Abstract
Skin and wound infections are serious medical problems, and the diversity of bacteria makes such infections difficult to treat. Bacteria possess many virulence factors, among which motility plays a key role in skin infections. This feature allows for movement over the skin surface and relocation into the wound. The aim of this paper is to review the type of bacterial movement and to indicate the underlying mechanisms than can serve as a target for developing or modifying antibacterial therapies applied in wound infection treatment. Five types of bacterial movement are distinguished: appendage-dependent (swimming, swarming, and twitching) and appendage-independent (gliding and sliding). All of them allow bacteria to relocate and aid bacteria during infection. Swimming motility allows bacteria to spread from ‘persister cells’ in biofilm microcolonies and colonise other tissues. Twitching motility enables bacteria to press through the tissues during infection, whereas sliding motility allows cocci (defined as non-motile) to migrate over surfaces. Bacteria during swarming display greater resistance to antimicrobials. Molecular motors generating the focal adhesion complexes in the bacterial cell leaflet generate a ‘wave’, which pushes bacterial cells lacking appendages, thereby enabling movement. Here, we present the five main types of bacterial motility, their molecular mechanisms, and examples of bacteria that utilise them. Bacterial migration mechanisms can be considered not only as a virulence factor but also as a target for antibacterial therapy.
1. Introduction
The life and proper functioning of the human body would not be possible without having extensive bacterial flora, which is present not only in the digestive and reproductive systems but also on the skin, which primarily has protective functions and is responsible for maintaining the proper homeostasis of the body. The skin microflora, also termed the skin microbiome, varies according to the sex and age of the host and is topographically diverse due to anatomical differences in the human body. The development of the microbiome on the skin in different bodily regions mainly depends on oxygen availability, the presence of sebaceous glands, humidity, and the temperature of the individual zones. It includes not only bacteria but also viruses, fungi, and bacteriophages [1,2]. The basic barrier, which is the skin, protects against mechanical and thermal factors, harmful radiation, and infections. The disruption of skin homeostasis, in combination with pathogen appearance, may lead to the development of various skin infections. Infection development is fostered by various factors, which include being immunocompromised, vascular insufficiency, poor lymphatic or venous drainage, sensory neuropathies, diabetes, obesity, poor hygiene, and surgical procedures [3]. Infection occurrence is also influenced by the virulence of microorganisms and the degree of their presence. The simplest division of bacterial infections is acute and chronic infections. Pathogenic bacteria cause a wide variety of acute skin infections with varied clinical appearances depending on the aetiological factor, location, and immune status of the host. The most common acute infections are staphylococcal, streptococcal, and mixed staphylococcal–streptococcal infections [4,5]. In addition, other species also cause skin infections, including Borrelia spp., Pseudomonas aeruginosa, Corynebacterium spp., Actinomyces spp., Bacillus anthracis, Listeria monocytogenes, Nocardia spp., Erysipelothrix rhusiopathiae, Bartonella spp., Klebsiella pneumoniae, Klebsiella rhinoscleromatis, Yersinia spp., Clostridium spp., Francisella tularensis, Brucellae spp., and Mycobacterium spp. [6]. Moreover, bacteria that would not be pathogenic with an undamaged skin barrier can penetrate through damaged skin and become virulent. In general, most acute infections are superficial and relatively easy to treat with appropriate antibiotics [7], whereas chronic infections, in particular chronic wound infections, often pose many difficulties in effective treatment, and in the era of multi-drug resistance, they are a major social and economic problem [8]. The most common aetiological factors in chronic wounds are Staphylococcus aureus, P. aeruginosa, Acinetobacter baumannii, Proteus mirabilis, Escherichia coli, and Corynebacterium spp. [9,10,11]. Among them, there are many antibiotic-resistant species included in the “ESKAPE” cluster composed of Enterococcus faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter species [12]. Due to the increasing drug resistance of bacteria responsible for skin infections, various studies have concentrated on the treatment of infections of wounds using various methods to reduce virulence factors [13,14,15]. Some virulence factors include biofilm formation, exotoxins and endotoxins, proteases, quorum sensing, and motility [14,16].
Microbes enter the wound in three main ways: bacterial deposition from air or water, direct contact with infected material, and through the transfer of one’s own physiological flora [17]. There are four microbial states of the infected wound (Table 1).

Table 1.
Microbial states of the infected wound [18,19].
The basic methods for treating chronic wounds are the use of antiseptics, a great variety of wound dressings, antibiotic therapy, compression therapy, and debridement. Other popular treatment options are hyperbaric oxygen therapy, negative pressure wound therapy, ultrasound, and electromagnetic therapy [20]. Hyperbaric oxygen therapy is widely in use for more than 20 years in diabetic foot ulcers, and this therapy may improve wound healing by stimulating angiogenesis. The mechanism of action of hyperbaric oxygen therapy on tissues has not yet been fully elucidated [21]. Another promising method for skin and wound treatment is negative pressure wound therapy, where the use of a vacuum reduces oedema and infection and increases local blood flow, which promotes healing [22]. This method is widely used as an alternative to surgery for a wide range of wounds [23]. Ultrasound therapy is the use of mechanical energy at high frequency (between 1 and 3 MHz) to induce thermal energy into musculoskeletal tissues and low frequency (between 22.5 and 40 kHz) to generate the cavitation effect, which, if delivered in waves, debride necrotic tissue from wound surface [24]. Interestingly, skin wounds generate large and endogenous electric fields termed “current of injury” [25]. This electrical skin activity is involved in the process of wound healing, so it has led to the hypothesis that applied electrical stimulation may support the healing of chronic wounds by imitating the natural electrical current [26]. Several electrical stimulation methods are available, including pulsed current, direct current, and rhythmic electric frequency. However, the best strategy of electrical stimulation is still unachieved [26]. Unfortunately, the above-mentioned methods frequently fail; thus, chronic wound treatment is difficult, long-lasting, and sometimes ineffective. As the problem of chronic wounds affects approximately 1–2% of the global population, innovative and more effective methods for treating hard-to-heal wounds are constantly being sought [27,28].
