Microbiome and Metabolome Variation as Indicator of Social Stress in Female Prairie Voles
Abstract
1. Introduction
2. Results
2.1. Weight and Anatomical Measures
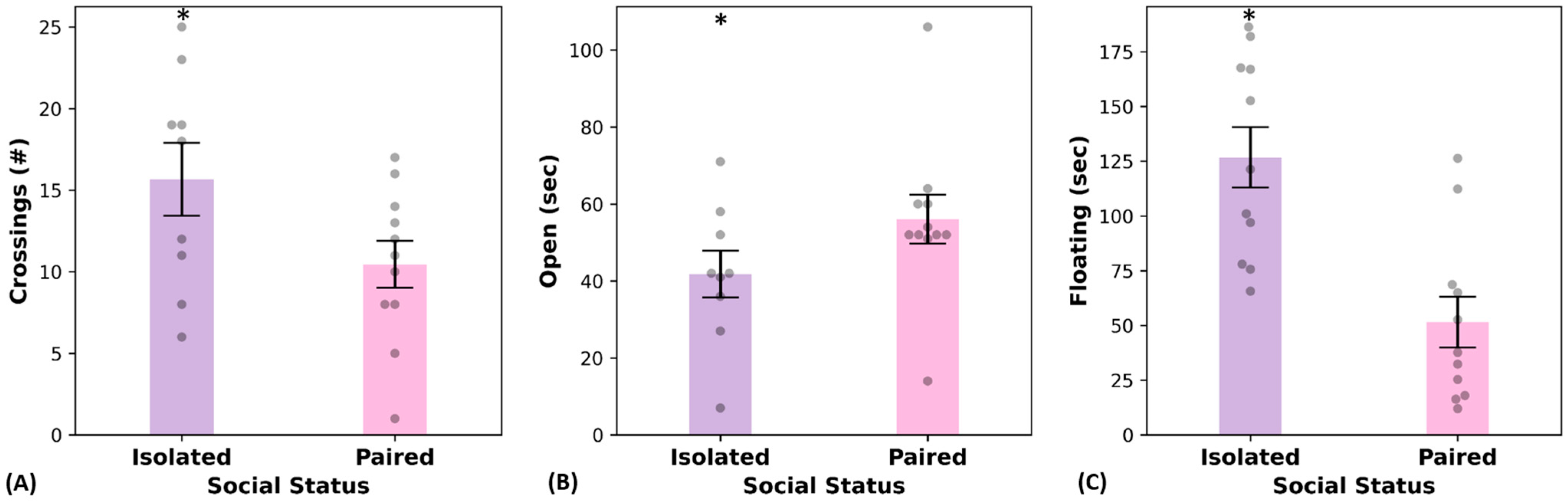
2.2. Behavioral Tests
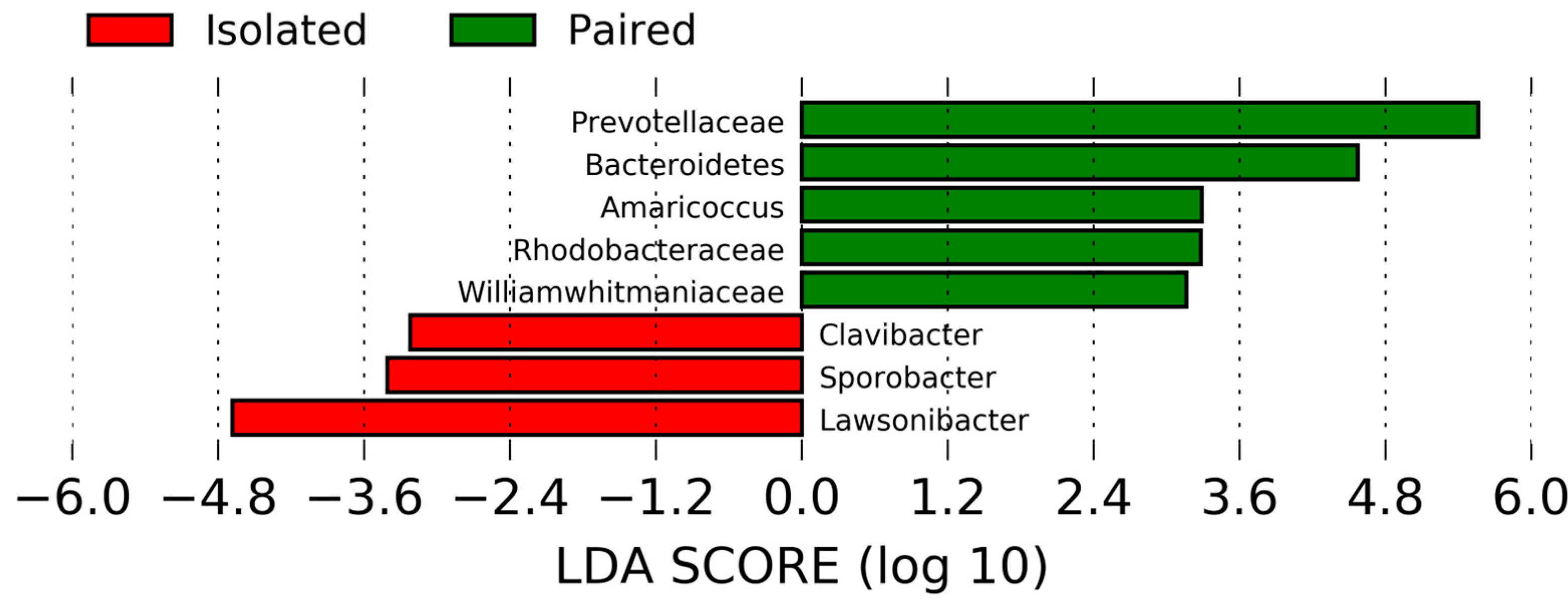
2.3. Microbiome Analyses
2.4. Fecal Time Point Analysis
2.5. Stress-Associated Fecal Community Changes with Time
2.6. Colon Microbial Community Analysis
2.7. Metabolite Profiling
2.8. Associations of Anxiety and Depressive-like Behaviors with Multi-Omics Data
3. Discussion
3.1. Social Isolation-Induced Behaviors Associated with Anxiety and Depression
3.2. Prairie Vole Fecal Microbiome Changes with Time
3.3. Isolation-Induced Changes in Gut Taxa
3.4. Prairie Vole Colon Communities Differ from Fecal Communities
3.5. Isolated Voles Exhibit Serum and Fecal Metabolomes Similar to Those of Animal Models for Type Two Diabetes and Colitis
3.6. Links between Metabolite Concentrations and Animal Behavior
3.7. Limitations and Future Directions
4. Conclusions
5. Methods
5.1. Animals and Condition Assignment
5.2. Sample Collection
5.3. Behavioral Testing
5.3.1. Elevated Plus Maze (EPM)
5.3.2. Forced Swim Test
- i.
- Swimming: coordinated fore- and hind- limb movements without breaking the surface of the water
- ii.
- Struggling: the movement of the forelimbs in a manner that broke the surface of the water
- iii.
- Climbing: attempts to scratch at or climb up the side of the FST apparatus.
- iv.
- Immobility: floating with minimal or no limb movement
5.4. Serum and Tissue Collection
5.5. Microbiome Analysis
5.6. Sequencing
5.7. Microbial Community Analyses
5.8. Metabolite Identification and Profiling
5.9. Statistical Analysis
5.9.1. Analysis of Behavioral Measures
5.9.2. Comparison of Microbial Communities
5.9.3. Comparison of Metabolites
5.9.4. Correlations between Behavior, Microbial Communities, and Metabolites
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Cacioppo, J.T.; Cacioppo, S.; Capitanio, J.P.; Cole, S.W. The neuroendocrinology of social isolation. Annu. Rev. Psychol. 2015, 66, 733–767. [Google Scholar] [CrossRef] [PubMed]
- Cacioppo, S.; Grippo, A.J.; London, S.; Goossens, L.; Cacioppo, J.T. Loneliness: Clinical import and interventions. Perspect. Psychol. Sci. 2015, 10, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Cacioppo, J.T.; Hawkley, L.C. Perceived social isolation and cognition. Trends Cognit. Sci. 2009, 13, 447–454. [Google Scholar] [CrossRef]
- Holt-Lunstad, J. Why social relationships are important for physical health: A systems approach to understanding and modifying risk and protection. Annu. Rev. Psychol. 2018, 69, 437–458. [Google Scholar] [CrossRef] [PubMed]
- Gjerstad, J.K.; Lightman, S.L.; Spiga, F. Role of glucocorticoid negative feedback in the regulation of HPA axis pulsatility. Stress 2018, 21, 403–416. [Google Scholar] [CrossRef]
- McEwen, B.S. Physiology and neurobiology of stress and adaptation: Central role of the brain. Physiol. Rev. 2007, 87, 873–904. [Google Scholar] [CrossRef] [PubMed]
- Edwards, P.D.; Mooney, S.J.; Bosson, C.O.; Toor, I.; Palme, R.; Holmes, M.M.; Boonstra, R. The stress of being alone: Removal from the colony, but not social subordination, increases fecal cortisol metabolite levels in eusocial naked mole-rats. Horm. Behav. 2020, 121, 104720. [Google Scholar] [CrossRef]
