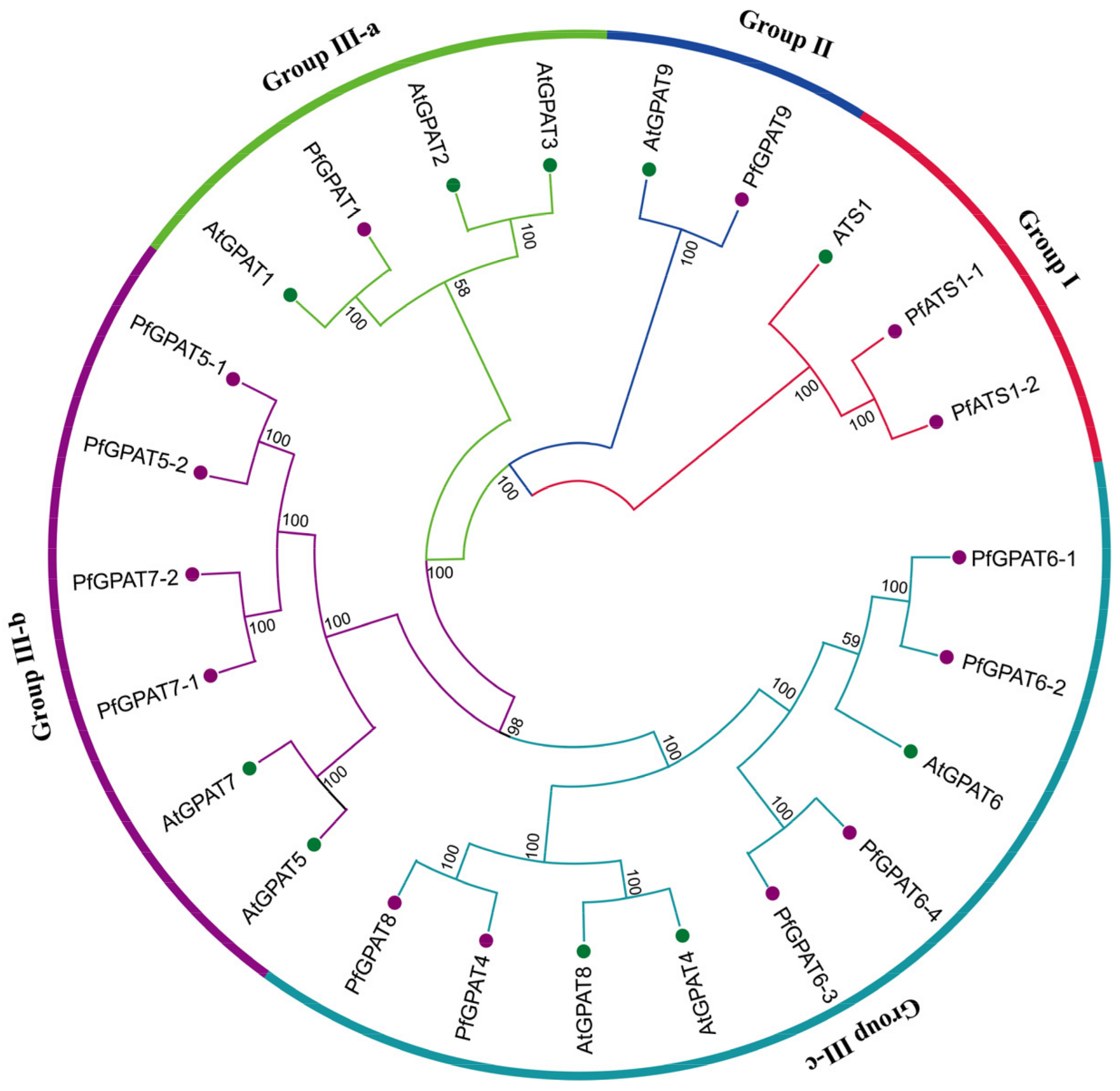
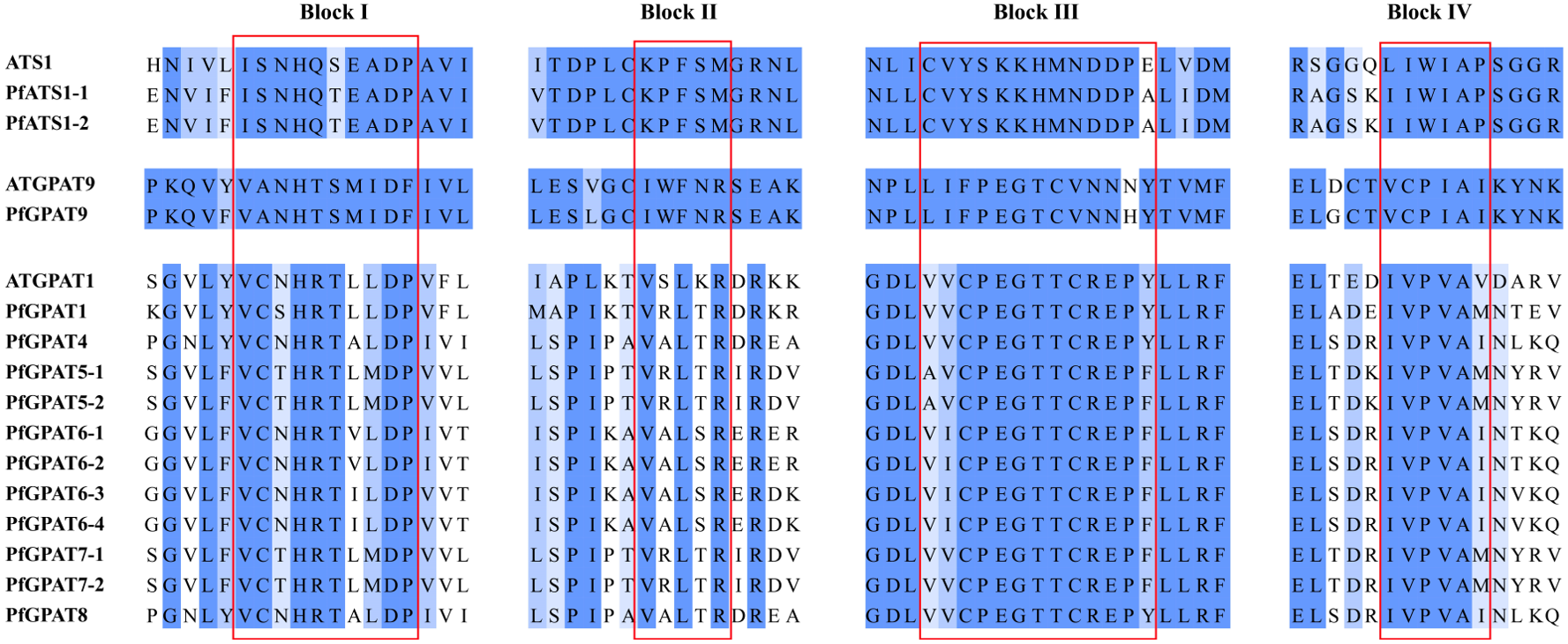
2.2. Phylogenetic Analysis and Multiple Sequence Alignment of PfGPAT Proteins
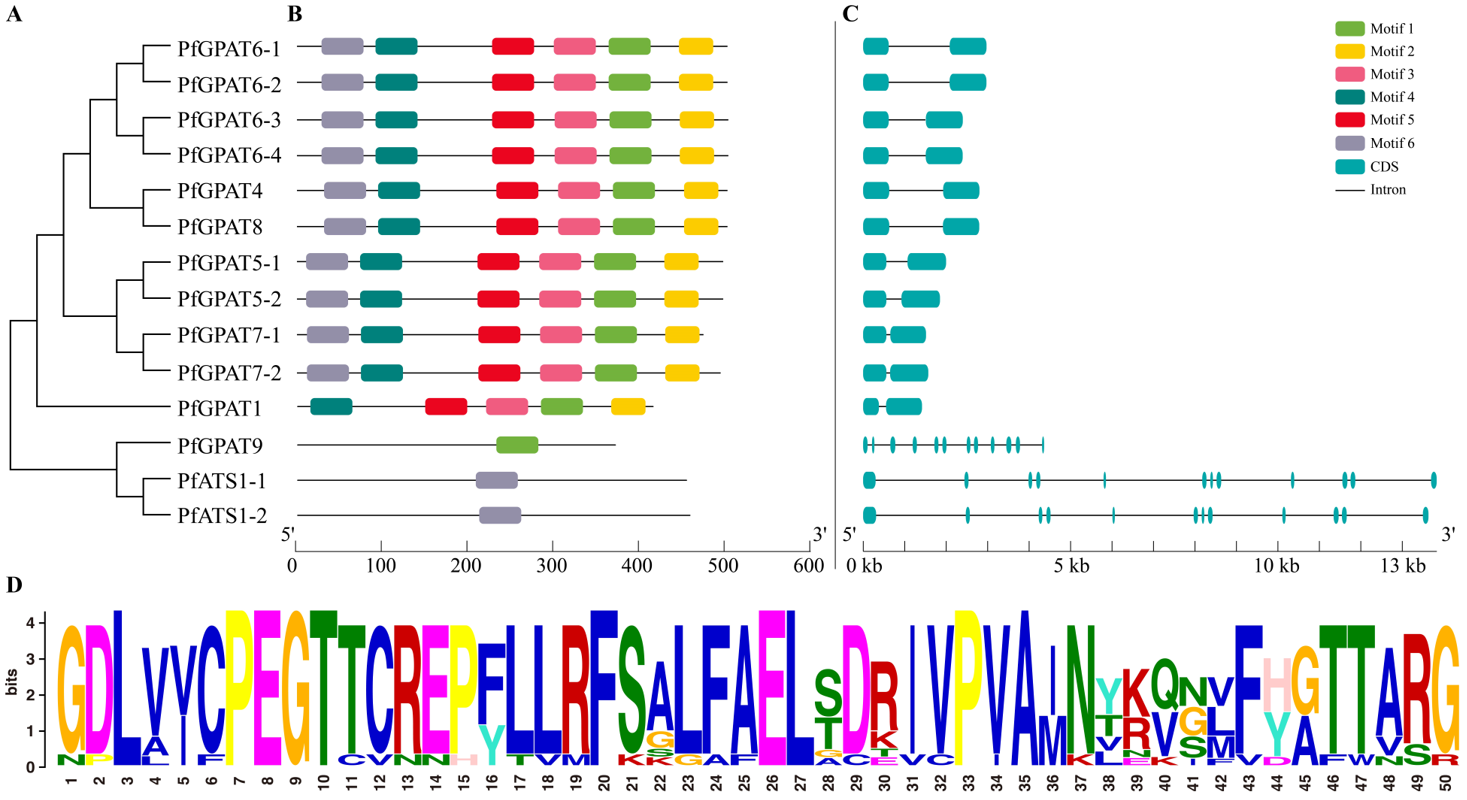
2.3. Gene Structure and Conserved Motif Analysis Further Confirmed the Subgroup Classification
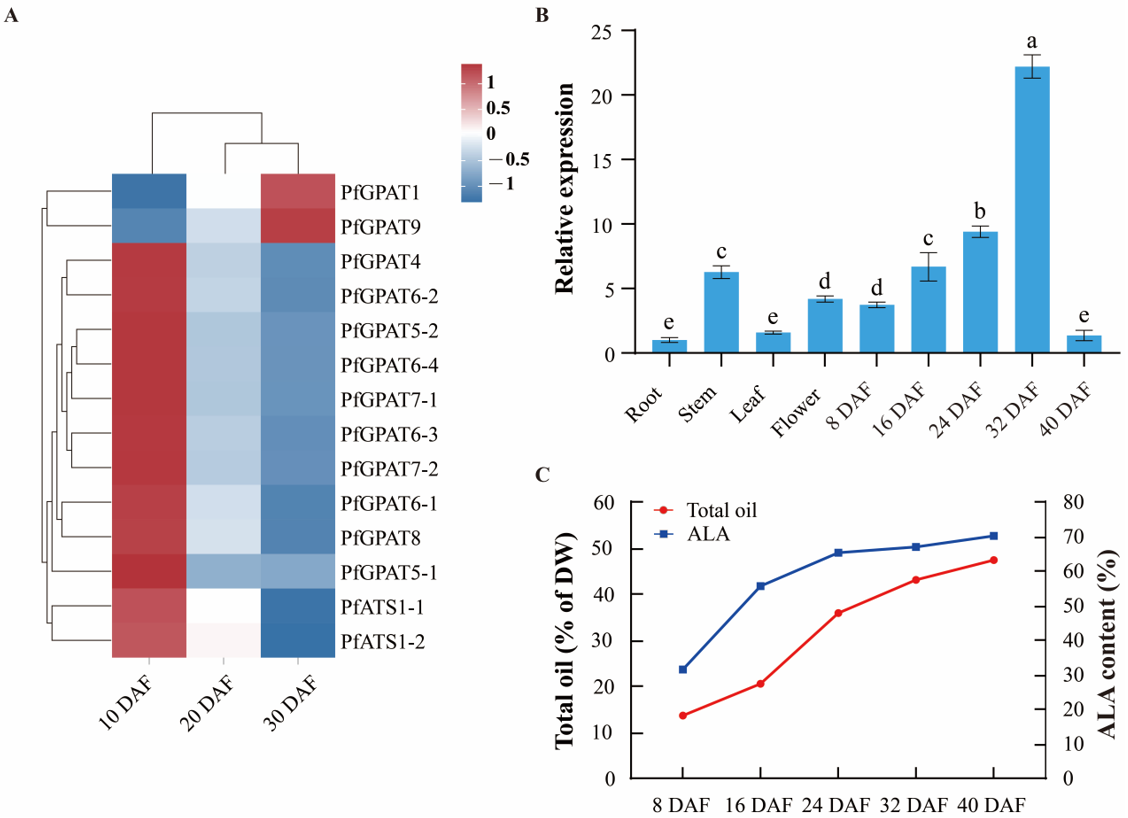
2.4. Expression Pattern of PfGPAT9 Gene Is Consistent with Oil Accumulation during Developing Seeds of P. frutescens
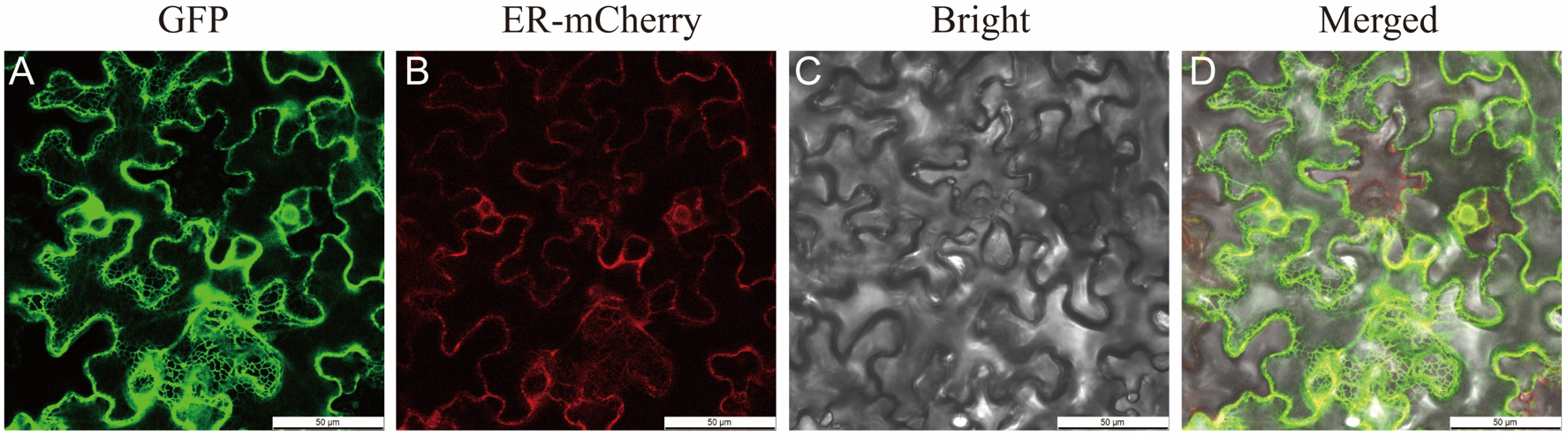
2.5. Isolation and Subcellular Localization of PfGPAT9 from P. frutescens
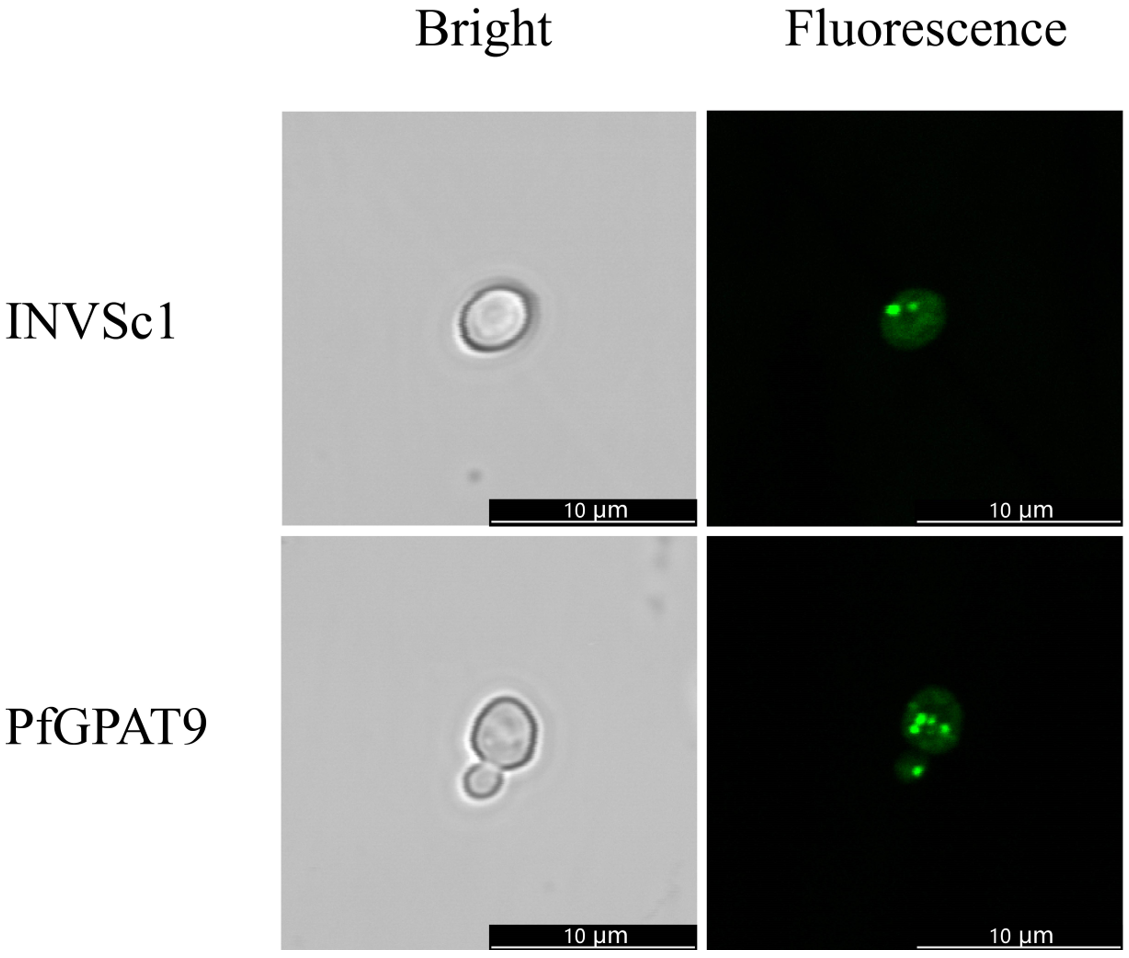
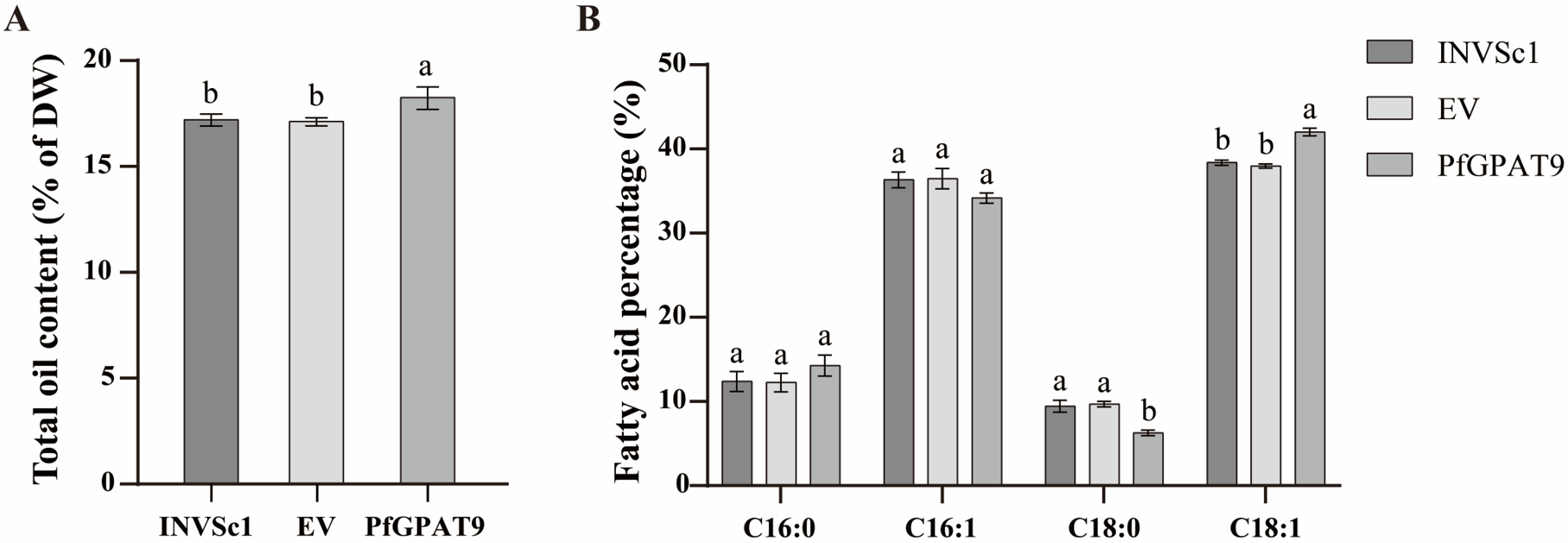
2.6. Heterogenous Expression of PfGPAT9 Promotes Oil Accumulation in Yeast
2.7. PfGPAT9 Has Higher Substrate Preference towards Oleic Acid Than ALA
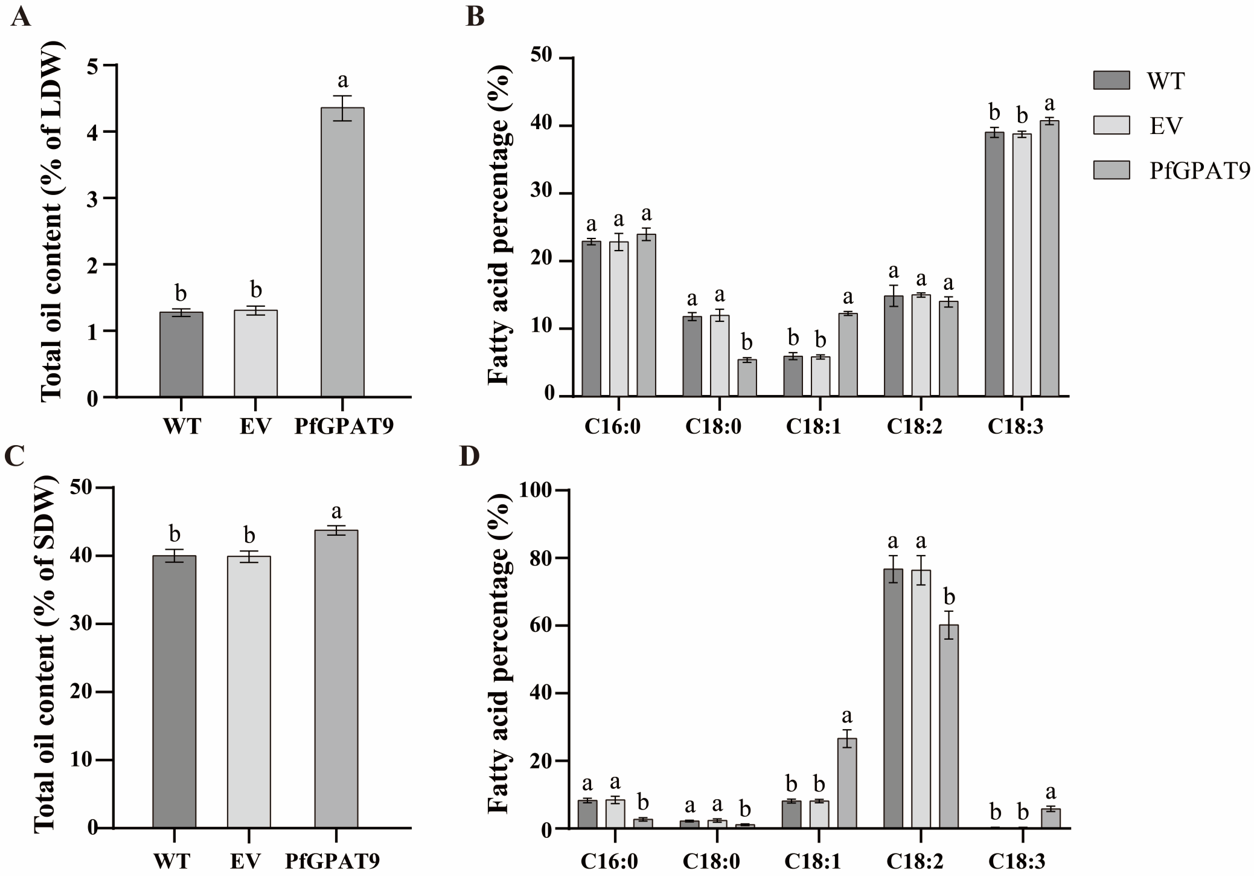
2.8. Heterogeneous Expression of PfGPAT9 Significantly Increases Contents of Total Oil and the Target FAs in Seeds and Non-Seed Tissues of Tobacco Plants
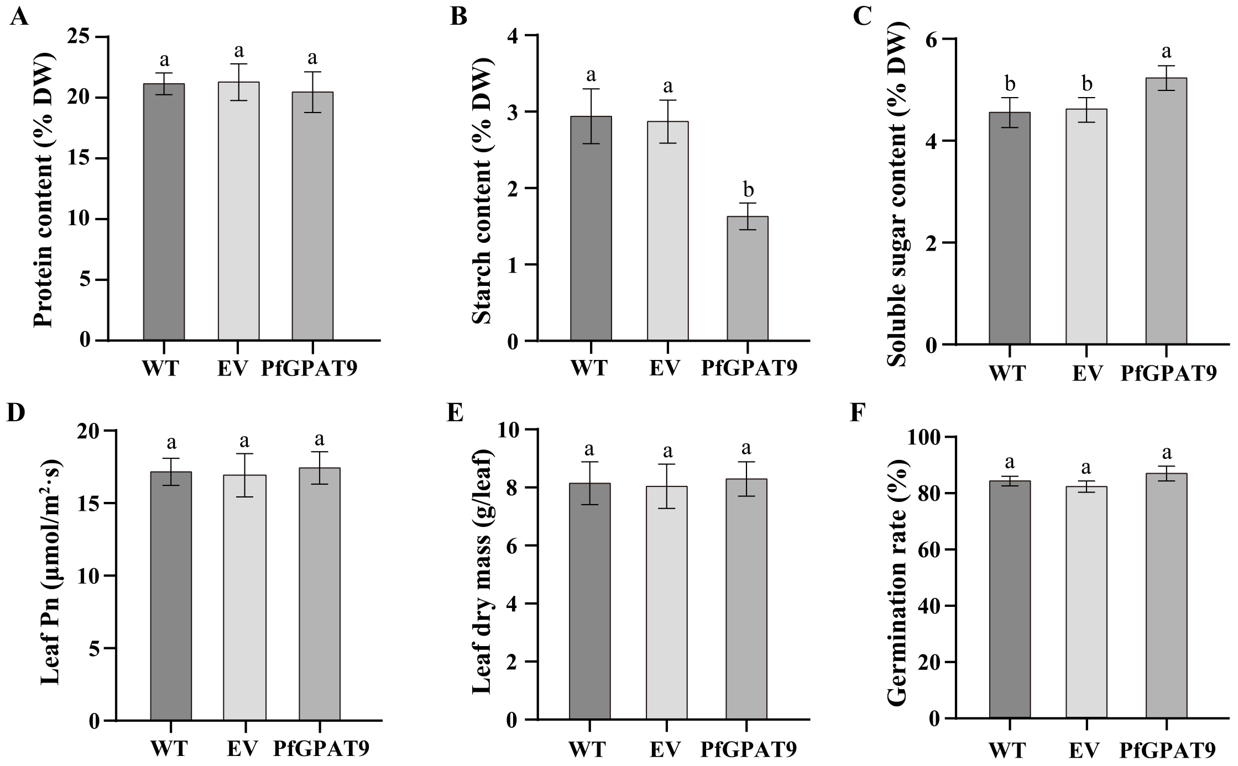
2.9. Overexpression of PfGPAT9 Leads to No Significant Negative Effects on Other Agronomic Traits of Tobacco Plants
3. Discussion
3.1. Fourteen PfGPATs Classified to Three Distinct Groups May Function Diversely
3.2. ER-Localized PfGPAT9 Functions Crucially in TAG Assembly, with Substrate Preference to UFAs (OA > ALA)
3.3. PfGPAT9 Is a Desirable Target in Metabolic Engineering to Increase Production of Storage Oils Enriched with Valuable UFAs in Plant Seed and Non-Seed Tissues
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification and Sequence Characterization of GPAT Genes from P. frutescens
4.2.1. Identification of GPAT Genes in the P. frutescens Genome and Chromosomal Localization
4.2.2. Multiple Sequence Alignment and Phylogenetic Analysis
4.2.3. Gene Structure and Conserved Motif Analysis
4.3. Expression Analysis of PfGPAT Genes in Different Tissues and Correlation with Oil Synthesis
4.3.1. Expression Profiles Analysis of PfGPAT Genes Based on RNA-Sequencing Data
4.3.2. Total RNA Extraction and RT-qPCR Analysis
4.3.3. Total Oil Extraction and Fatty Acid Analysis
4.4. PfGPAT9 Gene Cloning and Vectors Construction
4.5. Subcellular Localization Analysis of PfGPAT9 Protein
4.6. Overexpression of PfGPAT9 in the S. cerevisiae INVSc1
4.7. Heterologous Overexpression of PfGPAT9 in Tobacco Plant
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chapman, K.D.; Ohlrogge, J.B. Compartmentation of Triacylglycerol Accumulation in Plants. J. Biol. Chem. 2012, 287, 2288–2294. [Google Scholar] [CrossRef]
- Covas, M.-I.; de la Torre, R.; Fitó, M. Virgin Olive Oil: A Key Food for Cardiovascular Risk Protection. Br. J. Nutr. 2015, 113 (Suppl. S2), S19–S28. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Yang, Y.; Guo, L.; Yuan, L.; Ma, W. Molecular Basis of Plant Oil Biosynthesis: Insights Gained from Studying the WRINKLED1 Transcription Factor. Front. Plant Sci. 2020, 11, 24. [Google Scholar] [CrossRef]
