Reviewing the Potential Links between Viral Infections and TDP-43 Proteinopathies
Abstract
1. Introduction
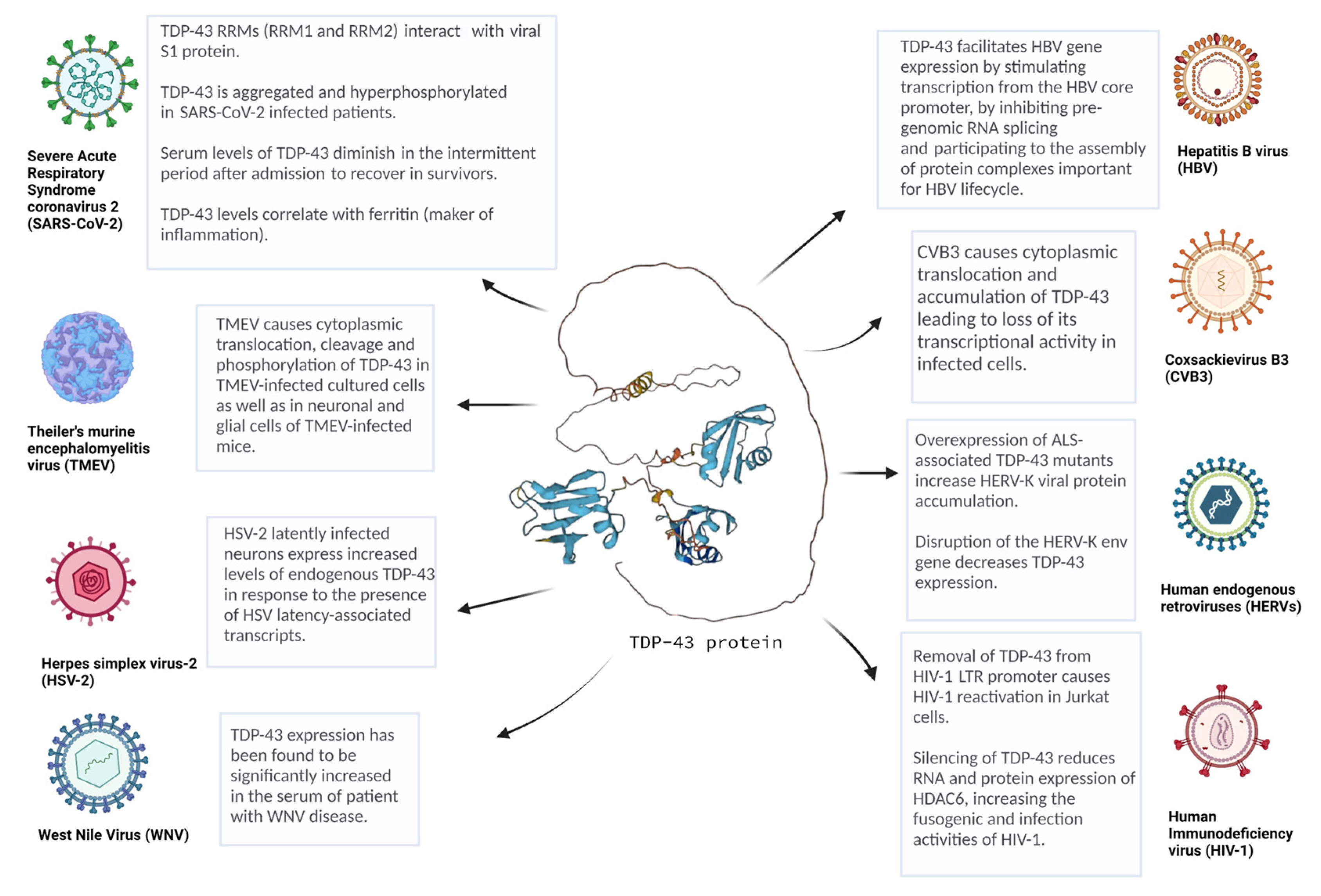
2. Viral Infections and TDP-43
2.1. Enteroviruses (EVs)
2.2. Theiler’s Murine Encephalomyelitis Virus (TMEV)
2.3. Human Immunodeficiency Virus (HIV)
2.4. Human Endogenous Retrovirus K (HERV-K)
2.5. Hepatitis B Virus (HBV)
2.6. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)
2.7. West Nile Virus (WNV)
2.8. Herpes Simplex Virus-2 (HSV-2)
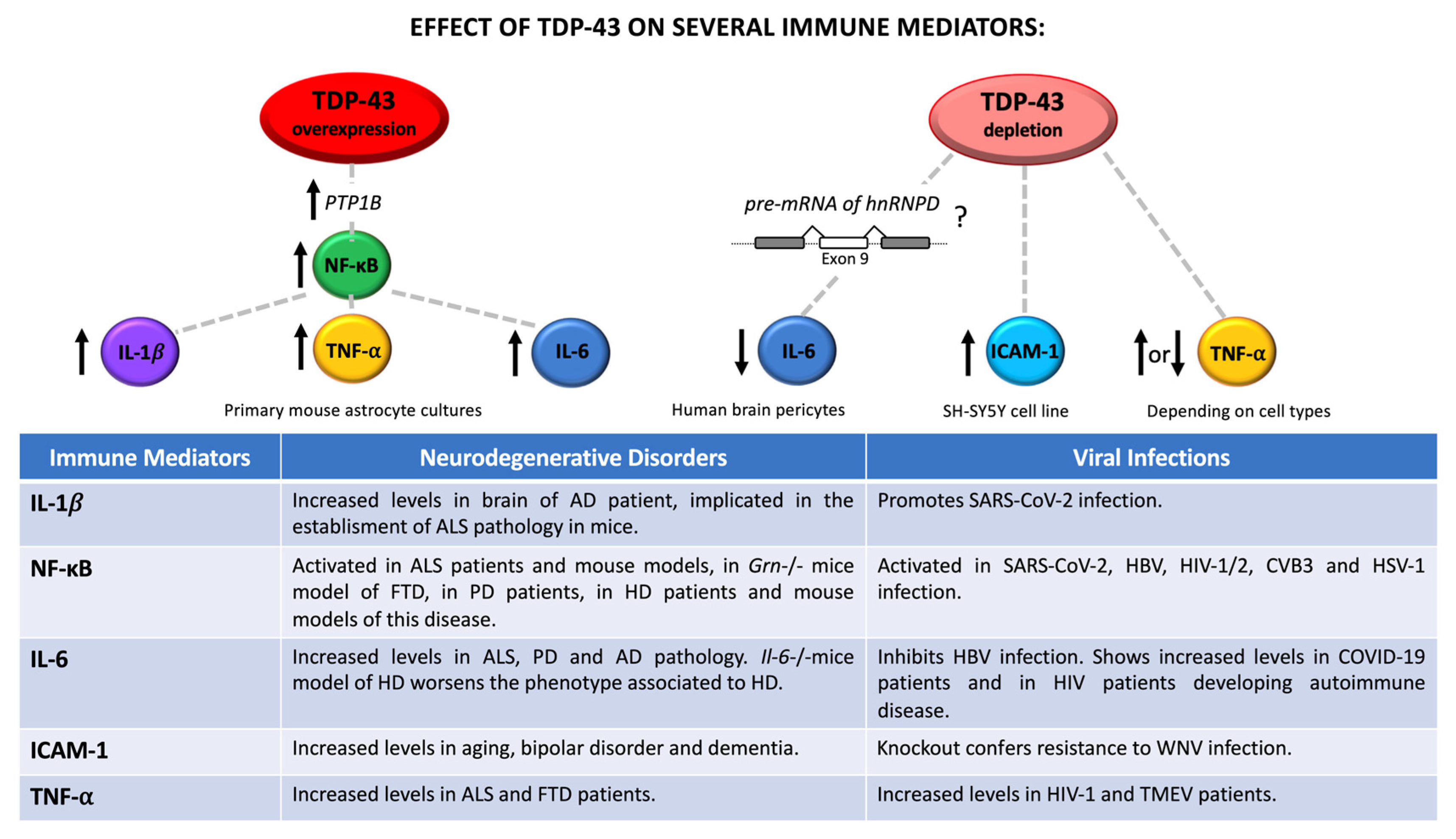
3. The Role of Neuroinflammation in Viral Infection and Neurodegeneration: A Potential Link with TDP-43 Dysfunction
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ou, S.H.; Wu, F.; Harrich, D.; García-Martínez, L.F.; Gaynor, R.B. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 1995, 69, 3584–3596. [Google Scholar] [CrossRef] [PubMed]
- Avendaño-Vázquez, S.E.; Dhir, A.; Bembich, S.; Buratti, E.; Proudfoot, N.; Baralle, F.E. Autoregulation of TDP-43 mRNA levels involves interplay between transcription, splicing, and alternative polyA site selection. Genes Dev. 2012, 26, 1679–1684. [Google Scholar] [CrossRef] [PubMed]