- Nonogaki, K.; Nozue, K.; Oka, Y. Social isolation affects the development of obesity and type 2 diabetes in mice. Endocrinology 2007, 148, 4658–4666. [Google Scholar] [CrossRef]
- Dunphy-Doherty, F.; O’Mahony, S.M.; Peterson, V.L.; O’Sullivan, O.; Crispie, F.; Cotter, P.D.; Wigmore, P.; King, M.V.; Cryan, J.F.; Fone, K.C.F. Post-weaning social isolation of rats leads to long-term disruption of the gut microbiota-immune-brain axis. Brain Behav. Immun. 2008, 68, 261–273. [Google Scholar] [CrossRef]
- Carter, C.S.; Getz, L.L. Monogamy and the prairie vole. Sci. Am. 1993, 268, 100–106. [Google Scholar] [CrossRef]
- Kenkel, W.M.; Gustison, M.L.; Beery, A.K. A neuroscientist’s guide to the vole. Curr. Protoc. 2021, 1, e175. [Google Scholar] [CrossRef] [PubMed]
- McGraw, L.A.; Young, L.J. The prairie vole: An emerging model organism for understanding the social brain. Trends Neurosci. 2010, 33, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Pohl, T.T.; Young, L.J.; Bosch, O.J. Lost connections: Oxytocin and the neural, physiological, and behavioral consequences of disrupted relationships. Int. J. Psychophysiol. 2019, 136, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Scotti, M.A.; Carlton, E.D.; Demas, G.E.; Grippo, A.J. Social isolation disrupts innate immune responses in both male and female prairie voles and enhances agonistic behavior in female prairie voles (Microtus ochrogaster). Horm. Behav. 2015, 70, 7–13. [Google Scholar] [CrossRef]
- Sun, P.; Smith, A.S.; Lei, K.; Liu, Y.; Wang, Z. Breaking bonds in male prairie vole: Long-term effects on emotional and social behavior, physiology, and neurochemistry. Behav. Brain Res. 2014, 265, 22–31. [Google Scholar] [CrossRef]
- Grippo, A.J.; Lamb, D.G.; Carter, C.S.; Porges, S.W. Social isolation disrupts autonomic regulation of the heart and influences negative affective behaviors. Biol. Psychiatry 2007, 62, 1162–1170. [Google Scholar] [CrossRef]
- Grippo, A.J.; Trahanas, D.M.; Zimmerman, R.R.; Porges, S.W.; Carter, C.S. Oxytocin protects against negative behavioral and autonomic consequences of long-term social isolation. Psychoneuroendocrinology 2009, 34, 1542–1553. [Google Scholar] [CrossRef]
- Donovan, M.; Mackey, C.S.; Platt, G.N.; Rounds, J.; Brown, A.N.; Trickey, D.J.; Liu, Y.; Jones, K.M.; Wang, Z. Social isolation alters behavior, the gut-immune-brain axis, and neurochemical circuits in male and female prairie voles. Neurobiol. Stress 2020, 13, 100278. [Google Scholar] [CrossRef]
- Grippo, A.J.; Cushing, B.S.; Carter, C.S. Depression-like behavior and stressor-induced neuroendocrine activation in female prairie voles exposed to chronic social isolation. Psychosom. Med. 2007, 69, 149–157. [Google Scholar] [CrossRef]
- Grippo, A.J.; Gerena, D.; Huang, J.; Kumar, N.; Shah, M.; Ughreja, R.; Carter, C.S. Social isolation induces behavioral and neuroendocrine disturbances relevant to depression in female and male prairie voles. Psychoneuroendocrinology 2007, 32, 966–980. [Google Scholar] [CrossRef]
