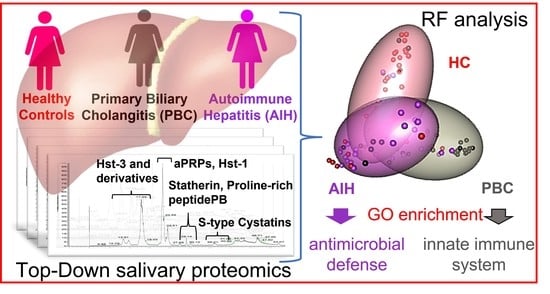
Top-Down Proteomics Detection of Potential Salivary Biomarkers for Autoimmune Liver Diseases Classification
Abstract
1. Introduction
2. Results
2.1. Top-Down Mass Spectrometry Pipeline
2.2. Characteristics of the Participants and Saliva Sampling
2.3. Statistical Analysis of the Protein/Peptide Abundances between Groups
2.4. Correlation between Protein Levels within Groups
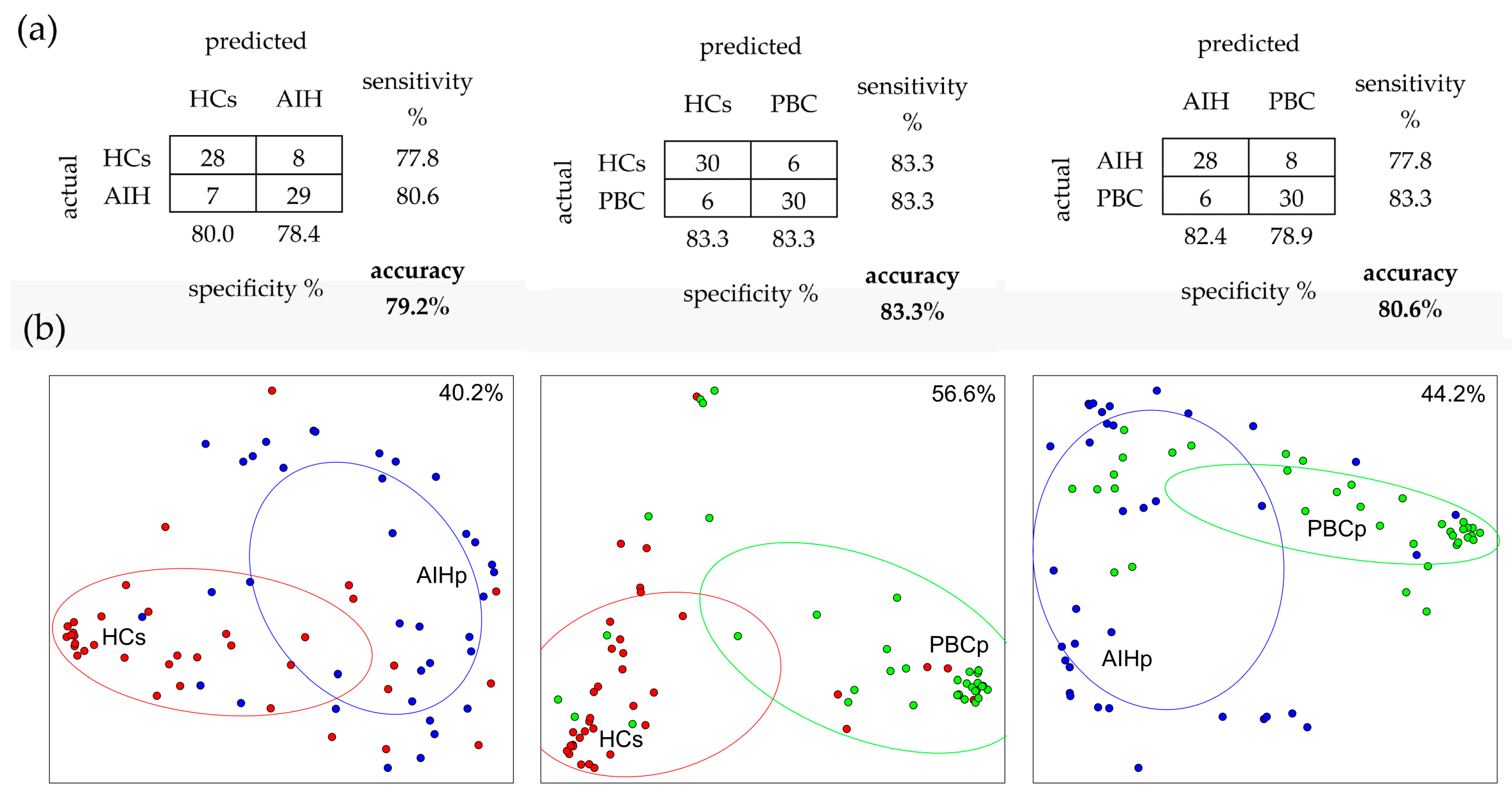
2.5. Random Forest (RF) Analysis
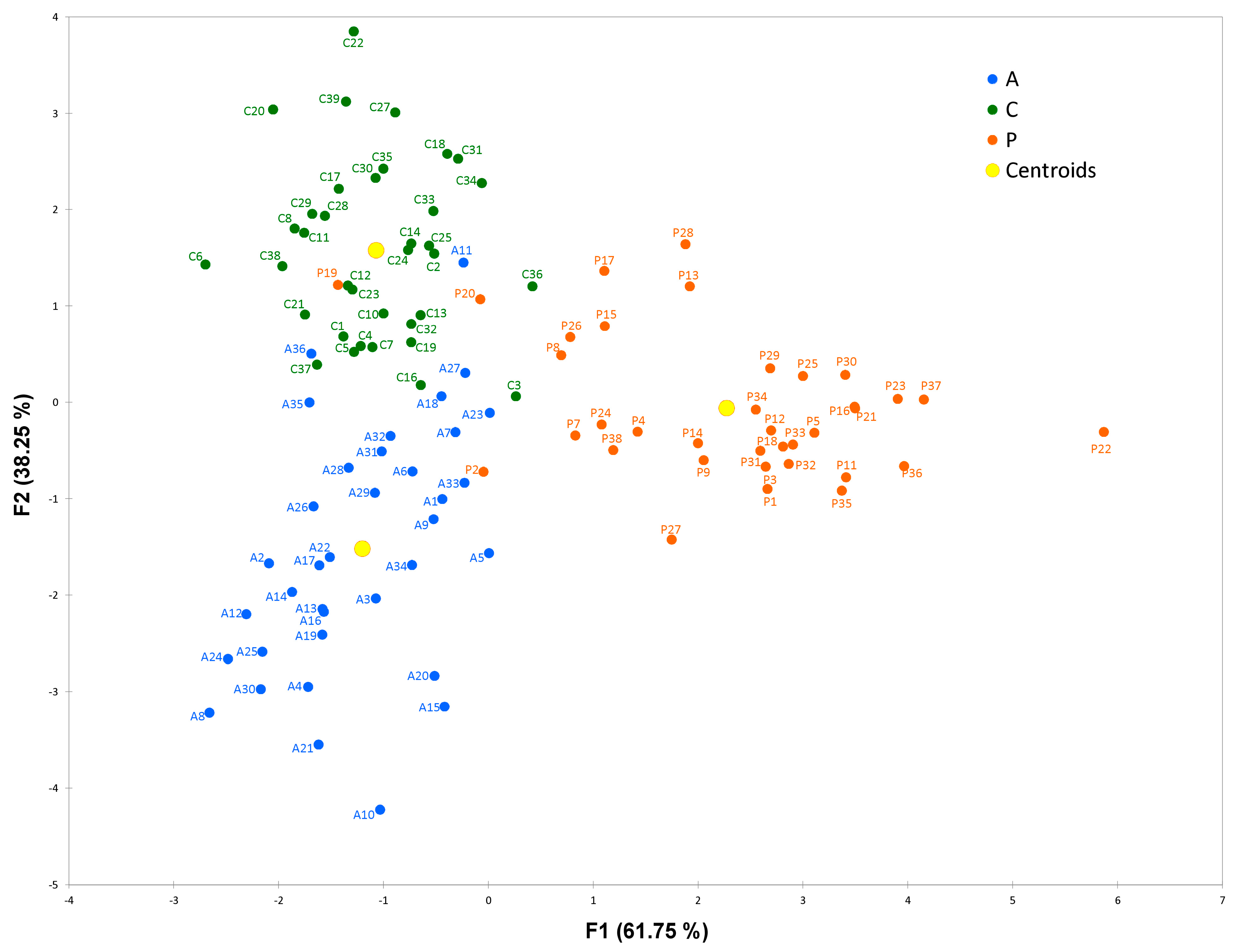
2.6. Linear Discriminant Analysis (LDA)
2.7. Enrichment Analyses
3. Discussion
3.1. Potential Salivary Biomarkers in AIHp
3.2. Potential Salivary Biomarkers in PBCp
3.3. Classification AIH and PBC Subjects from HCs
3.4. Functional Characterization of Proteins Most Discriminating AIHp from PBCp
3.5. Study Limitation
4. Materials and Methods
4.1. Ethical Statement
4.2. Study Subjects and Clinical Studies
4.3. Sample Collection and Treatment
4.4. RP-HPLC ESI-MS Analysis
4.5. Data Analysis and Quantification
4.6. Statistical Analysis
4.7. Gene Ontology Enrichment Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anand, L.; Choudhury, A.; Bihari, C.; Sharma, B.C.; Kumar, M.; Maiwall, R.; Siam Tan, S.; Shah, S.R.; Hamid, S.; Butt, A.S.; et al. Flare of Autoimmune Hepatitis Causing Acute on Chronic Liver Failure: Diagnosis and Response to Corticosteroid Therapy. Hepatology 2019, 70, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Heneghan, M.A.; Yeoman, A.D.; Verma, S.; Smith, A.D.; Longhi, M.S. Autoimmune hepatitis. Lancet 2013, 382, 1433–1444. [Google Scholar] [CrossRef] [PubMed]
- Manns, M.P.; Czaja, A.J.; Gorham, J.D.; Krawitt, E.L.; Mieli-Vergani, G.; Vergani, D.; Vierling, J.M. American Association for the Study of Liver Diseases. Diagnosis and management of autoimmune hepatitis. Hepatology 2010, 51, 2193–2213. [Google Scholar] [CrossRef] [PubMed]
- Liberal, R.; Krawitt, E.L.; Vierling, J.M.; Manns, M.P.; Mieli-Vergani, G.; Vergani, D. Cutting edge issues in autoimmune hepatitis. J. Autoimmun. 2016, 75, 6–19. [Google Scholar] [CrossRef]
- Mack, C.L.; Adams, D.; Assis, D.N.; Kerkar, N.; Manns, M.P.; Mayo, M.J.; Vierling, J.M.; Alsawas, M.; Murad, M.H.; Czaja, A.J. Diagnosis and Management of Autoimmune Hepatitis in Adults and Children: 2019 Practice Guidance and Guidelines From the American Association for the Study of Liver Diseases. Hepatology 2020, 72, 671–722. [Google Scholar] [CrossRef]
- Manns, M.P.; Lohse, A.W.; Vergani, D. Autoimmune hepatitis—Update 2015. J. Hepatol. 2015, 62, S100–S111. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: The diagnosis and management of patients with primary biliary cholangitis. J. Hepatol. 2017, 67, 145–172. [Google Scholar] [CrossRef]
- Lleo, A.; Leung, P.S.C.; Hirschfield, G.M.; Gershwin, E.M. The Pathogenesis of Primary Biliary Cholangitis: A Comprehensive Review. Semin. Liver Dis. 2020, 40, 34–48. [Google Scholar] [CrossRef]
- Marchioni Beery, R.M.; Vaziri, H.; Forouhar, F. Primary Biliary Cirrhosis and Primary Sclerosing Cholangitis: A Review Featuring a Women’s Health Perspective. J. Clin. Transl. Hepatol. 2014, 2, 266–284. [Google Scholar] [CrossRef]
- Floreani, A.; Franceschet, I.; Cazzagon, N.; Spinazzè, A.; Buja, A.; Furlan, P.; Baldo, V.; Gershwin, M.E. Extrahepatic autoimmune conditions associated with primary biliary cirrhosis. Clin. Rev. Allergy Immunol. 2015, 48, 192–197. [Google Scholar] [CrossRef]