- Song, J.-M.; Zhang, Y.; Zhou, Z.-W.; Lu, S.; Ma, W.; Lu, C.; Chen, L.-L.; Guo, L. Oil Plant Genomes: Current State of the Science. J. Exp. Bot. 2022, 73, 2859–2874. [Google Scholar] [CrossRef]
- Slocombe, S.P.; Cornah, J.; Pinfield-Wells, H.; Soady, K.; Zhang, Q.; Gilday, A.; Dyer, J.M.; Graham, I.A. Oil Accumulation in Leaves Directed by Modification of Fatty Acid Breakdown and Lipid Synthesis Pathways. Plant Biotechnol. J. 2009, 7, 694–703. [Google Scholar] [CrossRef]
- Gao, Y.; Sun, Y.; Gao, H.; Chen, Y.; Wang, X.; Xue, J.; Jia, X.; Li, R. Correction to: Ectopic Overexpression of a Type-II DGAT (CeDGAT2-2) Derived from Oil-Rich Tuber of Cyperus Esculentus Enhances Accumulation of Oil and Oleic Acid in Tobacco Leaves. Biotechnol. Biofuels 2021, 14, 139. [Google Scholar] [CrossRef] [PubMed]
- Kaup, M.T.; Froese, C.D.; Thompson, J.E. A Role for Diacylglycerol Acyltransferase during Leaf Senescence. Plant Physiol. 2002, 129, 1616–1626. [Google Scholar] [CrossRef] [PubMed]
- Theodoulou, F.L.; Eastmond, P.J. Seed Storage Oil Catabolism: A Story of Give and Take. Curr. Opin. Plant Biol. 2012, 15, 322–328. [Google Scholar] [CrossRef]
- Fan, J.; Yan, C.; Xu, C. Phospholipid:Diacylglycerol Acyltransferase-Mediated Triacylglycerol Biosynthesis Is Crucial for Protection against Fatty Acid-Induced Cell Death in Growing Tissues of Arabidopsis. Plant J. Cell Mol. Biol. 2013, 76, 930–942. [Google Scholar] [CrossRef]
- Yang, Y.; Kong, Q.; Lim, A.R.Q.; Lu, S.; Zhao, H.; Guo, L.; Yuan, L.; Ma, W. Transcriptional Regulation of Oil Biosynthesis in Seed Plants: Current Understanding, Applications, and Perspectives. Plant Commun. 2022, 3, 100328. [Google Scholar] [CrossRef]
- Pfleger, B.F.; Gossing, M.; Nielsen, J. Metabolic Engineering Strategies for Microbial Synthesis of Oleochemicals. Metab. Eng. 2015, 29, 1–11. [Google Scholar] [CrossRef]
- Ramos, M.J.; Fernández, C.M.; Casas, A.; Rodríguez, L.; Pérez, A. Influence of Fatty Acid Composition of Raw Materials on Biodiesel Properties. Bioresour. Technol. 2009, 100, 261–268. [Google Scholar] [CrossRef]
- Abdukeyum, G.G.; Owen, A.J.; McLennan, P.L. Dietary (n-3) Long-Chain Polyunsaturated Fatty Acids Inhibit Ischemia and Reperfusion Arrhythmias and Infarction in Rat Heart Not Enhanced by Ischemic Preconditioning. J. Nutr. 2008, 138, 1902–1909. [Google Scholar] [CrossRef]
- Prado, E.L.; Dewey, K.G. Nutrition and Brain Development in Early Life. Nutr. Rev. 2014, 72, 267–284. [Google Scholar] [CrossRef]
- Xin, Y.; Shen, C.; She, Y.; Chen, H.; Wang, C.; Wei, L.; Yoon, K.; Han, D.; Hu, Q.; Xu, J. Biosynthesis of Triacylglycerol Molecules with a Tailored PUFA Profile in Industrial Microalgae. Mol. Plant 2019, 12, 474–488. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Li, R. Metabolic Engineering a Model Oilseed Camelina Sativa for the Sustainable Production of High-Value Designed Oils. Front. Plant Sci. 2020, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Lan, C.; Wang, S.; Zhang, H.; Wang, Z.; Wan, W.; Liu, H.; Hu, Y.; Cui, Q.; Song, X. Cocktail Biosynthesis of Triacylglycerol by Rational Modulation of Diacylglycerol Acyltransferases in Industrial Oleaginous Aurantiochytrium. Biotechnol. Biofuels 2021, 14, 246. [Google Scholar] [CrossRef]