- McNeal, N.; Appleton, K.M.; Johnson, A.K.; Scotti, M.L.; Wardwell, J.; Murphy, R.; Bishop, C.; Knecht, A.; Grippo, A.J. The protective effects of social bonding on behavioral and pituitary-adrenal axis reactivity to chronic mild stress in prairie voles. Stress 2017, 20, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Cushing, B.S.; Carter, C.S. Peripheral pulses of oxytocin increase partner preferences in female, but not male, prairie voles. Horm. Behav. 2000, 37, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Assefa, S.; Ahles, K.; Bigelow, S.; Curtis, J.T.; Köhler, G.A. Lactobacilli with probiotic potential in the prairie vole (Microtus ochrogaster). Gut Pathog. 2015, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Curtis, J.T.; Assefa, S.; Francis, A.; Köhler, G.A. Fecal microbiota in the female prairie vole (Microtus ochrogaster). PLoS ONE 2018, 13, e0190648. [Google Scholar] [CrossRef] [PubMed]
- Donovan, M.; Lynch, M.; Mackey, C.S.; Platt, G.N.; Washburn, B.K.; Vera, D.L.; Trickey, D.J.; Charles, T.C.; Wang, Z.; Jones, K.M. Metagenome-Assembled Genome Sequences of Five Strains from the Microtus ochrogaster (Prairie Vole) Fecal Microbiome. Microbiol. Resour. Announce. 2020, 9, e01310-19. [Google Scholar] [CrossRef]
- Bravo, J.A.; Forsythe, P.; Chew, M.V.; Escaravage, E.; Savignac, H.M.; Dinan, T.G.; Bienenstock, J.; Cryan, J.F. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. USA 2011, 108, 16050–16055. [Google Scholar] [CrossRef]
- Dinan, T.G.; Cryan, J.F. Microbes, immunity, and behavior: Psychoneuroimmunoloyg meets the microbiome. Neuropsychopharmachology 2017, 42, 178–192. [Google Scholar] [CrossRef]
- El Aidy, S.; Dinan, T.G.; Cryan, J.F. Immune modulation of the brain-gut-microbe axis. Front. Microbiol. 2014, 5, 146. [Google Scholar] [CrossRef]
- Lyte, M. Microbial endocrinology in the microbiome-gut-brain axis: How bacterial production and utilization of neurochemicals influence behavior. PLoS Pathog. 2013, 9, e1003726. [Google Scholar] [CrossRef]
- Normann, M.C.; Cox, M.; Akinbo, O.I.; Watanasriyakul, W.T.; Kovalev, D.; Ciosek, S.; Miller, T.; Grippo, A.J. Differential paraventricular nucleus activation and behavioral responses to social isolation in prairie voles following environmental enrichment with and without physical exercise. Soc. Neurosci. 2021, 16, 375–390. [Google Scholar] [CrossRef]
- Grippo, A.J.; Wu, K.D.; Hassan, I.; Carter, C.S. Social isolation in prairie voles induces behaviors relevant to negative affect: Toward the development of a rodent model focused on co-occurring depression and anxiety. Depress. Anxiety 2008, 25, E17–E26. [Google Scholar] [CrossRef] [PubMed]
- Shetty, S.A.; Boeren, S.; Bui, T.P.N.; Smidt, H.; de Vos, W.M. Unravelling lactate-acetate and sugar conversion into butyrate by intestinal Anaerobutyricum and Anaerostipes species by comparative proteogenomics. Environ. Microbiol. 2020, 22, 4863–4875. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Duncan, S.H.; Scott, K.P.; Louis, P. Interactions and competition within the microbial community of the human colon: Links between diet and health. Environ. Microbiol. 2007, 9, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Lau, W.L.; Vaziri, N.D. Gut microbial short-chain fatty acids and the risk of diabetes. Nat. Rev. Nephrol. 2019, 15, 389–390. [Google Scholar] [CrossRef]