Consequently, it is imperative to study the bacterial colonisation of poorly healing wounds and assign them to specific taxonomic units, together with analysing their numerous functions and relationships. These factors underlie their unsurpassable ability to evolve and adapt to their environment, which has led to the rising issue of antibiotic resistance. Understanding this will bring us closer to a future with increasingly effective and less invasive therapies and improvement in the entire process of patient convalescence. Most wound-infecting bacteria are motile. Bacterial motility plays a key role in skin and wound infection, which leads to hindering therapy and full recovery. The molecular mechanisms underlying bacterial motility may be an attractive target for new therapies being developed, where the inhibition of motility will not only reduce the spread of infection but also facilitate bacterial eradication. Here, we present an overview of the types of bacterial movement along with a description of their molecular machinery, to indicate them as a potential target of interest.
2. Types of Bacterial Movement
One of the main characteristics of microbes adapting to and surviving in the external environment is how they move and grow, whether in liquid, semi-liquid, or solid environments. Notably, not all bacteria are capable of active movement. In most cases, the flagella enable motion. Bacterial cells have flagella in variable quantities and locations, which is one of the features that enable their classification. However, different movement phenomena, such as the result of type IV pili (T4P), are still being investigated. In 1972, the different types of microbial translocation were characterised and named [29]. Five of the original terms (swimming, swarming, twitching, gliding, and sliding) are still widely used [30]. Differences in bacterial types of movement are shown in Figure 1.
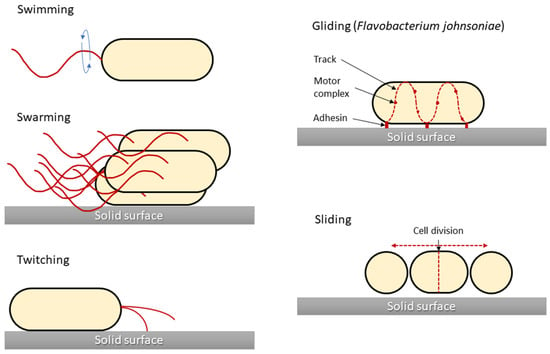
Figure 1.
Graphical presentation of five bacterial movement types. Structures responsible for generating bacterial movement are marked in red. Sliding motility is presented for Flavobacterium johnsoniae as a model for this type of motility, although for other bacteria (i.e., Myxococcus xanthus) there are differences in the cell propulsion mechanism. The figure was prepared based on the data and figures presented in [31].
2.1. Swimming Motility
Swimming motility is one of the basic skills in the life cycle of bacteria, giving them an advantage in surviving unfavourable environmental conditions [32]. Swimming motility is an individual cell movement that uses flagella rotation to move through aqueous environments [33,34]. Flagellated bacteria typically swim in a series of more or less straight runs interrupted by short reorientations [35]. Each flagellum is rotated by a motor that is embedded in the cell membrane. Flagella are highly complex bacterial organelles that are unusually well conserved among diverse bacterial species. Over 60 structural and regulatory proteins are involved in flagellum synthesis and function [36]. There are three elements in the structure of flagella found in prokaryotes: the fibre, the hook, and the basal body. A cell may have one, two, or more flagella on the front or back of the cell, over part of it, or covering its entire surface. The outer part of the cilia consists of the flagellin protein, of which there are approximately 20,000 subunits in a single cilium. The flagellum is anchored in the bacterial cell by a basic body consisting of the L, P, S, M, and C rings surrounding the cylindrical part. Gram-positive bacteria do not have the two outer L and P rings, which distinguishes them from Gram-negative bacteria with five rings [37]. The flagella fibre is connected to the basic body with a flexible section called a hook, the length of which is approximately 55 nm [38]. The direction and regulation of flagellar rotation enable bacteria to move in chemical gradients, termed chemotaxis [35,39]. This complex behaviour begins in cell membrane chemoreceptors, which detect chemical compounds and respond to them by changing their conformation. E. coli has five such receptors (Tar, Tsr, Tap, Trg, and Aer), which are arranged in chemoreceptor clusters, together with two cytoplasmic proteins, the adaptor CheW and the histidine autokinase CheA [40]. Motility regulation by the chemotaxis system is well investigated for a variety of bacterial species. Chemical gradients are sensed by chemoreceptors, which trigger the autophosphorylation of the cytoplasmic CheA, which forms a complex with the receptor through the coupling protein CheW [33,41]. In response to these changes, CheA transfers its phosphate group to CheY, a diffusible cytoplasmic response regulator that interacts with the flagellar motor and leads to a modulation of motility characteristics [33,41]. This signalling core is highly conserved among all chemotaxis pathways [42].
Although only a fraction of bacteria associated with animal hosts are motile, flagellar motility and chemotaxis are important for the successful colonisation and virulence of many pathogens, for example, gastrointestinal Campylobacter jejuni, Salmonella enterica serovar Typhimurium, Helicobacter pylori, and Vibrio cholerae [41]. In several cases, it has also been shown that the same flagellin, the protein subunit comprising the bacterial flagellum, can be a major driver of inflammation [35].
Motility might have several functions in the host–microbe interactions. For example, in the early stages of biofilm formation, planktonic bacteria swim close to the surface by rotating their flagella and attaching to the surface using their pili [43]. The most common opportunistic pathogen in wound infections is P. aeruginosa. Many studies have demonstrated that P. aeruginosa biofilms are the key factors for exacerbating the skin inflammatory response and its resistance to antimicrobial agents [13,36].
Bacteria are inextricably associated with wound healing. However, the latest research indicates that they are not always associated with infection and worse wound healing. It has been reported that the skin’s natural microbiome can contribute to more rapid wound healing. Several studies were performed with mice infected with staphylococci. Interestingly, S. aureus was found to be the best healing inducer.
Sometimes, swimming bacteria are used by non-flagellated bacteria that do not have the capacity to independently translocate with this mechanism. The genus Staphylococcus, for example, is classically considered non-motile in fluid environments due to a lack of flagella. Despite their motility limitations, staphylococcal species effectively reach and thrive in their preferred ecological niches. Data suggested that S. aureus has acquired, through P. aeruginosa, an increased capacity to travel longer distances, allowing it to colonise niches that are relatively inaccessible in the absence of swimming carrier bacteria, which may be due to the hitchhiking of S. aureus on P. aeruginosa. It was also observed that P. aeruginosa can carry another staphylococcal cargo, Staphylococcus epidermidis, which may also be important in wound infection [44].