- Bates, P.D.; Stymne, S.; Ohlrogge, J. Biochemical Pathways in Seed Oil Synthesis. Curr. Opin. Plant Biol. 2013, 16, 358–364. [Google Scholar] [CrossRef]
- Xu, R.; Yang, T.; Wang, R.; Liu, A. Characterisation of DGAT1 and DGAT2 from Jatropha Curcas and Their Functions in Storage Lipid Biosynthesis. Funct. Plant Biol. FPB 2014, 41, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-Lipid Metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef]
- Allen, D.K. Assessing Compartmentalized Flux in Lipid Metabolism with Isotopes. Biochim. Biophys. Acta 2016, 1861, 1226–1242. [Google Scholar] [CrossRef]
- Bates, P.D. Understanding the Control of Acyl Flux through the Lipid Metabolic Network of Plant Oil Biosynthesis. Biochim. Biophys. Acta 2016, 1861, 1214–1225. [Google Scholar] [CrossRef]
- Zhang, M.; Fan, J.; Taylor, D.C.; Ohlrogge, J.B. DGAT1 and PDAT1 Acyltransferases Have Overlapping Functions in Arabidopsis Triacylglycerol Biosynthesis and Are Essential for Normal Pollen and Seed Development. Plant Cell 2009, 21, 3885–3901. [Google Scholar] [CrossRef]
- Murphy, D.J. Storage Lipid Bodies in Plants and Other Organisms. Prog. Lipid Res. 1990, 29, 299–324. [Google Scholar]
- Lin, L.-J.; Tai, S.S.K.; Peng, C.-C.; Tzen, J.T.C. Steroleosin, a Sterol-Binding Dehydrogenase in Seed Oil Bodies. Plant Physiol. 2002, 128, 1200–1211. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.D.; Dyer, J.M.; Mullen, R.T. Biogenesis and Functions of Lipid Droplets in Plants: Thematic Review Series: Lipid Droplet Synthesis and Metabolism: From Yeast to Man. J. Lipid Res. 2012, 53, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Xia, Q.; Dauk, M.; Shen, W.; Selvaraj, G.; Zou, J. Arabidopsis AtGPAT1, a Member of the Membrane-Bound Glycerol-3-Phosphate Acyltransferase Gene Family, Is Essential for Tapetum Differentiation and Male Fertility. Plant Cell 2003, 15, 1872–1887. [Google Scholar] [CrossRef]
- Gidda, S.K.; Shockey, J.M.; Rothstein, S.J.; Dyer, J.M.; Mullen, R.T. Arabidopsis Thaliana GPAT8 and GPAT9 Are Localized to the ER and Possess Distinct ER Retrieval Signals: Functional Divergence of the Dilysine ER Retrieval Motif in Plant Cells. Plant Physiol. Biochem. 2009, 47, 867–879. [Google Scholar] [CrossRef]
- Yang, W.; Simpson, J.P.; Li-Beisson, Y.; Beisson, F.; Pollard, M.; Ohlrogge, J.B. A Land-Plant-Specific Glycerol-3-Phosphate Acyltransferase Family in Arabidopsis: Substrate Specificity, Sn-2 Preference, and Evolution. Plant Physiol. 2012, 160, 638–652. [Google Scholar] [CrossRef]