- Wu, X.; Wu, Y.; He, L.; Wu, L.; Wang, X.; Liu, Z. Effects of the intestinal microbial metabolite butyrate on the development of colorectal cancer. J. Cancer 2018, 9, 2510–2517. [Google Scholar] [CrossRef]
- Aljutaily, T.; Consuegra-Fernández, M.; Aranda, F.; Lozano, F.; Huarte, E. Gut microbiota metabolites for sweetening type I diabetes. Cell. Mol. Immunol. 2018, 15, 92–95. [Google Scholar] [CrossRef]
- Soo, R.M.; Woodcroft, B.J.; Parks, D.H.; Tyson, G.W.; Hugenholtz, P. Back from the dead: The curious tale of the predatory cyanobacterium Vampirovibrio chlorellavorus. PeerJ 2015, 3, e968. [Google Scholar] [CrossRef]
- Dwidar, M.; Monnappa, A.K.; Mitchell, R.J. The dual probiotic and antibiotic nature of Bdellovibrio bacteriovorus. BMB Rep. 2012, 45, 71–78. [Google Scholar] [CrossRef]
- Kadouri, D.E.; To, K.; Shanks, R.M.; Doi, Y. Predatory bacteria: A potential ally against multidrug-resistant Gram-negative pathogens. PLoS ONE 2013, 8, e63397. [Google Scholar] [CrossRef]
- Di Rienzi, S.C.; Sharon, I.; Wrighton, K.C.; Koren, O.; Hug, L.A.; Thomas, B.C.; Goodrich, J.K.; Bell, J.T.; Spector, T.D.; Banfield, J.F.; et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife 2013, 2, e01102. [Google Scholar] [CrossRef]
- Hu, C.; Rzymski, P. Non-photosynthetic Melainabacteria (Cyanobacteria) in human gut: Characteristics and association with health. Life 2022, 12, 476. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.; Song, J.; Pei, L.; Ling, Y. Comparison of the gut microbiota disturbance in rat models of irritable bowel syndrome induced by maternal separation and multiple early-life adversity. Front. Cell. Infect. Microbiol. 2021, 14, 581974. [Google Scholar] [CrossRef]
- Bui, T.; Mannerås-Holm, L.; Puschmann, R.; Wu, H.; Troise, A.D.; Nijsse, B.; Boeren, S.; Bäckhed, F.; Fiedler, D.; Devos, W.M. Conversion of dietary inositol into propionate and acetate by commensal Anaerostipes associates with host health. Nat. Commun. 2021, 12, 4798. [Google Scholar] [CrossRef] [PubMed]
- Chia, L.W.; Mank, M.; Blijenberg, B.; Aalvink, S.; Bongers, R.S.; Stahl, B.; Knol, J.; Belzer, C. Bacteroides thetaiotaomicron fosters the growth of butyrate-producing Anaerostipes caccae in the presence of lactose and total human milk carbohydrates. Microorganisms 2020, 8, 1513. [Google Scholar] [CrossRef] [PubMed]
- Kant, R.; Rasinkangas, P.; Satokari, R.; Pietilä, T.E.; Palva, A. Genome sequence of the butyrate-producing anaerobic bacterium Anaerostipes hadrus PEL 85. Genome Announc. 2015, 3, e00224-15. [Google Scholar] [CrossRef] [PubMed]
- Zawistowska-Rojek, A.; Kośmider, A.; Stępień, K.; Tyski, S. Adhesion and aggregation properties of Lactobacillaceae strains as protection ways against enteropathogenic bacteria. Arch. Microbiol. 2022, 204, 285. [Google Scholar] [CrossRef]
- Fiore, E.; Van Tyne, D.; Gilmore, M.S. Pathogenicity of Enterococci. Microbiol. Spectr. 2019, 7, 4. [Google Scholar] [CrossRef]
- Ricaboni, D.; Mailhe, M.; Cadoret, F.; Benezech, A.; Fournier, P.E.; Raoult, D. ‘Metaprevotella massiliensis’ gen. nov., sp. nov., isolated from human ileum. New Microbes New Infect. 2016, 17, 33–35. [Google Scholar] [CrossRef]