- Wong, G.W.; Yeong, T.; Lawrence, D.; Yeoman, A.D.; Verma, S.; Heneghan, M.A. Concurrent extrahepatic autoimmunity in autoimmune hepatitis: Implications for diagnosis, clinical course and long-term outcomes. Liver Int. 2017, 37, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Marciano, F.; Savoia, M.; Vajro, P. Celiac disease-related hepatic injury: Insights into associated conditions and underlying pathomechanisms. Dig. Liver Dis. 2012, 48, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Chazouillères, O.; Wendum, D.; Serfaty, L.; Montembault, S.; Rosmorduc, O.; Poupon, R. Primary biliary cirrhosis-autoimmune hepatitis overlap syndrome: Clinical features and response to therapy. Hepatology 1998, 28, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Czaja, A.J. Diagnosis and management of the overlap syndromes of autoimmune hepatitis. Can. J. Gastroenterol. 2013, 27, 417–423. [Google Scholar] [CrossRef]
- Freedman, B.L.; Danford, C.J.; Patwardhan, V.; Bonder, A. Treatment of Overlap Syndromes in Autoimmune Liver Disease: A Systematic Review and Meta-Analysis. J. Clin. Med. 2020, 9, 1449. [Google Scholar] [CrossRef]
- Czaja, A.J. Emerging therapeutic biomarkers of autoimmune hepatitis and their impact on current and future management. Expert Rev. Gastroenterol. Hepatol. 2018, 12, 547–564. [Google Scholar] [CrossRef]
- Wang, H.; Yan, W.; Feng, Z.; Gao, Y.; Zhang, L.; Feng, X.; Tian, D. Plasma proteomic analysis of autoimmune hepatitis in an improved AIH mouse model. J. Transl. Med. 2020, 18, 3. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Fritzler, M.J.; Swain, G.G. A Review on Biomarkers for the Evaluation of Autoimmune Cholestatic Liver Diseases and Their Overlap Syndromes. Front. Mol. Med. 2022, 2, 914505. [Google Scholar] [CrossRef]
- Harrington, C.; Krishnan, S.; Mack, C.L.; Cravedi, P.; Assis, D.N.; Levitsky, J. Noninvasive biomarkers for the diagnosis and management of autoimmune hepatitis. Hepatology 2022, 76, 1862–1879. [Google Scholar] [CrossRef]
- Cabras, T.; Pisano, E.; Montaldo, C.; Giuca, M.R.; Iavarone, F.; Zampino, G.; Castagnola, M.; Messana, I. Significant modifications of the salivary proteome potentially associated with complications of Down syndrome revealed by top-down proteomics. Mol. Cell. Proteom. 2013, 12, 1844–1852. [Google Scholar] [CrossRef]
- Vierling, J.M. Diagnosis and treatment of autoimmune hepatitis. Curr. Gastroenterol. Rep. 2012, 14, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Norman, G.L.; Shums, Z.; Worman, H.J.; Krawitt, E.L.; Bizzaro, N.; Vergani, D.; Bogdanos, D.P.; Dalekos, G.N.; Milkiewicz, P.; et al. PBC screen: An IgG/IgA dual isotype ELISA detecting multiple mitochondrial and nuclear autoantibodies specific for primary biliary cirrhosis. J. Autoimmun. 2010, 35, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Granito, A.; Muratori, L.; Muratori, P.; Pappas, G.; Guidi, M.; Cassani, F.; Volta, U.; Ferri, A.; Lenzi, M.; Bianchi, F.B. Antibodies to filamentous actin (F-actin) in type 1 autoimmune hepatitis. J. Clin. Pathol. 2006, 59, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Wichmann, I.; Montes-Cano, M.A.; Respaldiza, N.; Alvarez, A.; Walter, K.; Franco, E.; Sanchez-Roman, J.; Núñez-Roldán, A. Clinical significance of anti-multiple nuclear dots/Sp100 autoantibodies. Scand. J. Gastroenterol. 2003, 38, 996–999. [Google Scholar] [CrossRef] [PubMed]
- Castagnola, M.; Messana, I.; Inzitari, R.; Fanali, C.; Cabras, T.; Morelli, A.; Pecoraro, A.M.; Neri, G.; Torrioli, M.G.; Gurrieri, F. Hypo-phosphorylation of salivary peptidome as a clue to the molecular pathogenesis of autism spectrum disorders. J. Proteome Res. 2008, 7, 5327–5332. [Google Scholar] [CrossRef] [PubMed]
- Sanna, M.; Firinu, D.; Manconi, P.E.; Pisanu, M.; Murgia, G.; Piras, V.; Castagnola, M.; Messana, I.; Del Giacco, S.; Cabras, T. The salivary proteome profile in patients affected by SAPHO syndrome characterized by a top-down RP-HPLC-ESI-MS platform. Mol. Biosyst. 2015, 11, 1552–1562. [Google Scholar] [CrossRef] [PubMed]