- Sui, N.; Li, M.; Zhao, S.-J.; Li, F.; Liang, H.; Meng, Q.-W. Overexpression of Glycerol-3-Phosphate Acyltransferase Gene Improves Chilling Tolerance in Tomato. Planta 2007, 226, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Tamada, T.; Feese, M.D.; Ferri, S.R.; Kato, Y.; Yajima, R.; Toguri, T.; Kuroki, R. Substrate Recognition and Selectivity of Plant Glycerol-3-Phosphate Acyltransferases (GPATs) from Cucurbita Moscata and Spinacea Oleracea. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 13–21. [Google Scholar] [CrossRef]
- Payá-Milans, M.; Aznar-Moreno, J.A.; Balbuena, T.S.; Haslam, R.P.; Gidda, S.K.; Pérez-Hormaeche, J.; Mullen, R.T.; Thelen, J.J.; Napier, J.A.; Salas, J.J.; et al. Sunflower HaGPAT9-1 Is the Predominant GPAT during Seed Development. Plant Sci. Int. J. Exp. Plant Biol. 2016, 252, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Waschburger, E.; Kulcheski, F.R.; Veto, N.M.; Margis, R.; Margis-Pinheiro, M.; Turchetto-Zolet, A.C. Genome-Wide Analysis of the Glycerol-3-Phosphate Acyltransferase (GPAT) Gene Family Reveals the Evolution and Diversification of Plant GPATs. Genet. Mol. Biol. 2018, 41, 355–370. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Ma, J.; Liu, G.; Wang, N.; Pei, W.; Wu, M.; Li, X.; Zhang, J.; Yu, J. Genome-Wide Identification, Sequence Variation, and Expression of the Glycerol-3-Phosphate Acyltransferase (GPAT) Gene Family in Gossypium. Front. Genet. 2019, 10, 116. [Google Scholar] [CrossRef]
- Chen, X.; Snyder, C.L.; Truksa, M.; Shah, S.; Weselake, R.J. Sn-Glycerol-3-Phosphate Acyltransferases in Plants. Plant Signal. Behav. 2011, 6, 1695–1699. [Google Scholar] [CrossRef] [PubMed]
- Nishida, I.; Tasaka, Y.; Shiraishi, H.; Murata, N. The Gene and the RNA for the Precursor to the Plastid-Located Glycerol-3-Phosphate Acyltransferase of Arabidopsis Thaliana. Plant Mol. Biol. 1993, 21, 267–277. [Google Scholar] [CrossRef]
- Beisson, F.; Li, Y.; Bonaventure, G.; Pollard, M.; Ohlrogge, J.B. The Acyltransferase GPAT5 Is Required for the Synthesis of Suberin in Seed Coat and Root of Arabidopsis. Plant Cell 2007, 19, 351–368. [Google Scholar] [CrossRef]
- Zhu, T.; Wu, S.; Zhang, D.; Li, Z.; Xie, K.; An, X.; Ma, B.; Hou, Q.; Dong, Z.; Tian, Y.; et al. Genome-Wide Analysis of Maize GPAT Gene Family and Cytological Characterization and Breeding Application of ZmMs33/ZmGPAT6 Gene. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2019, 132, 2137–2154. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Yu, B.; Cornish, A.J.; Froehlich, J.E.; Benning, C. Phosphatidylglycerol Biosynthesis in Chloroplasts of Arabidopsis Mutants Deficient in Acyl-ACP Glycerol-3- Phosphate Acyltransferase. Plant J. Cell Mol. Biol. 2006, 47, 296–309. [Google Scholar] [CrossRef]
- Jayawardhane, K.N.; Singer, S.D.; Weselake, R.J.; Chen, G. Plant Sn-Glycerol-3-Phosphate Acyltransferases: Biocatalysts Involved in the Biosynthesis of Intracellular and Extracellular Lipids. Lipids 2018, 53, 469–480. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Pollard, M.; Sauveplane, V.; Pinot, F.; Ohlrogge, J.; Beisson, F. Nanoridges That Characterize the Surface Morphology of Flowers Require the Synthesis of Cutin Polyester. Proc. Natl. Acad. Sci. USA 2009, 106, 22008–22013. [Google Scholar] [CrossRef]