- Scheperjans, F.; Aho, V.; Pereira, P.A.; Koskinen, K.; Paulin, L.; Pekkonen, E.; Haapaniemi, E.; Kaakkola, S.; Eerola-Rautio, J.; Pohja, M.; et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 2015, 30, 350–358. [Google Scholar] [CrossRef]
- Doumatey, A.P.; Adeyemo, A.; Zhou, J.; Lei, L.; Adebamowo, S.N.; Adebamowo, C.; Rotimi, C.N. Gut microbiome profiles are associated with type 2 diabetes in urban Africans. Front. Cell. Infect. Microbiol. 2020, 10, 63. [Google Scholar] [CrossRef]
- Ley, R. Prevotella in the gut: Choose carefully. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 69–70. [Google Scholar] [CrossRef] [PubMed]
- Bowerman, K.L.; Rehman, S.F.; Vaughan, A.; Lachner, N.; Budden, K.F.; Kim, R.Y.; Wood, D.; Gellatly, S.L.; Shukla, S.D.; Wood, L.G.; et al. Disease-associated gut microbiome and metabolome changes in patients with chronic obstructive pulmonary disease. Nat. Commun. 2020, 11, 5886. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Jung, Y.H.; Jin, Y.; Kang, S.; Jang, C.; Lee, J. A comprehensive metabolomics investigation of hippocampus, serum, and feces affected by chronic fluoxetine treatment using the chronic unpredictable mild stress mouse model of depression. Sci. Rep. 2019, 9, 7566. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, X.; Zhang, Y.; Zheng, K.; Xiang, Q.; Chen, N.; Chen, Z.; Zhang, N.; Zhu, J.; He, Q. Antibiotic-induced disruption of gut microbiota alters local metabolomes and immune responses. Front. Cell. Infect. Microbiol. 2019, 9, 99. [Google Scholar] [CrossRef]
- Li, J.; Jia, X.; Cui, W.; Wu, C.; Qin, X. Altered gut metabolome contributes to depression-like behaviors in rats exposed to chronic unpredictable mild stress. Transl. Psychiatry 2019, 9, 40. [Google Scholar] [CrossRef]
- Shi, X.; Wei, X.; Yin, X.; Wang, Y.; Zhang, M.; Zhao, C.; Zhao, H.; McClain, C.J.; Feng, W.; Zhang, X. Hepatic and fecal metabolomic analysis of the effects of Lactobacillus rhamnosus GG on alcoholic fatty liver disease in mice. J. Proteome Res. 2015, 14, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Osaka, T.; Moriyama, E.; Arai, S.; Date, Y.; Yagi, J.; Kikuchi, J.; Tsuneda, S. Meta-analysis of fecal microbiota and metabolites in experimental colitic mice during the inflammatory and healing phases. Nutrients 2017, 9, 1329. [Google Scholar] [CrossRef]
- Robinson, A.M.; Gondalia, S.V.; Karpe, A.V.; Eri, R.; Beale, D.J.; Morrison, P.D.; Palombo, E.A.; Nurgali, K. Fecal microbiota and metabolome in a mouse model of spontaneous chronic colitis: Relevance to human inflammatory bowel disease. Inflamm. Bowel Dis. 2016, 22, 2767–2787. [Google Scholar] [CrossRef]
- Mora-Ortiz, M.; Nuñez Ramos, P.; Oregioni, A.; Claus, S.P. NMR metabolomics identifies over 60 biomarkers associated with Type II Diabetes impairment in db/db mice. Metabolomics 2019, 15, 89. [Google Scholar] [CrossRef]
- Zhao, L.; Lou, H.; Peng, Y.; Chen, S.; Zhang, Y.; Li, X. Comprehensive relationship between gut microbiome and faecal metabolome in individuals with type 2 diabetes and its complications. Endocrine 2019, 66, 526–537. [Google Scholar] [CrossRef]