- Rao, B.; Lou, J.; Lu, H.; Liang, H.; Li, J.; Zhou, H.; Fan, Y.; Zhang, H.; Sun, Y.; Zou, Y.; et al. Oral Microbiome Characteristics in Patients with Autoimmune Hepatitis. Front. Cell. Infect. Microbiol. 2021, 11, 656674. [Google Scholar] [CrossRef]
- Messana, I.; Cabras, T.; Pisano, E.; Sanna, M.T.; Olianas, A.; Manconi, B.; Pellegrini, M.; Paludetti, G.; Scarano, E.; Fiorita, A.; et al. Trafficking and postsecretory events responsible for the formation of secreted human salivary peptides. Mol. Cell. Proteom. 2008, 7, 911–926. [Google Scholar] [CrossRef]
- Ghallab, N.A. Diagnostic potential and future directions of biomarkers in gingival crevicular fluid and saliva of periodontal diseases: Review of the current evidence. Arch. Oral Biol. 2018, 87, 115–124. [Google Scholar] [CrossRef]
- Iavarone, F.; Olianas, A.; Patini, R.; Gallenzi, P.; Di Tonno, L.; Desiderio, C.; Cabras, T.; Manconi, B.; Vincenzoni, F.; Cordaro, M.; et al. Top down proteomic analysis of gingival crevicular fluid in deciduous, exfoliating and permanent teeth in children. J. Proteomics 2020, 226, 103890. [Google Scholar] [CrossRef]
- Grassl, N.; Kulak, N.A.; Pichler, G.; Geyer, P.E.; Jung, J.; Schubert, S.; Sinitcyn, P.; Cox, J.; Mann, M. Ultra-deep and quantitative saliva proteome reveals dynamics of the oral microbiome. Genome Med. 2016, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Cabras, T.; Sanna, M.; Manconi, B.; Fanni, D.; Demelia, L.; Sorbello, O.; Iavarone, F.; Castagnola, M.; Faa, G.; Messana, I. Proteomic investigation of whole saliva in Wilson’s disease. J. Proteom. 2015, 128, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Manconi, B.; Liori, B.; Cabras, T.; Vincenzoni, F.; Iavarone, F.; Lorefice, L.; Cocco, E.; Castagnola, M.; Messana, I.; Olianas, A. Top-down proteomic profiling of human saliva in multiple sclerosis patients. J. Proteom. 2018, 187, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Boroumand, M.; Olianas, A.; Cabras, T.; Manconi, B.; Fanni, D.; Faa, G.; Desiderio, C.; Messana, I.; Castagnola, M. Saliva, a bodily fluid with recognized and potential diagnostic applications. J. Sep. Sci. 2021, 44, 3677–3690. [Google Scholar] [CrossRef] [PubMed]
- Stafford, I.S.; Kellermann, M.; Mossotto, E.; Beattie, R.M.; MacArthur, B.D.; Ennis, S. A systematic review of the applications of artificial intelligence and machine learning in autoimmune diseases. NPJ Digit. Med. 2020, 3, 30. [Google Scholar] [CrossRef]
- MacEachern, S.J.; Forkert, N.D. Machine learning for precision medicine. Genome 2021, 64, 416–425. [Google Scholar] [CrossRef]
- Seyed Tabib, N.S.; Madgwick, M.; Sudhakar, P.; Verstockt, B.; Korcsmaros, T.; Vermeire, S. Big data in IBD: Big progress for clinical practice. Gut 2020, 69, 1520–1532. [Google Scholar] [CrossRef]
- Nam, D.; Chapiro, J.; Paradis, V.; Seraphin, T.P.; Kather, J.N. Artificial intelligence in liver diseases: Improving diagnostics, prognostics and response prediction. JHEP Rep. 2022, 4, 100443. [Google Scholar] [CrossRef]
- Xia, Z.; Qin, L.; Ning, Z.; Zhang, X. Deep learning time series prediction models in surveillance data of hepatitis incidence in China. PLoS ONE 2022, 17, e0265660. [Google Scholar] [CrossRef]
- Candles, L.S.; Rahim, M.N.; Shah, S.; Heneghan, M.A. Towards personalised medicine in autoimmune hepatitis: Measurement of thiopurine metabolites results in higher biochemical response rates. J. Hepatol. 2021, 75, 324–332. [Google Scholar] [CrossRef]