- Shockey, J.; Regmi, A.; Cotton, K.; Adhikari, N.; Browse, J.; Bates, P.D. Identification of Arabidopsis GPAT9 (At5g60620) as an Essential Gene Involved in Triacylglycerol Biosynthesis. Plant Physiol. 2016, 170, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Singer, S.D.; Chen, G.; Mietkiewska, E.; Tomasi, P.; Jayawardhane, K.; Dyer, J.M.; Weselake, R.J. Arabidopsis GPAT9 Contributes to Synthesis of Intracellular Glycerolipids but Not Surface Lipids. J. Exp. Bot. 2016, 67, 4627–4638. [Google Scholar] [CrossRef]
- Yang, S.U.; Kim, J.; Kim, H.; Suh, M.C. Functional Characterization of Physcomitrella Patens Glycerol-3-Phosphate Acyltransferase 9 and an Increase in Seed Oil Content in Arabidopsis by Its Ectopic Expression. Plants 2019, 8, 284. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Zhang, X.; Luo, L.; Yang, H.; Li, P.; Zhang, K.; Liu, F.; Wan, Y. Characterization of Glycerol-3-Phosphate Acyltransferase 9 (AhGPAT9) Genes, Their Allelic Polymorphism and Association with Oil Content in Peanut (Arachis Hypogaea L.). Sci. Rep. 2020, 10, 14648. [Google Scholar] [CrossRef]
- Fukuda, S.; Hirasawa, E.; Takemura, T.; Takahashi, S.; Chokshi, K.; Pancha, I.; Tanaka, K.; Imamura, S. Accelerated Triacylglycerol Production without Growth Inhibition by Overexpression of a Glycerol-3-Phosphate Acyltransferase in the Unicellular Red Alga Cyanidioschyzon Merolae. Sci. Rep. 2018, 8, 12410. [Google Scholar] [CrossRef] [PubMed]
- Iskandarov, U.; Sitnik, S.; Shtaida, N.; Didi-Cohen, S.; Leu, S.; Khozin-Goldberg, I.; Cohen, Z.; Boussiba, S. Cloning and Characterization of a GPAT-like Gene from the Microalga Lobosphaera Incisa (Trebouxiophyceae): Overexpression in Chlamydomonas Reinhardtii Enhances TAG Production. J. Appl. Phycol. 2016, 28, 907–919. [Google Scholar] [CrossRef]
- Tsakraklides, V.; Kamineni, A.; Consiglio, A.L.; MacEwen, K.; Friedlander, J.; Blitzblau, H.G.; Hamilton, M.A.; Crabtree, D.V.; Su, A.; Afshar, J.; et al. High-Oleate Yeast Oil without Polyunsaturated Fatty Acids. Biotechnol. Biofuels 2018, 11, 131. [Google Scholar] [CrossRef]
- Nitta, M.; Lee, J.K.; Kang, C.W.; Katsuta, M.; Yasumoto, S.; Liu, D.; Nagamine, T.; Ohnishi, O. The Distribution of Perilla Species. Genet. Resour. Crop Evol. 2005, 52, 797–804. [Google Scholar] [CrossRef]
- Kim, H.U.; Lee, K.-R.; Shim, D.; Lee, J.H.; Chen, G.Q.; Hwang, S. Transcriptome Analysis and Identification of Genes Associated with ω-3 Fatty Acid Biosynthesis in Perilla Frutescens (L.) Var. Frutescens. BMC Genom. 2016, 17, 474. [Google Scholar] [CrossRef]