2.2. Swarming Motility
Swarming motility is mainly based on the differentiation of vegetative cells into swarming cells with a large number of flagella, which can migrate rapidly in a coordinated manner over solid surfaces. Bacteria display different phenotypes during swarming, but not all of them are equal: swarming P. aeruginosa do not develop typical swarmer cell phenotypes (elongated, hyper-flagellated cells) similar to other swarming bacteria, and individual cells swarming in rafts also reveal great variance and the lack of a highly differentiated swarmer cell phenotype [45]. Swarming phenotypes vary not only within a species but also within a strain, whereby the different P. aeruginosa strains grown on identical medium conditions will display different swarming patterns [46]. A swarm of migrating bacteria moves forward and traps a water reservoir, and in this moist region, individual cell speed is comparable to swimming speeds in bulk liquid, typically in the order of 20 μm/sec [47]. Bacterial swarming plays a crucial role in many pathogen–host interactions, and is considered an important virulence factor. The differentiation during swarming into the hyper-flagellated elongated cell of P. mirabilis is coupled to the ability of this bacterium to enter host cells, with the manifestation of upregulated virulence protein expression (haemolysin, urease, and protease) [48,49].
The stimuli that are necessary to control this movement are responsive to cell density, surface contact, and physiological signals. The swarming motility needs signals from the quorum sensing system and the cyclic di-GMP network that regulates the transitions between motile and biofilm modes across many bacterial species [50]. These signals activate flagella biogenesis via the main flagellar operon flhDC, which is the main point of the regulatory network of cell differentiation and migration and is crucial for swarming motility [48,51]. Surface sensing occurs in two proposed ways, through the inhibition of flagellar rotation and/or through the detection of the O antigen contact of lipopolysaccharide (LPS) with a solid surface [51] in Gram-negative bacteria. In addition, LPS is suggested to act as an osmolarity agent and facilitate swarming [52].
The swarming phenomenon is characterised by bacterial growth in the form of zones with clearly delimited darker peripheral circles on an agar medium. In a typical swarming, after contact with a surface, microbial cells are able to extend up to 40 times and increase the number of flagella. In the following stage, cells divide into short cells that are unable to move. The cycles of cell elongation, movement, and division are repeated constantly, resulting in a characteristic “pattern” of their growth on the substrate, in which the darker rings reflect the periods of bacterial division. Changes in cell length and shape result from changes in the hardness of the surface on which the cells grow, and the presence of LPS, which participates in the regulation of osmolarity, helps them overcome the environmental barriers that inhibit bacterial mobility. Among others, E. coli, Salmonella spp., P. mirabilis, Bacillus subtilis, and P. aeruginosa are able to migrate in this way. Compared with swimming, chemotaxis is unnecessary here, and the oscillating motion is mainly based on the flagellar drive and the mechanical interactions taking place [53]. The swarming motility is also closely associated with the resistance of bacterial cells, which are antibiotic-sensitive, providing cells with greater availability of nutrients and competitive advantage due to secreted surfactants [48]. For example, the multi-resistant phenotype of P. aeruginosa is closely associated with swarming growth and has a transient form [54]. Currently, research on this motility is focused on obtaining more detailed information about their lipids, proteins, and enzymes, as well as their relationships, the metabolic pathways involved, and methods of regulation at the molecular level [55].
2.3. Twitching Motility
When a single cell adheres to a solid surface, it may initiate a type of surface-associated motility termed twitching. This is a movement across abiotic and biotic surfaces such as glass, plastic, or skin tissues, which is mediated by hair-like, multi-subunit compounds T4P [56,57,58]. T4P machinery is a subclass of the type IV filament superfamily closely associated with the type II secretion system (T2SS) of Gram-negative bacteria as well as the competence pilus of Gram-positive bacteria and the archaellum in Archaea [59,60]. T4P are widespread in bacteria, including important human pathogens such as P. aeruginosa, Neisseria gonorrhoeae, and V. cholerae [61], and play an important role in, for example, cell–cell adherence, cell-surface adherence, DNA transfer in the environment, predation, surface sensing, and motility [31].
The mechanism of twitching motility is based on cycles of rapid extension, adhesion to the surface, and the retraction of T4P at rates of approximately 1 μm/s [31]. Although the T4P assembly system is conserved between bacterial species, the nomenclature of T4P proteins is very heterogeneous [62]. Ten conserved proteins have been described for Myxococcus xanthus, as revealed by cryo-electron tomography. Nine of these form an envelope-spanning complex termed the basal body [63]. On the cytoplasmic side, the basal body consists of a cytoplasmic dome (PilC) separating two antagonistically acting ATPases (PilT and PilB) forming a disk-like structure and acting as a motor, providing energy for pilus assembly. The cytoplasmic disk is surrounded by a cytoplasmic ring (PilM) and is interconnected with two distinct periplasmic rings, the lower periplasmic ring (PilN and PilO) and the mid periplasmic ring (PilP), and with one ring immediately below the OM (TsaP) surrounding the channel (PilQ) of the OM pore. The final protein PilA is the major N-terminal hydrophobic α-helix protein that forms the extracellular pilus fibre [63]. PilA is not the only protein that forms the extracellular fibre, because there are some minor pilins that form a priming complex and, therefore, localise at the pilus tip. Different minor pilins are involved in different biological functions, for example, aggregation via pilus–pilus interactions and the acquisition of external DNA and motility (the structure and function of minor pilins of T4P) [58,62,63]. Extracellular fibre filaments range from thin (6–9 nm in diameter) to long (several microns) and can expand to be many times longer than the cell [64].
Although the term twitching refers to fast incoherent movements, there are studies that show that single cells moving with T4P can move in one direction for over an hour, and individual T4P compounds generate a force of approximately 100–150 pN [31,65,66,67]. Moreover, the cell speed during twitching may vary from approximately 0.02 µm/s to 0.16 µm/s, which depends on environmental conditions, for example, the number of pili involved, the surface, oxygen conditions, and the chemotactic factor [33,40,41,43]. Studies indicated that twitching can be induced chemotactically and/or mechanotactically; in P. aeruginosa, this signalling is suggested in both cases to be attributed to the Chp operon [66,68].
Spherical bacterial forms distribute the T4P over the entire cell surface, and movement is coordinated through a tug-of-war mechanism, whereas rod-shaped bacteria have T4P located on the poles [31,67]. On the basis of a time-averaged tilt angle of the bacterial long axis with respect to the surface, and the slope of the mean-squared displacement curve of the bacterial trajectory, three types of twitching motility can be distinguished. Walking upright is a movement of a cell attached to the surface with one pole. The frequent alteration of their twitching directions results in a high angular speed movement. Wiggling is a movement when bacterial cells display bipolar attachment, and this normally generates low net translational motion. Crawling is a movement when bacteria also display bipolar attachment and tends to generate high net translational motion. In comparison to walking, both crawling and wiggling cells have low angular speeds [69].