- Serrao, S.; Firinu, D.; Olianas, A.; Deidda, M.; Contini, C.; Iavarone, F.; Sanna, M.T.; Boroumand, M.; Amado, F.; Castagnola, M.; et al. Top-Down Proteomics of Human Saliva Discloses Significant Variations of the Protein Profile in Patients with Mastocytosis. J. Proteome Res. 2020, 19, 3238–3253. [Google Scholar] [CrossRef] [PubMed]
- Contini, C.; Olianas, A.; Serrao, S.; Deriu, C.; Iavarone, F.; Boroumand, M.; Bizzarro, A.; Lauria, A.; Faa, G.; Castagnola, M.; et al. Top-Down Proteomics of Human Saliva Highlights Anti-inflammatory, Antioxidant, and Antimicrobial Defense Responses in Alzheimer Disease. Front. Neurosci. 2021, 15, 668852, Erratum in Front. Neurosci. 2021, 15, 743596. [Google Scholar] [CrossRef] [PubMed]
- Oppenheim, F.G.; Xu, T.; McMillian, F.M.; Levitz, S.M.; Diamond, R.D.; Offner, G.D.; Troxler, R.F. Histatins, a novel family of histidine-rich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Candida albicans. J. Biol. Chem. 1988, 263, 7472–7477. [Google Scholar] [CrossRef] [PubMed]
- Castagnola, M.; Cabras, T.; Iavarone, F.; Vincenzoni, F.; Vitali, A.; Pisano, E.; Nemolato, S.; Scarano, E.; Fiorita, A.; Vento, G.; et al. Top-down platform for deciphering the human salivary proteome. J. Matern. Fetal. Neonatal. Med. 2012, 25, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Castagnola, M.; Inzitari, R.; Rossetti, D.V.; Olmi, C.; Cabras, T.; Piras, V.; Nicolussi, P.; Sanna, M.T.; Pellegrini, M.; Giardina, B.; et al. A cascade of 24 histatins (histatin 3 fragments) in human saliva. Suggestions for a pre-secretory sequential cleavage pathway. J. Biol. Chem. 2004, 279, 41436–41443. [Google Scholar] [CrossRef]
- Wang, G. Human antimicrobial peptides and proteins. Pharmaceuticals 2014, 7, 545–594. [Google Scholar] [CrossRef]
- Peters, T., Jr. Interaction of one mole of copper with the alpha amino group of bovine serum albumin. Biochim. Biophys. Acta 1960, 39, 546–547. [Google Scholar] [CrossRef]
- Cabras, T.; Patamia, M.; Melino, S.; Inzitari, R.; Messana, I.; Castagnola, M.; Petruzzelli, R. Pro-oxidant activity of histatin 5 related Cu(II)-model peptide probed by mass spectrometry. Biochem. Biophys. Res. Commun. 2007, 358, 277–284. [Google Scholar] [CrossRef]
- Harford, C.; Sarkar, B. Amino Terminal Cu(II)- and Ni(II)-Binding (ATCUN) Motif of Proteins and Peptides: Metal Binding, DNA Cleavage, and Other Properties. Acc. Chem. Res. 1997, 30, 123–130. [Google Scholar] [CrossRef]
- Grogan, J.; McKnight, C.J.; Troxler, R.F.; Oppenheim, F.G. Zinc and copper bind to unique sites of histatin 5. FEBS Lett. 2001, 491, 76–80. [Google Scholar] [CrossRef]
- Johnson, D.A.; Yeh, C.K.; Dodds, M.W. Effect of donor age on the concentrations of histatins in human parotid and submandibular/sublingual saliva. Arch. Oral Biol. 2000, 45, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Liu, X.; Cheng, Y.; Jiang, X.; Jiang, H.; Wang, Y.; Li, L. Decreased Diversity of the Oral Microbiota of Patients with Hepatitis B Virus-Induced Chronic Liver Disease: A Pilot Project. Sci. Rep. 2015, 5, 17098. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, J.S.; Betrapally, N.S.; Hylemon, P.B.; Heuman, D.M.; Daita, K.; White, M.B.; Unser, A.; Thacker, L.R.; Sanyal, A.J.; Kang, D.J.; et al. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology 2015, 62, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, Y.; Amir, A.; Ben-Simon, S.; Veitsman, E.; Cohen-Ezra, O.; Davidov, Y.; Weiss, P.; Bradichevski, T.; Segev, S.; Koren, O.; et al. Alterations of the salivary and fecal microbiome in patients with primary sclerosing cholangitis. Hepatol. Int. 2021, 15, 191–201. [Google Scholar] [CrossRef]
- Ren, Z.; Li, A.; Jiang, J.; Zhou, L.; Yu, Z.; Lu, H.; Xie, H.; Chen, X.; Shao, L.; Zhang, R.; et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2019, 68, 1014–1023. [Google Scholar] [CrossRef]