- Nikiforova, V.J.; Giesbertz, P.; Wiemer, J.; Bethan, B.; Looser, R.; Liebenberg, V.; Noppinger, P.R.; Daniel, H.; Rein, D. Glyoxylate, a new marker metabolite of type 2 diabetes. J. Diabetes Res. 2014, 2014, 685204. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Fritsche, J.; Wang, J.; Chen, J.; Rittig, K.; Schmitt-Kopplin, P.; Fritsche, A.; Häring, H.; Schleicher, E.D.; Xu, G.; et al. Metabonomic fingerprints of fasting plasma and spot urine reveal human pre-diabetic metabolic traits. Metabolomics 2010, 6, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Preston, G.M.; Calle, R.A. Elevated serum sorbitol and not fructose in type 2 diabetic patients. Biomark. Insights 2010, 4, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.M.; Hu, Z.; Klimko, C.; Narayanan, S.; Deberardinis, R.; Sperandio, V. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 2014, 16, 759–769. [Google Scholar] [CrossRef]
- Ferreyra, J.A.; Wu, K.J.; Hryckowian, A.J.; Bouley, D.M.; Weimer, B.C.; Sonnenburg, J.L. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 2014, 16, 770–777. [Google Scholar] [CrossRef]
- Khan, S.; Waliullah, S.; Godfrey, V.; Khan, M.A.W.; Ramachandran, R.A.; Cantarel, B.L.; Behrendt, C.; Peng, L.; Hooper, L.V.; Zaki, H. Dietary simple sugars alter microbial ecology in the gut and promote colitis in mice. Sci. Transl. Med. 2020, 12, eaay6218. [Google Scholar] [CrossRef]
- Karen, C.; Shyu, D.; Rajan, K.E. Lactobacillus paracasei supplementation prevents early life stress-induced anxiety and depressive-like behavior in maternal separation model-possible involvement of microbiota-gut-brain axis in differential regulation of MicroRNA124a/132 and glutamate receptors. Front. Neurosci. 2021, 15, 719933. [Google Scholar] [CrossRef]
- Contreras, C.M.; Rodríguez-Landa, J.F.; Gutiérrez-García, A.G.; Mendoza-López, M.R.; García-Ríos, R.I.; Cueto-Escobedo, J. Anxiolytic-like effects of human amniotic fluid and its fatty acids in Wistar rats. Behav. Pharmacol. 2011, 22, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Contreras, C.M.; Rodríguez-Landa, J.F.; García-Ríos, R.I.; Cueto-Escobedo, J.; Guillen-Ruiz, G.; Bernal-Morales, B. Myristic acid produces anxiolytic-like effects in Wistar rats in the elevated plus maze. Biomed Res. Int. 2014, 492141. [Google Scholar] [CrossRef]
- Van de Wouw, M.; Boehme, M.; Lyte, J.M.; Wiley, N.; Strain, C.; O’Sullivan, O.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Short-chain fatty acids: Microbial metabolites that alleviate stress-induced brain-gut axis alterations. J. Physiol. 2018, 596, 4923–4944. [Google Scholar] [CrossRef]
- Gundersen, B.B.; Blendy, J.A. Effects of the histone deacetylase inhibitor sodium butyrate in models of depression and anxiety. Neuropharmacology 2009, 57, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Cranford, J.A.; Johnson, E.O. Effects of coprophagy and diet quality on two microtine rodents (Microtus pennsylvanicus and Microtus pinetorum). J. Mammal. 1989, 70, 494–502. [Google Scholar] [CrossRef]
- Lee, W.B.; Houston, D.C. The role of coprophagy in digestion in voles (Microtus agrestis and Clethrionomys glareolus). Funct. Ecol. 1993, 7, 427–432. [Google Scholar] [CrossRef]
- National Research Council. Nutrient Requirements of Laboratory Animals, 4th ed.; National Academies Press: Washington, DC, USA, 1995.