Twitching is associated with tissue invasion and virulence [70]. A correlation between wound area and bacterial twitching has been demonstrated [16]. The clinical strains of P. aeruginosa isolated from burn wound patients possess T4P as the acute virulence factor required for the colonisation and establishment of burn wound infections [14]. Moreover, twitching promotes microcolonies and biofilm formation, which influences chronic wound infection development [71]. Data from the literature indicated that T4P mutations result in the formation of incorrect mature forms of biofilm. In addition, it has been suggested that twitching can be used by bacterial cells to climb on microcolonies formed by a subpopulation of immobile cells attached to the surface, creating a mushroom-like architecture of mature biofilm [72]. Negative pressure wound therapy may reduce the motility of P. aeruginosa and biofilm formation and thereby enhance wound healing [13]. Twitching also promotes the dispersal of biofilm cells, making the released bacteria susceptible to the action of antibacterial substances [73]. There are known anti-biofilm peptides stimulating twitching motility, including synthetic 1037 and natural LL-37. Low levels of iron stimulate bacterial twitching and biofilm dispersion. Lactoferrin increases twitching motility by limiting the iron level in the bacterial environment [73].
2.4. Gliding Motility
Gliding and sliding forms of motility are considered to be the ability to translocate on solid surfaces by a flagellum-independent mechanism of smooth motion along the long axes of rod-shaped bacteria [74]. The physical principles behind gliding motility involve no external appendage utilisation, and they remain poorly understood. Gliding motility is known as a horizontal translation of bacteria under zero net force [75]. On soft substrates, the thrust of bacterial cells arises from bacterial shape deformations, which result in a flow of slime along the bacterial length [75]. Gliding motility, as observed in Myxococcus xanthus, can be distinguished from A-motility, which allows individual cells to glide on a surface, and S-motility, dependent on the functional T4P movement of groups of bacterial cells that are in close proximity to each other [76]. In recent decades, a model for gliding motility was proposed where focal-adhesion complexes (FACs) that penetrate the cell envelope and are anchored on the cell to the substratum were believed to be responsible for this motility [77]. Unfortunately, this idea assumed that FACs would migrate along the cell, leading to cell wall reassembly, including peptidoglycan, which appeared unlikely to occur [78]. Other studies revealed that FACs are generated by molecular motors (AglR, AglQ, and AglS) located in the inner membrane of the cell and are proton-conducting channels similar to the MotAB flagellar stator complex and the transport channel TolQR. Moreover, further studies showed that the FACs could be considered a fluid aggregate formed by motors and cargo proteins that accumulate at a contact site with a solid surface, which generates a slight deformation of the cell envelope and subsequently generates the force that pushes the cell forward [74,79,80]. While gliding motility was intensely studied and characterised in M. xanthus, this type of motility was also reported for Bacteroidetes and the genus Mycoplasma [30], but the gliding mechanism in other bacteria may be different from that determined in myxococci [81].
2.5. Sliding Motility
Sliding motility is considered to be a passive form of bacterial movement on a solid surface, where the motion force mainly originates from the force of dividing cells pushing other cells forward. Sliding is promoted by several factors reducing the friction between cells and the substratum, including exopolysaccharides, hydrophobic proteins, and glycopeptides. This form of motility is considered to play an important role in surface colonisation by mycobacteria in the environment as well as in the host [82]. Sliding bacteria were classified by the current knowledge of different sliding mechanisms. The first group of sliding bacteria requires only the pushing force of cell division and secreted surfactant. This first group of sliding bacteria includes Serratia marcescens, P. aeruginosa, Pseudomonas syringae, Pseudomonas fluorescens, and Legionella pneumophila. L. pneumophila exhibits a form of surface translocation that is the most similar to sliding motility and is dependent on the T2SS [83]. The second group of sliding bacteria is that which requires additional secreted components such as exopolysaccharides. This type of movement is characteristic of B. subtilis and Sinorhizobium meliloti species. The bacteria belonging to the third group require a factor other than a surfactant component for sliding, and this motility occurs in S. enterica serovar Typhimurium and Mycobacterium smegmatis species [84]. A recent study showed that S. enterica serovar Typhimurium exhibits sliding motility under magnesium-limited conditions, which induces PagM protein secretion [30]. S. aureus, common in wound and skin infections, is historically defined as non-motile, but recent studies proved the passive motion abilities of this species. This passive motion was termed spreading and was distinguished from sliding by the production of specific phenol-soluble modulin (PSM) surfactants. S. aureus spreading manifests as a finger-like dendrite that emerges from the central colony on a solid agar medium. The spreading is followed by the formation of a comet-like structure, which first emerges from the central colony, and the comet heads are composed of aggregated cells joined together through a matrix of slime. PSM production is considered to be a virulence factor in S. aureus [85].
3. Conclusions
Skin and wound infections are serious and unsolved medical problems. Bacterial translocation favours recurrent infections, and bacterial motility is considered to be a major virulence factor, where the relationship between bacterial migration and chemotaxis enables bacteria to sense the source of nutrients and subsequently translocate into the niches for colonisation [48]. Most bacteria display at least one form of motility, either involving appendages (swimming, swarming, and twitching motilities) or without appendages (gliding and sliding motilities). Bacterial movement involves a number of molecular mechanisms that could serve as targets for antibacterial therapy during skin and wound treatment, enhancing the action of antibacterial and/or disinfectant agents. The main bacterial factors involved in bacterial motility are appendages such as flagella and pili, and recent studies indicate that bacterial type IV pili are a promising therapeutic target. Moreover, it was shown that hydrogel coatings can inhibit swarming and swimming motilities by distorting E. coli flagella functions, which leads to the inhibition of biofilm formation. By contrast, appendage-less bacteria need to produce large amounts of lubricants to reduce the frictional force, which could also serve as a useful target for antibacterial therapies. In addition, the chemical modification of the interaction between the bacterial cell and the substrate may affect the gliding motility by disrupting the interaction of focal adhesion sites (FACs) with the substrate, thereby inhibiting the movement of bacteria and facilitating its eradication.