- Cabras, T.; Iavarone, F.; Manconi, B.; Olianas, A.; Sanna, M.T.; Castagnola, M.; Messana, I. Top-down analytical platforms for the characterization of the human salivary proteome. Bioanalysis 2014, 6, 563–581. [Google Scholar] [CrossRef]
- Kirschke, H.; Schmidt, I.; Wiederanders, B.; Cathepsin, S. The cysteine proteinase from bovine lymphoid tissue is distinct from cathepsin L (EC 3.4.22.15). Biochem. J. 1986, 240, 455–459. [Google Scholar] [CrossRef]
- Wilkinson, R.D.; Williams, R.; Scott, C.J.; Burden, R.E. Cathepsin S: Therapeutic, diagnostic, and prognostic potential. Biol. Chem. 2015, 396, 867–882. [Google Scholar] [CrossRef]
- Rupanagudi, K.V.; Kulkarni, O.P.; Lichtnekert, J.; Darisipudi, M.N.; Mulay, S.R.; Schott, B.; Gruner, S.; Haap, W.; Hartmann, G.; Anders, H.J. Cathepsin S inhibition suppresses systemic lupus erythematosus and lupus nephritis because cathepsin S is essential for MHC class II-mediated CD4 T cell and B cell priming. Ann. Rheum. Dis. 2015, 74, 452–463. [Google Scholar] [CrossRef]
- Klinngam, W.; Janga, S.R.; Lee, C.; Ju, Y.; Yarber, F.; Shah, M.; Guo, H.; Wang, D.; MacKay, J.A.; Edman, M.C.; et al. Inhibition of Cathepsin S Reduces Lacrimal Gland Inflammation and Increases Tear Flow in a Mouse Model of Sjögren’s Syndrome. Sci. Rep. 2019, 9, 9559. [Google Scholar] [CrossRef]
- Allan, E.R.; Yates, R.M. Redundancy between Cysteine Cathepsins in Murine Experimental Autoimmune Encephalomyelitis. PLoS ONE 2015, 10, e0128945. [Google Scholar] [CrossRef] [PubMed]
- Thanei, S.; Theron, M.; Silva, A.P.; Reis, B.; Branco, L.; Schirmbeck, L.; Kolb, F.A.; Haap, W.; Schindler, T.; Trendelenburg, M. Cathepsin S inhibition suppresses autoimmune-triggered inflammatory responses in macrophages. Biochem. Pharmacol. 2017, 146, 151–164. [Google Scholar] [CrossRef]
- de Mingo Pulido, Á.; de Gregorio, E.; Chandra, S.; Colell, A.; Morales, A.; Kronenberg, M.; Marí, M. Differential Role of Cathepsins S and B In Hepatic APC-Mediated NKT Cell Activation and Cytokine Secretion. Front. Immunol. 2018, 9, 391. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Blázquez, P.; Pistorio, V.; Fernández-Fernández, M.; Moles, A. The multifaceted role of cathepsins in liver disease. J. Hepatol. 2021, 75, 1192–1202. [Google Scholar] [CrossRef] [PubMed]
- Baron, A.; DeCarlo, A.; Featherstone, J. Functional aspects of the human salivary cystatins in the oral environment. Oral Dis. 1999, 5, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Guicciardi, M.E.; Deussing, J.; Miyoshi, H.; Bronk, S.F.; Svingen, P.A.; Peters, C.; Kaufmann, S.H.; Gores, G.J. Cathepsin B contributes to TNF-alpha-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J. Clin. Investig. 2000, 106, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Feldstein, A.E.; Werneburg, N.W.; Canbay, A.; Guicciardi, M.E.; Bronk, S.F.; Rydzewski, R.; Burgart, L.J.; Gores, G.J. Free fatty acids promote hepatic lipotoxicity by stimulating TNF-alpha expression via a lysosomal pathway. Hepatology 2004, 40, 185–194. [Google Scholar] [CrossRef]
- Baskin-Bey, E.S.; Canbay, A.; Bronk, S.F.; Werneburg, N.; Guicciardi, M.E.; Nyberg, S.L.; Gores, G.J. Cathepsin B inactivation attenuates hepatocyte apoptosis and liver damage in steatotic livers after cold ischemia-warm reperfusion injury. Am. J. Physiol.-Gastrointest. Liver Physiol. 2005, 288, G396–G402. [Google Scholar] [CrossRef][Green Version]
- Canbay, A.; Guicciardi, M.E.; Higuchi, H.; Feldstein, A.; Bronk, S.F.; Rydzewski, R.; Taniai, M.; Gores, G.J. Cathepsin B inactivation attenuates hepatic injury and fibrosis during cholestasis. J. Clin. Investig. 2003, 112, 152–159. [Google Scholar] [CrossRef]