- Zhang, Y.; Shen, Q.; Leng, L.; Zhang, D.; Chen, S.; Shi, Y.; Ning, Z.; Chen, S. Incipient Diploidization of the Medicinal Plant Perilla within 10,000 Years. Nat. Commun. 2021, 12, 5508. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, K.H.; Lee, M.-H.; Kim, H.-T.; Seo, W.D.; Kim, J.Y.; Baek, I.-Y.; Jang, D.S.; Ha, T.J. Identification, Characterisation, and Quantification of Phenolic Compounds in the Antioxidant Activity-Containing Fraction from the Seeds of Korean Perilla (Perilla Frutescens) Cultivars. Food Chem. 2013, 136, 843–852. [Google Scholar] [CrossRef]
- Igarashi, M.; Miyazaki, Y. A Review on Bioactivities of Perilla: Progress in Research on the Functions of Perilla as Medicine and Food. Evid. Based Complement. Alternat. Med. 2013, 2013, 925342. [Google Scholar] [CrossRef]
- Mungmai, L.; Preedalikit, W.; Aunsri, N.; Amornlerdpison, D. Efficacy of Cosmetic Formulation Containing Perilla Frutescens Leaves Extract for Irritation and Aging Skin. Biomed. Pharmacol. J. 2020, 13, 779–787. [Google Scholar] [CrossRef]
- Ciftci, O.N.; Przybylski, R.; Rudzińska, M. Lipid Components of Flax, Perilla, and Chia Seeds. Eur. J. Lipid Sci. Technol. 2012, 114, 794–800. [Google Scholar] [CrossRef]
- Wijendran, V.; Hayes, K.C. Dietary N-6 and n-3 Fatty Acid Balance and Cardiovascular Health. Annu. Rev. Nutr. 2004, 24, 597–615. [Google Scholar] [CrossRef]
- Müller-Waldeck, F.; Sitzmann, J.; Schnitzler, W.H.; Grassmann, J. Determination of Toxic Perilla Ketone, Secondary Plant Metabolites and Antioxidative Capacity in Five Perilla Frutescens L. Varieties. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2010, 48, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-Y.; Hong, C.-O.; Lee, H.; Park, S.-Y.; Park, B.-G.; Lee, K.-W. Protective Effect of Extracts of Perilla Frutescens Treated with Sucrose on Tert-Butyl Hydroperoxide-Induced Oxidative Hepatotoxicity in Vitro and in Vivo. Food Chem. 2012, 133, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Manaf, A.M.; Harwood, J.L. Purification and Characterisation of Acyl-CoA: Glycerol 3-Phosphate Acyltransferase from Oil Palm (Elaeis Guineensis) Tissues. Planta 2000, 210, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Lewin, T.M.; Wang, P.; Coleman, R.A. Analysis of Amino Acid Motifs Diagnostic for the Sn-Glycerol-3-Phosphate Acyltransferase Reaction. Biochemistry 1999, 38, 5764–5771. [Google Scholar] [CrossRef]
- Asif, M. Health Effects of Omega-3,6,9 Fatty Acids: Perilla Frutescens Is a Good Example of Plant Oils. Orient. Pharm. Exp. Med. 2011, 11, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhao, Y.; Wang, Y.; Liu, Z.; Ijaz, B.; Huang, Y.; Hua, J. Genome-Wide Identification and Expression Analysis of the Biotin Carboxyl Carrier Subunits of Heteromeric Acetyl-CoA Carboxylase in Gossypium. Front. Plant Sci. 2017, 8, 624. [Google Scholar] [CrossRef]
- Payá-Milans, M.; Venegas-Calerón, M.; Salas, J.J.; Garcés, R.; Martínez-Force, E. Cloning, Heterologous Expression and Biochemical Characterization of Plastidial Sn-Glycerol-3-Phosphate Acyltransferase from Helianthus Annuus. Phytochemistry 2015, 111, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.P.; Kroon, J.T.M.; Swarbreck, D.; Febrer, M.; Larson, T.R.; Graham, I.A.; Caccamo, M.; Slabas, A.R. Tissue-Specific Whole Transcriptome Sequencing in Castor, Directed at Understanding Triacylglycerol Lipid Biosynthetic Pathways. PLoS ONE 2012, 7, e30100. [Google Scholar] [CrossRef] [PubMed]