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. PNAS 2009, 106, 3698–3703. [Google Scholar] [CrossRef]
- Carter, C.S.; Witt, D.M.; Schneider, J.; Harris, Z.L.; Volkening, D. Male stimuli are necessary for female sexual behavior and uterine growth in prairie voles (Microtus ochrogaster). Horm. Behav. 1987, 21, 74–82. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Van Rossum, G.; Drake, F.L. Python 3 Reference Manual; CreateSpace: Scotts Valley, CA, USA, 2009. [Google Scholar]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- McKinney, W. Data structures for statistical computing in python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 51–56. [Google Scholar]
- Scikit-Bio Development Team. Scikit-Bio: A Bioinformatics Library for Data Scientists, Students, and Developers, Version 0.5.6. 2020. Available online: http://scikit-bio.org (accessed on 5 March 2022).
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M.L. Seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]

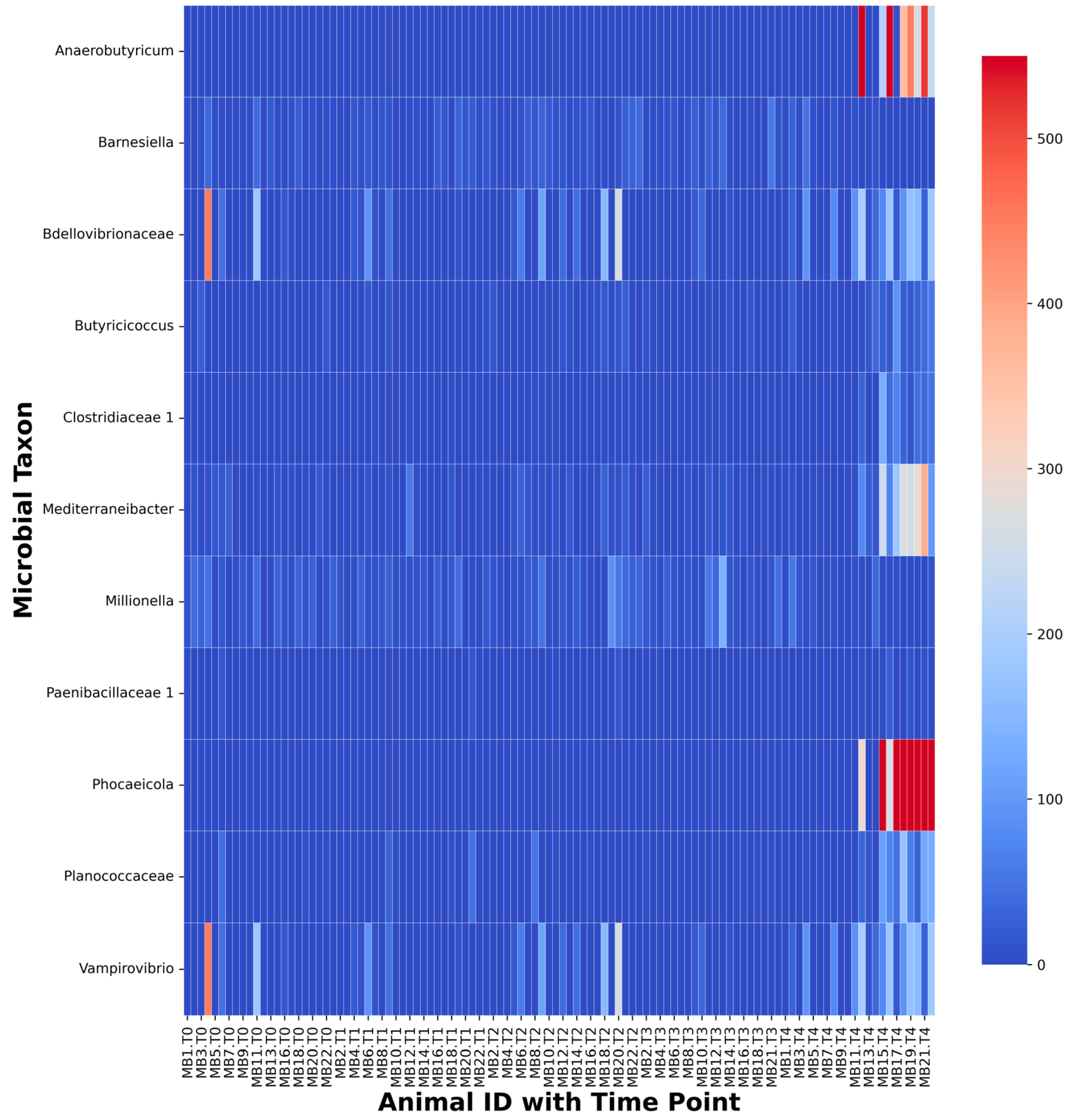

| Taxon | Stats | Proportional Abundances | |||||
|---|---|---|---|---|---|---|---|
| Q Test | p-Value | Median T0 | Median T1 | Median T2 | Median T3 | Median T4 | |
| Clostridiaceae 1 | 21.726 | <0.001 | 0 | 1.0185E − 05 | 4.0718E − 06 | 0 | 4.8021E − 05 |
| Paenibacillaceae 1 | 19.84 | 0.001 | 0 | 0 | 0 | 0 | 1.4675E − 05 |
| Planococcaceae | 16.416 | 0.003 | 0 | 0 | 0 | 0 | 0.00018553 |
| Bdellovibrionaceae | 16.486 | 0.002 | 1.0146E − 05 | 0 | 0 | 0 | 0.00027892 |
| Ruminococcus | 14.8 | 0.005 | 0.17876649 | 0.12738538 | 0.141373 | 0.15814367 | 0.07193627 |