- Manchanda, M.; Das, P.; Gahlot, G.P.S.; Singh, R.; Roeb, E.; Roderfeld, M.; Datta Gupta, S.; Saraya, A.; Pandey, R.M.; Chauhan, S.S. Cathepsin L and B as Potential Markers for Liver Fibrosis: Insights From Patients and Experimental Models. Clin. Transl. Gastroenterol. 2017, 8, e99. [Google Scholar] [CrossRef]
- Miranda, L.P.; Tao, T.; Jones, A.; Chernushevich, I.; Standing, K.G.; Geczy, C.L.; Alewood, P.F. Total chemical synthesis and chemotactic activity of human S100A12 (EN-RAGE). FEBS Lett. 2001, 488, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.A.; Drury, S.; Fu, C.; Qu, W.; Taguchi, A.; Lu, Y.; Avila, C.; Kambham, N.; Bierhaus, A.; Nawroth, P.; et al. RAGE mediates a novel proinflammatory axis: A central cell surface receptor for S100/calgranulin polypeptides. Cell 1999, 97, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Li, X.; Zhang, L.; Deng, C.; Zhang, T.; Wang, L.; Hu, C.; Li, Y.; Zhang, F. S100A12 expression in patients with primary biliary cirrhosis. Immunol. Investig. 2015, 44, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; An, S.H.; Kang, S.W.; Kwon, K. Ursodeoxycholic Acid (UDCA) Exerts Anti-Atherogenic Effects by Inhibiting RAGE Signaling in Diabetic Atherosclerosis. PLoS ONE 2016, 11, e0147839. [Google Scholar] [CrossRef] [PubMed]
- Contini, C.; Serrao, S.; Manconi, B.; Olianas, A.; Iavarone, F.; Bizzarro, A.; Masullo, C.; Castagnola, M.; Messana, I.; Diaz, G.; et al. Salivary Proteomics Reveals Significant Changes in Relation to Alzheimer’s Disease and Aging. J. Alzheimers Dis. 2022, 89, 605–622. [Google Scholar] [CrossRef]
- Amaratunga, D.; Cabrera, J.; Lee, Y.S. Enriched random forests. Bioinformatics 2008, 24, 2010–2014. [Google Scholar] [CrossRef] [PubMed]
- Kochańska, B.; Kedzia, A.; Kamysz, W.; Maćkiewicz, Z.; Kupryszewski, G. The effect of statherin and its shortened analogues on anaerobic bacteria isolated from the oral cavity. Acta Microbiol. Pol. 2000, 49, 243–251. [Google Scholar]
- Amano, A.; Kataoka, K.; Raj, P.A.; Genco, R.J.; Shizukuishi, S. Binding sites of salivary statherin for Porphyromonas gingivalis recombinant fimbrillin. Infect. Immun. 1996, 64, 4249–4254. [Google Scholar] [CrossRef]
- Boberg, K.M. Prevalence and epidemiology of autoimmune hepatitis. Clin. Liver Dis. 2002, 6, 635–647. [Google Scholar] [CrossRef]
- Zhang, Z.; Marshall, A.G. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 1998, 9, 225–233. [Google Scholar] [CrossRef]
- Ong, S.E.; Mann, M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005, 1, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Messana, I.; Inzitari, R.; Fanali, C.; Cabras, T.; Castagnola, M. Facts and artifacts in proteomics of body fluids. What proteomics of saliva is telling us? J. Sep. Sci. 2008, 31, 1948–1963. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B Stat. Methodol. 1995, 57, 289–290. [Google Scholar] [CrossRef]
- Kursa, M.B.; Jankowski, A.; Rudnicki, W.R. Boruta—A System for Feature Selection. Fundam. Inform. 2010, 101, 271–285. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]


| Parameters | AIHp | PBCp | |
|---|---|---|---|
| Age, Average (range) | Years | 54.53 (29.81–74.89) | 65.13 (41.27–81.15) |
| Gender, n (%) | Female | 32 (88.8%) | 35 (97.2%) |
| BMI, Average (range) | Kg/m2 | 25.68 (17.57–38.45) | 24.72 (19.10–40.43) |
| Cirrhosis, n (%) | 7 (19.4%) | 4 (11.1%) | |
| Histological stage, n (%) | I | 9 (25%) | 15 (41.6%) |
| II | 4 (11.1%) | 8 (22.2%) | |
| III | 6 (16.6%) | 1 (2.7%) | |
| IV | 4 (11.1%) | 4 (11.1%) | |
| Not available | 11 (30.5%) | 10 (27.7%) | |
| Positivity to autoantibodies, n (%) | ANA | 25 (69.4%) | 30 (83.3%) |