Author Contributions
Conceptualisation, G.C.; writing—original draft preparation, M.G., K.Z., P.Ż., K.D.-P. and G.C.; writing—review and editing, P.Ż., K.D.-P. and G.C.; supervision, B.K. and W.K.; funding acquisition, G.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Jan Kochanowski University, grant number SUPB.RN.22.128.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schommer, N.N.; Gallo, R.L. Structure and Function of the Human Skin Microbiome. Trends Microbiol. 2013, 21, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The Human Skin Microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- DiNubile, M.J.; Lipsky, B.A. Complicated Infections of Skin and Skin Structures: When the Infection Is More than Skip Deep. J. Antimicrob. Chemother. 2004, 53, ii37–ii50. [Google Scholar] [CrossRef] [PubMed]
- Kaye, K.S.; Petty, L.A.; Shorr, A.F.; Zilberberg, M.D. Current Epidemiology, Etiology, and Burden of Acute Skin Infections in the United States. Clin. Infect. Dis. 2019, 68, S193–S199. [Google Scholar] [CrossRef] [PubMed]
- Tognetti, L.; Martinelli, C.; Berti, S.; Hercogova, J.; Lotti, T.; Leoncini, F.; Moretti, S. Bacterial Skin and Soft Tissue Infections: Review of the Epidemiology, Microbiology, Aetiopathogenesis and Treatment: A Collaboration between Dermatologists and Infectivologists. J. Eur. Acad. Dermatol. Venereol. 2012, 26, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Plewig, G.; French, L.; Ruzicka, T.; Kaufmann, R.; Hertl, M. Braun-Falco’s Dermatology, 2020th ed.; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Cates, J.E.; Mitrani-Gold, F.S.; Li, G.; Mundy, L.M. Systematic Review and Meta-Analysis to Estimate Antibacterial Treatment Effect in Acute Bacterial Skin and Skin Structure Infection. Antimicrob. Agents Chemother. 2015, 59, 4510–4520. [Google Scholar] [CrossRef]
- Olsson, M.; Järbrink, K.; Divakar, U.; Bajpai, R.; Upton, Z.; Schmidtchen, A.; Car, J. The Humanistic and Economic Burden of Chronic Wounds: A Systematic Review: The Humanistic and Economic Burden of Chronic Wounds. Wound Repair Regen. 2019, 27, 114–125. [Google Scholar] [CrossRef]
- Bessa, L.J.; Fazii, P.; di Giulio, M.; Cellini, L. Bacterial Isolates from Infected Wounds and Their Antibiotic Susceptibility Pattern: Some Remarks about Wound Infection. Int. Wound J. 2015, 12, 47–52. [Google Scholar] [CrossRef]
- Puca, V.; Marulli, R.Z.; Grande, R.; Vitale, I.; Niro, A.; Molinaro, G.; Prezioso, S.; Muraro, R.; di Giovanni, P. Microbial Species Isolated from Infected Wounds and Antimicrobial Resistance Analysis: Data Emerging from a Three-Years Retrospective Study. Antibiotics 2021, 10, 1162. [Google Scholar] [CrossRef]
- Wolcott, R.D.; Hanson, J.D.; Rees, E.J.; Koenig, L.D.; Phillips, C.D.; Wolcott, R.A.; Cox, S.B.; White, J.S. Analysis of the Chronic Wound Microbiota of 2,963 Patients by 16S RDNA Pyrosequencing. Wound Repair Regen. 2016, 24, 163–174. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Guoqi, W.; Zhirui, L.; Song, W.; Tongtong, L.; Lihai, Z.; Licheng, Z.; Peifu, T. Negative Pressure Wound Therapy Reduces the Motility of Pseudomonas Aeruginosa and Enhances Wound Healing in a Rabbit Ear Biofilm Infection Model. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2018, 111, 1557–1570. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, M.; Behrouz, B.; Irajian, G.; Mahdavi, M.; Korpi, F.; Motamedifar, M. Passive Immunization against Pseudomonas Aeruginosa Recombinant PilA in a Murine Burn Wound Model. Microb. Pathog. 2016, 101, 83–88. [Google Scholar] [CrossRef]
- Pfalzgraff, A.; Brandenburg, K.; Weindl, G. Antimicrobial Peptides and Their Therapeutic Potential for Bacterial Skin Infections and Wounds. Front. Pharmacol. 2018, 9, 281. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, G.; Morohoshi, T.; Ikeda, T.; Ohta, Y.; Sagara, H.; Huang, L.; Nagase, T.; Sugama, J.; Sanada, H. Contribution of Quorum Sensing to the Virulence of Pseudomonas Aeruginosa in Pressure Ulcer Infection in Rats. Wound Repair Regen. 2011, 19, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Clinmicrorevs, M.; Bowler, P.; Armstrong, D.G. Wound Microbiology and Associated Approaches to Wound. Clin. Microbiol. Rev. 2016, 14, 244–269. [Google Scholar] [CrossRef]
- Jawień, A.; Bartoszewicz, M.; Przondo-Mordarska, A.; Szewczyk, M.T.; Kaszuba, A.; Urbanek, T.; Staszkiewicz, W.; Sopata, M.; Kucharzewski, M.; Korzon-Burakowska, A.; et al. Wytyczne Postępowania Miejscowego i Ogólnego w Ranach Objętych Procesem Infekcji. Leczenie Ran 2012, 9, 59–75. [Google Scholar]
- White, R.; Cutting, K. Critical Colonisation of Chronic Wounds: Microbial Mechanisms. Wounds 2008, 4, 70–78. [Google Scholar]
- Han, G.; Ceilley, R. Chronic Wound Healing: A Review of Current Management and Treatments. Adv. Ther. 2017, 34, 599–610. [Google Scholar] [CrossRef]