- Khozin-Goldberg, I.; Cohen, Z. Unraveling Algal Lipid Metabolism: Recent Advances in Gene Identification. Biochimie 2011, 93, 91–100. [Google Scholar] [CrossRef]
- Zou, S.; Lu, Y.; Ma, H.; Li, Y.; Chen, G.; Han, D.; Hu, Q. Microalgal Glycerol-3-Phosphate Acyltransferase Role in Galactolipids and High-Value Storage Lipid Biosynthesis. Plant Physiol. 2023, 192, 426–441. [Google Scholar] [CrossRef]
- Strotbek, C.; Krinninger, S.; Frank, W. The Moss Physcomitrella Patens: Methods and Tools from Cultivation to Targeted Analysis of Gene Function. Int. J. Dev. Biol. 2013, 57, 553–564. [Google Scholar] [CrossRef]
- Arroyo-Caro, J.M.; Chileh, T.; Kazachkov, M.; Zou, J.; Alonso, D.L.; García-Maroto, F. The Multigene Family of Lysophosphatidate Acyltransferase (LPAT)-Related Enzymes in Ricinus Communis. Cloning and Molecular Characterization of Two LPAT Genes That Are Expressed in Castor Seeds. Plant Sci. 2013, 199–200, 29–40. [Google Scholar] [CrossRef]
- Burgal, J.; Shockey, J.; Lu, C.; Dyer, J.; Larson, T.; Graham, I.; Browse, J. Metabolic Engineering of Hydroxy Fatty Acid Production in Plants: RcDGAT2 Drives Dramatic Increases in Ricinoleate Levels in Seed Oil. Plant Biotechnol. J. 2008, 6, 819–831. [Google Scholar] [CrossRef]
- Kim, H.U.; Lee, K.-R.; Go, Y.S.; Jung, J.H.; Suh, M.-C.; Kim, J.B. Endoplasmic Reticulum-Located PDAT1-2 from Castor Bean Enhances Hydroxy Fatty Acid Accumulation in Transgenic Plants. Plant Cell Physiol. 2011, 52, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Van Erp, H.; Bates, P.D.; Burgal, J.; Shockey, J.; Browse, J. Castor Phospholipid:Diacylglycerol Acyltransferase Facilitates Efficient Metabolism of Hydroxy Fatty Acids in Transgenic Arabidopsis. Plant Physiol. 2011, 155, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.K.; Coffey, M.; Lai, K.; Kumar, A.; MacKenzie, S.L. Enhancement of Seed Oil Content by Expression of Glycerol-3-Phosphate Acyltransferase Genes. Biochem. Soc. Trans. 2000, 28, 958–961. [Google Scholar] [CrossRef]
- Misra, A.; Khan, K.; Niranjan, A.; Kumar, V.; Sane, V.A. Heterologous Expression of Two GPATs from Jatropha Curcas Alters Seed Oil Levels in Transgenic Arabidopsis Thaliana. Plant Sci. Int. J. Exp. Plant Biol. 2017, 263, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.-F.; Wang, X.; Hu, D.-X.; Balamurugan, S.; Li, D.-W.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. Molecular Characterization of a Glycerol-3-Phosphate Acyltransferase Reveals Key Features Essential for Triacylglycerol Production in Phaeodactylum Tricornutum. Biotechnol. Biofuels 2016, 9, 60. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- El Tahchy, A.; Reynolds, K.B.; Petrie, J.R.; Singh, S.P.; Vanhercke, T. Thioesterase Overexpression in Nicotiana Benthamiana Leaf Increases the Fatty Acid Flux into Triacylgycerol. FEBS Lett. 2017, 591, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, L.; Chen, H.; Chen, Y.Q.; Ratledge, C.; Song, Y.; Chen, W. Regulatory Properties of Malic Enzyme in the Oleaginous Yeast, Yarrowia Lipolytica, and Its Non-Involvement in Lipid Accumulation. Biotechnol. Lett. 2013, 35, 2091–2098. [Google Scholar] [CrossRef]