| SMA | 20 (55.5%) | 7 (19.4%) | |
| LKM | 3 (8.3%) | 1 (2.7%) | |
| AST, Median (range) | IU/L | 23.5 (13–57) | 27.0 (16–71) |
| ALT, Median (range) | IU/L | 21.0 (5–58) | 23.0 (11–78) |
| GGT, Median (range) | IU/L | 25.5 (6–167) | 42.0 (12–167) |
| ALP, Median (range) | IU/L | 67.0 (28–216) | 107.0 (52–222) |
| IgG, Median (range) | g/dL | 1.4 (0.69–2.51) | 1.4 (0.7–2.3) |
| Albumin, Median (range) | g/dL | 3.9 (1.2–4.83) | 3.9 (2.8–4.3) |
| Prothrombin time, Median (range) | INR | 0.97 (0.92–1.06) | 1.01(0.86–1.81) |
| TB, Median (range) | mg/dL | 0.7 (0.25–2.19) | 0.6 (0.34–2.95) |
| Platelets, Median (range) | 109/L | 217.5 (91–423) | 242 (46–418) |
| Pharmacological treatment (% treated) | Azathioprine + Steroids | 41% | n.a. |
| Steroids | 25.5% | n.a. | |
| Azathioprine | 17.6% | n.a. | |
| Naïve | 5.5% | n.a. | |
| UDCA | n.a. | 100% |
| Components | HCs vs. AIHp | HCs vs. PBCp | AIHp vs. PBCp | AIHp vs. PBCp vs. HCs | ||||
|---|---|---|---|---|---|---|---|---|
| N | Description | Mann—Whitney | Mann—Whitney | Mann—Whitney | Kruskal—Wallis | |||
| p-Value | Change | p-Value | Change | p-Value | Change | p-Value | ||
| 1 | S100A12 | <0.05 | PBC > AIH | <0.05 | ||||
| 2 | S100A8 | |||||||
| 3 | S100A7D27 | <0.05 | PBC > AIH | <0.01 | ||||
| 4 | S100A9_short | |||||||
| 5 | S100A9_short_ox | <0.05 | C > PBC | <0.05 | ||||
| 6 | S100A9_short_P | <0.05 | ||||||
| 7 | S100A9_short_P_ox | |||||||
| 8 | Sum_S100A9_short_and_ox | |||||||
| 9 | Sum_S100A9_s_and_s_P | |||||||
| 10 | Sum_S100A9_s_P_and_P_ox | |||||||
| 11 | Sum_S100A9_s_ox_and_P_ox | <0.05 | C > PBC | <0.05 | ||||
| 12 | Sum_S100A9_short | <0.05 | C > PBC | |||||
| 13 | S100A9_long_g | |||||||
| 14 | S100A9_long_g_p | |||||||
| 15 | S100A9_long_g_ox | <0.05 | ||||||
| 16 | Sum_S100A9_long_g | |||||||
| 17 | Cystatin_A | <0.05 | AIH > C | <0.05 | AIH > PBC | <0.05 | ||
| 18 | Cystatin_A_Acetyl | |||||||
| 19 | Cystatin_A_Acetyl_T96L | |||||||
| 20 | Sum_Cystatin_A | <0.05 | AIH > C | |||||
| 21 | Cystatin_B_S_glut | |||||||
| 22 | Cystatin_B_S_cyst | |||||||
| 23 | Cystatin_B_SSdimer | |||||||
| 24 | Cystatin_B_S_CMC | |||||||
| 25 | Sum_Cystatin_B | |||||||
| 26 | Cystatin_C | <0.05 | PBC > C | <0.01 | ||||
| 27 | Cystatin_D_des_1_5 | |||||||
| 28 | Cystatin_S | |||||||
| 29 | Cystatin_S1 | <0.001 | PBC > C | <0.01 | PBC > AIH | <0.001 | ||
| 30 | Cystatin_S2 | <0.0001 | PBC > C | <0.01 | PBC > AIH | <0.0001 | ||
| 31 | Cystatin_SN | <0.01 | PBC > C | <0.05 | PBC > AIH | <0.05 | ||
| 32 | Cystatin_SN_des_1_4 | |||||||
| 33 | Cystatin_SA | |||||||
| 34 | Cystatin_S1_ox | <0.001 | AIH > C | <0.01 | ||||
| 35 | Cystatin_S2_ox | |||||||
| 36 | Cystatin_SN_ox | |||||||
| 37 | Sum_Cystatin_S1 | <0.0001 | PBC > C | <0.01 | PBC > AIH | <0.0001 | ||
| 38 | Sum_Cystatin_S2 | <0.0001 | PBC > C | <0.01 | PBC > AIH | <0.0001 | ||
| 39 | Sum_Cystatin_S_S1_S2 | <0.0001 | PBC > C | <0.001 | PBC > AIH | <0.0001 | ||
| 40 | Sum_Cystatin_SN | <0.01 | PBC > C | <0.05 | ||||
| 41 | Sum_Cystatin_SA | |||||||
| 42 | Hst_1 | |||||||
| 43 | Hst_1_0P | |||||||
| 44 | Sum_Hst_1 | |||||||
| 45 | Hst_6 | <0.01 | AIH > C | <0.05 | AIH > PBC | |||
| 46 | Hst_5 | <0.05 | AIH > C | <0.05 | AIH > PBC | <0.05 | ||
| 47 | Hst_3 | <0.05 | AIH > C | <0.01 | AIH > PBC | <0.01 | ||
| 48 | Sum_Hst_3 | <0.01 | AIH > C | <0.05 | AIH > PBC | <0.05 | ||
| 49 | Sum_Hst | <0.05 | AIH > C | <0.05 | ||||
| 50 | α_defensin_1 | |||||||
| 51 | α_defensin_2 | |||||||
| 52 | α_defensin_3 | |||||||
| 53 | α_defensin_4 | |||||||
| 54 | Sum_α_defensins | |||||||
| 55 | PRP1_2P | <0.05 | AIH > PBC | |||||
| 56 | PRP1_1P | |||||||
| 57 | PRP1_0P | |||||||