- Huang, X.; Liang, P.; Jiang, B.; Zhang, P.; Yu, W.; Duan, M.; Guo, L.; Cui, X.; Huang, M.; Huang, X. Hyperbaric Oxygen Potentiates Diabetic Wound Healing by Promoting Fibroblast Cell Proliferation and Endothelial Cell Angiogenesis. Life Sci. 2020, 259, 118246. [Google Scholar] [CrossRef]
- Agarwal, P.; Kukrele, R.; Sharma, D. Vacuum Assisted Closure (VAC)/Negative Pressure Wound Therapy (NPWT) for Difficult Wounds: A Review. J. Clin. Orthop. Trauma 2019, 10, 845–848. [Google Scholar] [CrossRef]
- Gupta, S.; Gabriel, A.; Lantis, J.; Téot, L. Clinical Recommendations and Practical Guide for Negative Pressure Wound Therapy with Instillation. Int. Wound J. 2016, 13, 159–174. [Google Scholar] [CrossRef]
- Kloth, L.C. Discussion: Advanced Technologies to Improve Wound Healing: Electrical Stimulation, Vibration Therapy, and Ultrasound-What Is the Evidence? Plast. Reconstr. Surg. 2016, 138, 105–106. [Google Scholar] [CrossRef] [PubMed]
- Balakatounis, K.C.; Angoules, A.G. Low-Intensity Electrical Stimulation in Wound Healing: Review of the Efficacy of Externally Applied Currents Resembling the Current of Injury. Eplasty 2008, 8, e28. [Google Scholar]
- Ashrafi, M.; Alonso-Rasgado, T.; Baguneid, M.; Bayat, A. The Efficacy of Electrical Stimulation in Lower Extremity Cutaneous Wound Healing: A Systematic Review. Exp. Dermatol. 2017, 26, 171–178. [Google Scholar] [CrossRef]
- Singh, M.R.; Saraf, S.; Vyas, A.; Jain, V.; Singh, D. Innovative Approaches in Wound Healing: Trajectory and Advances. Artif. Cells Nanomed. Biotechnol. 2013, 41, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Bekara, F.; Vitse, J.; Fluieraru, S.; Masson, R.; de Runz, A.; Georgescu, V.; Bressy, G.; Labbé, J.L.; Chaput, B.; Herlin, C. New Techniques for Wound Management: A Systematic Review of Their Role in the Management of Chronic Wounds. Arch. Plast. Surg. 2018, 45, 102–110. [Google Scholar] [CrossRef]
- Henrichsen, J. Bacterial Surface Translocation: A Survey and a Classification. Bacteriol. Rev. 1972, 36, 478–503. [Google Scholar] [CrossRef]
- Shrout, J.D. A Fantastic Voyage for Sliding Bacteria. Trends Microbiol. 2015, 23, 244–246. [Google Scholar] [CrossRef]
- Wadhwa, N.; Berg, H.C. Bacterial Motility: Machinery and Mechanisms. Nat. Rev. Microbiol. 2022, 20, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Minnullina, L.; Kostennikova, Z.; Evtugin, V.; Akosah, Y.; Sharipova, M.; Mardanova, A. Diversity in the Swimming Motility and Flagellar Regulon Structure of Uropathogenic Morganella Morganii Strains. Int. Microbiol. 2022, 25, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Corral, J.; Sebastià, P.; Coll, N.S.; Barbé, J.; Aranda, J.; Valls, M. Twitching and Swimming Motility Play a Role in Ralstonia Solanacearum Pathogenicity. Msphere 2020, 5, e00740-19. [Google Scholar] [CrossRef] [PubMed]
- Miyata, M.; Robinson, R.C.; Uyeda, T.Q.P.; Fukumori, Y.; Fukushima, S.I.; Haruta, S.; Homma, M.; Inaba, K.; Ito, M.; Kaito, C.; et al. Tree of Motility—A Proposed History of Motility Systems in the Tree of Life. Genes Cells 2020, 25, 6–21. [Google Scholar] [CrossRef]
- Colin, R.; Ni, B.; Laganenka, L.; Sourjik, V. Multiple Functions of Flagellar Motility and Chemotaxis in Bacterial Physiology. FEMS Microbiol. Rev. 2021, 45, fuab038. [Google Scholar] [CrossRef]
- Li, Y.; Xia, H.; Bai, F.; Song, X.; Zhuang, L.; Xu, H.; Zhang, X.; Qiao, M. PA5001 Gene Involves in Swimming Motility and Biofilm Formation in Pseudomonas Aeruginosa. Microb. Pathog. 2020, 144, 103982. [Google Scholar] [CrossRef]
- Terashima, H.; Kawamoto, A.; Morimoto, Y.V.; Imada, K.; Minamino, T. Structural Differences in the Bacterial Flagellar Motor among Bacterial Species. Biophys. Physicobiol. 2017, 14, 191–198. [Google Scholar] [CrossRef]
- Bardy, S.L.; Ng, S.Y.M.; Jarrell, K.F. Prokaryotic Motility Structures. Microbiology 2003, 149, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Sun, E.; Liu, S.; Hancock, R.E.W. Surfing Motility: A Conserved yet Diverse Adaptation among Motile Bacteria. J. Bacteriol. 2018, 191, 5569–5576. [Google Scholar] [CrossRef]
- Schauer, O.; Mostaghaci, B.; Colin, R.; Hürtgen, D.; Kraus, D.; Sitti, M.; Sourjik, V. Motility and Chemotaxis of Bacteria-Driven Microswimmers Fabricated Using Antigen 43-Mediated Biotin Display. Sci. Rep. 2018, 8, 9801. [Google Scholar] [CrossRef]
- Schwanbeck, J.; Oehmig, I.; Groß, U.; Zautner, A.E.; Bohne, W. Clostridioides Difficile Single Cell Swimming Strategy: A Novel Motility Pattern Regulated by Viscoelastic Properties of the Environment. Front. Microbiol. 2021, 12, 715220. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Vu, A.; Lee, K.; Dahlquist, F.W. CheA-Receptor Interaction Sites in Bacterial Chemotaxis. J. Mol. Biol. 2012, 422, 282–290. [Google Scholar] [CrossRef]