| Strains | Fatty Acid Composition (% of Total FAs) | ||||||
|---|---|---|---|---|---|---|---|
| C16:0 | C16:1 | C18:0 | C18:1 | C18:2 | C18:3 | ||
| Unfed | INVSc1 | 12.38 ± 0.99 | 36.34 ± 1.52 | 9.44 ± 0.70 | 38.38 ± 1.94 | ||
| PfGPAT9 | 14.26 ± 1.95 | 34.18 ± 1.18 | 6.27 ± 0.63 ** | 42.02 ± 1.74 | |||
| C18:1-fed | INVSc1 | 11.45 ± 0.98 | 35.16 ± 1.85 | 8.22 ± 0.82 | 41.98 ± 2.11 | ||
| PfGPAT9 | 6.23 ± 0.88 ** | 16.52 ± 1.18 ** | 3.46 ± 0.44 ** | 70.54 ± 2.57 ** | |||
| C18:2-fed | INVSc1 | 12.01 ± 1.23 | 32.85 ± 1.70 | 10.49 ± 0.97 | 32.31 ± 1.67 | 9.16 ± 0.90 | |
| PfGPAT9 | 7.96 ± 0.94 ** | 31.28 ± 1.73 ** | 8.34 ± 0.75 | 36.09 ± 2.29 | 13.21 ± 1.16 ** | ||
| C18:3-fed | INVSc1 | 12.46 ± 1.21 | 34.19 ± 2.27 | 8.98 ± 0.42 | 35.15 ± 1.87 | 6.13 ± 0.33 | |
| PfGPAT9 | 10.63 ± 0.70 | 29.14 ± 1.12 ** | 6.27 ± 0.80 ** | 29.28 ± 0.96 ** | 21.65 ± 1.64 ** | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Huang, X.; Hu, T.; Chen, S.; Wang, Y.; Shi, X.; Yin, M.; Li, R.; Wang, J.; Jia, X. Genome-Wide Analysis of Glycerol-3-Phosphate Acyltransferase (GPAT) Family in Perilla frutescens and Functional Characterization of PfGPAT9 Crucial for Biosynthesis of Storage Oils Rich in High-Value Lipids. Int. J. Mol. Sci. 2023, 24, 15106. https://doi.org/10.3390/ijms242015106
Zhou Y, Huang X, Hu T, Chen S, Wang Y, Shi X, Yin M, Li R, Wang J, Jia X. Genome-Wide Analysis of Glycerol-3-Phosphate Acyltransferase (GPAT) Family in Perilla frutescens and Functional Characterization of PfGPAT9 Crucial for Biosynthesis of Storage Oils Rich in High-Value Lipids. International Journal of Molecular Sciences. 2023; 24(20):15106. https://doi.org/10.3390/ijms242015106
Chicago/Turabian StyleZhou, Yali, Xusheng Huang, Ting Hu, Shuwei Chen, Yao Wang, Xianfei Shi, Miao Yin, Runzhi Li, Jiping Wang, and Xiaoyun Jia. 2023. "Genome-Wide Analysis of Glycerol-3-Phosphate Acyltransferase (GPAT) Family in Perilla frutescens and Functional Characterization of PfGPAT9 Crucial for Biosynthesis of Storage Oils Rich in High-Value Lipids" International Journal of Molecular Sciences 24, no. 20: 15106. https://doi.org/10.3390/ijms242015106
APA StyleZhou, Y., Huang, X., Hu, T., Chen, S., Wang, Y., Shi, X., Yin, M., Li, R., Wang, J., & Jia, X. (2023). Genome-Wide Analysis of Glycerol-3-Phosphate Acyltransferase (GPAT) Family in Perilla frutescens and Functional Characterization of PfGPAT9 Crucial for Biosynthesis of Storage Oils Rich in High-Value Lipids. International Journal of Molecular Sciences, 24(20), 15106. https://doi.org/10.3390/ijms242015106