| Barnesiella | 16.364 | 0.003 | 0.0001477 | 0.0003441 | 0.00039466 | 0.00030383 | 8.3054E − 05 |
| Millionella | 13.88 | 0.008 | 0.00031359 | 0.0004766 | 0.00047283 | 0.00066886 | 0.00013524 |
| Butyricicoccus | 13.055 | 0.011 | 0 | 1.2102E − 05 | 7.9223E − 05 | 1.3882E − 05 | 0.00018001 |
| Vampirovibrio | 16.487 | 0.002 | 2.0047E − 05 | 0 | 0 | 0 | 0.00062135 |
| Mediterranea | 19.304 | 0.001 | 0 | 0 | 0 | 0 | 8.0281E − 05 |
| Anaerobutyricum | 27.333 | <0.001 | 0 | 0 | 0 | 0 | 2.9015E − 05 |
| Phocaeicola | 15.843 | 0.003 | 0 | 0 | 0 | 0 | 1.9131E − 05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nuccio, D.A.; Normann, M.C.; Zhou, H.; Grippo, A.J.; Singh, P. Microbiome and Metabolome Variation as Indicator of Social Stress in Female Prairie Voles. Int. J. Mol. Sci. 2023, 24, 1677. https://doi.org/10.3390/ijms24021677
Nuccio DA, Normann MC, Zhou H, Grippo AJ, Singh P. Microbiome and Metabolome Variation as Indicator of Social Stress in Female Prairie Voles. International Journal of Molecular Sciences. 2023; 24(2):1677. https://doi.org/10.3390/ijms24021677
Chicago/Turabian StyleNuccio, Daniel A., Marigny C. Normann, Haiming Zhou, Angela J. Grippo, and Pallavi Singh. 2023. "Microbiome and Metabolome Variation as Indicator of Social Stress in Female Prairie Voles" International Journal of Molecular Sciences 24, no. 2: 1677. https://doi.org/10.3390/ijms24021677
APA StyleNuccio, D. A., Normann, M. C., Zhou, H., Grippo, A. J., & Singh, P. (2023). Microbiome and Metabolome Variation as Indicator of Social Stress in Female Prairie Voles. International Journal of Molecular Sciences, 24(2), 1677. https://doi.org/10.3390/ijms24021677