- Khong, N.Z.J.; Zeng, Y.; Lai, S.K.; Koh, C.G.; Liang, Z.X.; Chiam, K.H.; Li, H.Y. Dynamic Swimming Pattern of Pseudomonas Aeruginosa near a Vertical Wall during Initial Attachment Stages of Biofilm Formation. Sci. Rep. 2021, 11, 1952. [Google Scholar] [CrossRef] [PubMed]
- Samad, T.; Billings, N.; Birjiniuk, A.; Crouzier, T.; Doyle, P.S.; Ribbeck, K. Swimming Bacteria Promote Dispersal of Non-Motile Staphylococcal Species. ISME J. 2017, 11, 1933–1937. [Google Scholar] [CrossRef] [PubMed]
- Madukoma, C.S.; Liang, P.; Dimkovikj, A.; Chen, J.; Lee, S.W.; Chen, D.Z.; Shrout, J.D. Single Cells Exhibit Differing Behavioral Phases during Early Stages of Pseudomonas Aeruginosa Swarming. J. Bacteriol. 2019, 201, e00184-19. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.D.; Hewitt, J.L.; Wolfe, L.G.; Kamatkar, N.G.; Chapman, S.M.; Diener, J.M.; Courtney, A.J.; Leevy, W.M.; Shrout, J.D. Imaging and Analysis of Pseudomonas Aeruginosa Swarming and Rhamnolipid Production. Appl. Environ. Microbiol. 2011, 77, 8310–8317. [Google Scholar] [CrossRef] [PubMed]
- Be’er, A.; Ariel, G. A Statistical Physics View of Swarming Bacteria. Mov. Ecol. 2019, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Kearns, D.B. A Field Guide to Bacterial Swarming Motility. Nat. Rev. Microbiol. 2010, 8, 634–644. [Google Scholar] [CrossRef]
- Walker, K.E.; Moghaddame-Jafari, S.; Lockatell, C.V.; Johnson, D.; Belas, R. ZapA, the IgA-Degrading Metalloprotease of Proteus Mirabilis, Is a Virulence Factor Expressed Specifically in Swarmer Cells. Mol. Microbiol. 1999, 32, 825–836. [Google Scholar] [CrossRef]
- Yan, J.; Monaco, H.; Xavier, J.B. The Ultimate Guide to Bacterial Swarming: An Experimental Model to Study the Evolution of Cooperative Behavior. Annu. Rev. Microbiol. 2019, 73, 293–312. [Google Scholar] [CrossRef]
- Morgenstein, R.M.; Szostek, B.; Rather, P.N. Regulation of Gene Expression during Swarmer Cell Differentiation in Proteus Mirabilis. FEMS Microbiol. Rev. 2010, 34, 753–763. [Google Scholar] [CrossRef]
- Toguchi, A.; Siano, M.; Burkart, M.; Harshey, R.M. Genetics of Swarming Motility in Salmonella Enterica Serovar Typhimurium: Critical Role for Lipopolysaccharide. J. Bacteriol. 2000, 182, 6308–6321. [Google Scholar] [CrossRef] [PubMed]
- Damton, N.C.; Turner, L.; Rojevsky, S.; Berg, H.C. Dynamics of Bacterial Swarming. Biophys. J. 2010, 98, 2082–2090. [Google Scholar] [CrossRef]
- Lai, S.; Tremblay, J.; Déziel, E. Swarming Motility: A Multicellular Behaviour Conferring Antimicrobial Resistance. Environ. Microbiol. 2009, 11, 126–136. [Google Scholar] [CrossRef]
- Poudel, S.; Giannone, R.J.; Farmer, A.T.; Campagna, S.R.; Bible, A.N.; Morrell-Falvey, J.L.; Elkins, J.G.; Hettich, R.L. Integrated Proteomics and Lipidomics Reveal That the Swarming Motility of Paenibacillus Polymyxa Is Characterized by Phospholipid Modification, Surfactant Deployment, and Flagellar Specialization Relative to Swimming Motility. Front. Microbiol. 2019, 10, 2594. [Google Scholar] [CrossRef] [PubMed]
- Carabelli, A.M.; Isgró, M.; Sanni, O.; Figueredo, G.P.; Winkler, D.A.; Burroughs, L.; Blok, A.J.; Dubern, J.F.; Pappalardo, F.; Hook, A.L.; et al. Single-Cell Tracking on Polymer Microarrays Reveals the Impact of Surface Chemistry on Pseudomonas Aeruginosa Twitching Speed and Biofilm Development. ACS Appl. Bio Mater. 2020, 3, 8471–8480. [Google Scholar] [CrossRef]
- Taylor, V.L.; Fitzpatrick, A.D.; Islam, Z.; Maxwell, K.L. The Diverse Impacts of Phage Morons on Bacterial Fitness and Virulence. In Advances in Virus Research; Academic Press Inc.: Cambridge, MA, USA, 2019; Volume 103, pp. 1–31. ISBN 9780128177228. [Google Scholar]
- Shanmugasundarasamy, T.; Karaiyagowder Govindarajan, D.; Kandaswamy, K. A Review on Pilus Assembly Mechanisms in Gram-Positive and Gram-Negative Bacteria. Cell Surf. 2022, 8, 100077. [Google Scholar] [CrossRef]
- Korotkov, K.V.; Sandkvist, M.; Hol, W.G.J. The Type II Secretion System: Biogenesis, Molecular Architecture and Mechanism. Nat. Rev. Microbiol. 2012, 10, 336–351. [Google Scholar] [CrossRef]
- Mercier, R.; Bautista, S.; Delannoy, M.; Gibert, M.; Guiseppi, A.; Herrou, J.; Mauriello, E.M.F.; Mignot, T. The Polar Ras-like GTPase MglA Activates Type IV Pilus via SgmX to Enable Twitching Motility in Myxococcus Xanthus. Proc. Natl. Acad. Sci. USA 2020, 117, 28366–28373. [Google Scholar] [CrossRef]
- Craig, L.; Forest, K.T.; Maier, B. Type IV Pili: Dynamics, Biophysics and Functional Consequences. Nat. Rev. Microbiol. 2019, 17, 429–440. [Google Scholar] [CrossRef]
- Jacobsen, T.; Bardiaux, B.; Francetic, O.; Izadi-Pruneyre, N.; Nilges, M. Structure and Function of Minor Pilins of Type IV Pili. Med. Microbiol. Immunol. 2020, 209, 301–308. [Google Scholar] [CrossRef]
- Chang, Y.W.; Rettberg, L.A.; Treuner-Lange, A.; Iwasa, J.; Søgaard-Andersen, L.; Jensen, G.J. Architecture of the Type IVa Pilus Machine. Science 2016, 351, aad2001. [Google Scholar] [CrossRef] [PubMed]