| 58 | PRP1_3P | <0.01 | PBC > C | <0.05 | ||||
| 59 | Sum_PRP1 | <0.05 | AIH > PBC | |||||
| 60 | PRP3_2P | |||||||
| 61 | PRP3_1P | <0.05 | C > PBC | <0.05 | AIH > PBC | <0.05 | ||
| 62 | PRP3_0P | |||||||
| 63 | PRP_3_diphos_Des_Arg106 | <0.05 | PBC > AIH | <0.05 | ||||
| 64 | Sum_PRP3 | |||||||
| 65 | P_C_peptide | |||||||
| 66 | Statherin_2P | <0.05 | AIH > C | |||||
| 67 | Statherin_1P | <0.05 | AIH > C | <0.05 | AIH > PBC | |||
| 68 | Statherin_0P | |||||||
| 69 | Sum_Statherin | <0.05 | AIH > C | |||||
| 70 | PB_peptide | <0.01 | AIH > PBC | <0.05 | ||||
| 71 | SLPI | <0.05 | PBC > C | <0.05 | ||||
| HCs-AIHp Mixed Data Set | ||
| GO Biological Process | No. Associated Proteins | Enrichment p-Value |
| defense response (GO:0006952) | 6/9 | 9.62 × 10−6 |
| antimicrobial humoral immune response mediated by antimicrobial peptide (GO:0061844) | 5/9 | 4.69 × 10−10 |
| antimicrobial humoral response (GO:0019730) | 5/9 | 2.28 × 10−9 |
| humoral immune response (GO:0006959) | 5/9 | 1.51 × 10−7 |
| defense response to bacterium (GO:0042742) | 5/9 | 2.50 × 10−7 |
| regulation of endopeptidase activity (GO:0052548) | 5/9 | 4.40 × 10−7 |
| regulation of peptidase activity (GO:0052547) | 5/9 | 6.16 × 10−7 |
| response to bacterium (GO:0009617) | 5/9 | 6.95 × 10−6 |
| regulation of proteolysis (GO:0030162) | 5/9 | 7.47 × 10−6 |
| regulation of hydrolase activity (GO:0051336) | 5/9 | 3.22 × 10−5 |
| HCs-PBCp Mixed Data Set | ||
| regulation of peptidase activity (GO:0052547) | 5/6 | 3.10 × 10−8 |
| regulation of endopeptidase activity (GO:0052548) | 5/6 | 2.21 × 10−8 |
| regulation of proteolysis (GO:0030162) | 5/6 | 3.63 × 10−7 |
| regulation of hydrolase activity (GO:0051336) | 5/6 | 1.74 × 10−6 |
| negative regulation of peptidase activity (GO:0010466) | 4/6 | 3.71 × 10−7 |
| negative regulation of endopeptidase activity (GO:0010951) | 4/6 | 3.22 × 10−7 |
| regulation of cysteine-type endopeptidase activity (GO:2000116) | 4/6 | 2.78 × 10−7 |
| negative regulation of proteolysis (GO:0045861) | 4/6 | 1.18 × 10−6 |
| negative regulation of hydrolase activity (GO:0051346) | 4/6 | 1.45 × 10−6 |
| negative regulation of catalytic activity (GO:0043086) | 4/6 | 2.96 × 10−5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olianas, A.; Guadalupi, G.; Cabras, T.; Contini, C.; Serrao, S.; Iavarone, F.; Castagnola, M.; Messana, I.; Onali, S.; Chessa, L.; et al. Top-Down Proteomics Detection of Potential Salivary Biomarkers for Autoimmune Liver Diseases Classification. Int. J. Mol. Sci. 2023, 24, 959. https://doi.org/10.3390/ijms24020959
Olianas A, Guadalupi G, Cabras T, Contini C, Serrao S, Iavarone F, Castagnola M, Messana I, Onali S, Chessa L, et al. Top-Down Proteomics Detection of Potential Salivary Biomarkers for Autoimmune Liver Diseases Classification. International Journal of Molecular Sciences. 2023; 24(2):959. https://doi.org/10.3390/ijms24020959
Chicago/Turabian StyleOlianas, Alessandra, Giulia Guadalupi, Tiziana Cabras, Cristina Contini, Simone Serrao, Federica Iavarone, Massimo Castagnola, Irene Messana, Simona Onali, Luchino Chessa, and et al. 2023. "Top-Down Proteomics Detection of Potential Salivary Biomarkers for Autoimmune Liver Diseases Classification" International Journal of Molecular Sciences 24, no. 2: 959. https://doi.org/10.3390/ijms24020959
APA StyleOlianas, A., Guadalupi, G., Cabras, T., Contini, C., Serrao, S., Iavarone, F., Castagnola, M., Messana, I., Onali, S., Chessa, L., Diaz, G., & Manconi, B. (2023). Top-Down Proteomics Detection of Potential Salivary Biomarkers for Autoimmune Liver Diseases Classification. International Journal of Molecular Sciences, 24(2), 959. https://doi.org/10.3390/ijms24020959