- Leong, C.G.; Bloomfield, R.A.; Boyd, C.A.; Dornbusch, A.J.; Lieber, L.; Liu, F.; Owen, A.; Slay, E.; Lang, K.M.; Lostroh, C.P. The Role of Core and Accessory Type IV Pilus Genes in Natural Transformation and Twitching Motility in the Bacterium Acinetobacter Baylyi. PLoS ONE 2017, 12, e0182139. [Google Scholar] [CrossRef] [PubMed]
- Zaburdaev, V.; Biais, N.; Schmiedeberg, M.; Eriksson, J.; Jonsson, A.B.; Sheetz, M.P.; Weitz, D.A. Uncovering the Mechanism of Trapping and Cell Orientation during Neisseria Gonorrhoeae Twitching Motility. Biophys. J. 2014, 107, 1523–1531. [Google Scholar] [CrossRef]
- Oliveira, N.M.; Foster, K.R.; Durham, W.M. Single-Cell Twitching Chemotaxis in Developing Biofilms. Proc. Natl. Acad. Sci. USA 2016, 113, 6532–6537. [Google Scholar] [CrossRef] [PubMed]
- Marathe, R.; Meel, C.; Schmidt, N.C.; Dewenter, L.; Kurre, R.; Greune, L.; Alexander Schmidt, M.; Müller, M.J.I.; Lipowsky, R.; Maier, B.; et al. Bacterial Twitching Motility Is Coordinated by a Two-Dimensional Tug-of-War with Directional Memory. Nat. Commun. 2014, 5, 3759. [Google Scholar] [CrossRef]
- Kühn, M.J.; Talà, L.; Inclan, Y.F.; Patino, R.; Pierrat, X.; Vos, I.; Al-Mayyah, Z.; Macmillan, H.; Negrete Jr, J.; Engel, J.N.; et al. Mechanotaxis Directs Pseudomonas Aeruginosa Twitching Motility. Proc. Natl. Acad. Sci. USA 2021, 118, e2101759118. [Google Scholar] [CrossRef]
- Xia, A.; Qian, M.; Wang, C.; Huang, Y.; Liu, Z.; Ni, L.; Jin, F. Optogenetic Modification of Pseudomonas Aeruginosa Enables Controllable Twitching Motility and Host Infection. ACS Synth. Biol. 2021, 10, 531–541. [Google Scholar] [CrossRef]
- Dalton, T.; Dowd, S.E.; Wolcott, R.D.; Sun, Y.; Watters, C.; Griswold, J.A.; Rumbaugh, K.P. An in Vivo Polymicrobial Biofilm Wound Infection Model to Study Interspecies Interactions. PLoS ONE 2011, 6, e27317. [Google Scholar] [CrossRef]
- Wu, Y.K.; Cheng, N.C.; Cheng, C.M. Biofilms in Chronic Wounds: Pathogenesis and Diagnosis. Trends Biotechnol. 2019, 37, 505–517. [Google Scholar] [CrossRef]
- Schulze, A.; Mitterer, F.; Pombo, J.P.; Schild, S. Biofilms by Bacterial Human Pathogens: Clinical Relevance—Development, Composition and Regulation—Therapeutical Strategies. Microb. Cell 2021, 8, 28–56. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Alford, M.A.; Haney, E.F. Antibiofilm Activity of Host Defence Peptides: Complexity Provides Opportunities. Nat. Rev. Microbiol. 2021, 19, 786–797. [Google Scholar] [CrossRef] [PubMed]
- Nan, B. Bacterial Gliding Motility: Rolling Out a Consensus Model. Curr. Biol. 2017, 27, R154–R156. [Google Scholar] [CrossRef]
- Tchoufag, J.; Ghosh, P.; Pogue, C.B.; Nan, B.; Mandadapu, K.K. Mechanisms for Bacterial Gliding Motility on Soft Substrates. Proc. Natl. Acad. Sci. USA 2019, 116, 25087–25096. [Google Scholar] [CrossRef] [PubMed]
- Mauriello, E.M.F.; Mignot, T.; Yang, Z.; Zusman, D.R. Gliding Motility Revisited: How Do the Myxobacteria Move without Flagella? Microbiol. Mol. Biol. Rev. 2010, 74, 229–249. [Google Scholar] [CrossRef]
- Faure, L.M.; Fiche, J.B.; Espinosa, L.; Ducret, A.; Anantharaman, V.; Luciano, J.; Lhospice, S.; Islam, S.T.; Tréguier, J.; Sotes, M.; et al. The Mechanism of Force Transmission at Bacterial Focal Adhesion Complexes. Nature 2016, 539, 530–535. [Google Scholar] [CrossRef]
- Islam, S.T.; My, L.; Jolivet, N.Y.; Belgrave, A.M.; Fleuchot, B.; Brasseur, G.; Faure, L.M.; Sharma, G.; Lemon, D.J.; Saïdi, F.; et al. CglB Adhesins Secreted at Bacterial Focal Adhesions Mediate Gliding Motility. bioRxiv 2020, 1–70. [Google Scholar] [CrossRef]
- Nan, B.; Bandaria, J.N.; Moghtaderi, A.; Sun, I.H.; Yildiz, A.; Zusman, D.R. Flagella Stator Homologs Function as Motors for Myxobacterial Gliding Motility by Moving in Helical Trajectories. Proc. Natl. Acad. Sci. USA 2013, 110, E1508–E1513. [Google Scholar] [CrossRef] [PubMed]
- Nan, B.; Zusman, D.R. Novel Mechanisms Power Bacterial Gliding Motility. Mol. Microbiol. 2016, 101, 186–193. [Google Scholar] [CrossRef]
- Spormann, A.M. Gliding Motility in Bacteria: Insights from Studies of Myxococcus xanthus. Microbiol. Mol. Biol. Rev. 1999, 63, 621–641. [Google Scholar] [CrossRef]
- Martinez, A.; Torello, S.; Kolter, R. Sliding Motility in Mycobacteria. J. Bacteriol. 1999, 181, 7331–7338. [Google Scholar] [CrossRef]
- Stewart, C.R.; Rossier, O.; Cianciotto, N.P. Surface Translocation by Legionella Pneumophila: A Form of Sliding Motility That Is Dependent upon Type II Protein Secretion. J. Bacteriol. 2009, 191, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Hölscher, T.; Kovács, Á.T. Sliding on the Surface: Bacterial Spreading without an Active Motor. Environ. Microbiol. 2017, 19, 2537–2545. [Google Scholar] [CrossRef] [PubMed]
- Pollitt, E.J.G.; Crusz, S.A.; Diggle, S.P. Staphylococcus Aureus Forms Spreading Dendrites That Have Characteristics of Active Motility. Sci. Rep. 2015, 5, 17698. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).