- Higashi, S.; Kabuta, T.; Nagai, Y.; Tsuchiya, Y.; Akiyama, H.; Wada, K. TDP-43 associates with stalled ribosomes and contributes to cell survival during cellular stress. J. Neurochem. 2013, 126, 288–300. [Google Scholar] [CrossRef]
- De Conti, L.; Akinyi, M.V.; Mendoza-Maldonado, R.; Romano, M.; Baralle, M.; Buratti, E. TDP-43 affects splicing profiles and isoform production of genes involved in the apoptotic and mitotic cellular pathways. Nucleic Acids Res. 2015, 43, 8990–9005. [Google Scholar] [CrossRef]
- Alami, N.H.; Smith, R.B.; Carrasco, M.A.; Williams, L.A.; Winborn, C.S.; Han, S.S.W.; Kiskinis, E.; Winborn, B.; Freibaum, B.D.; Kanagaraj, A.; et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron 2014, 81, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Fiesel, F.C.; Weber, S.S.; Supper, J.; Zell, A.; Kahle, P.J. TDP-43 regulates global translational yield by splicing of exon junction complex component SKAR. Nucleic Acids Res. 2012, 40, 2668–2682. [Google Scholar] [CrossRef]
- Chou, C.C.; Zhang, Y.; Umoh, M.E.; Vaughan, S.W.; Lorenzini, I.; Liu, F.; Sayegh, M.; Donlin-Asp, P.G.; Chen, Y.H.; Duong, D.M.; et al. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 2018, 21, 228–239. [Google Scholar] [CrossRef]
- Colombrita, C.; Zennaro, E.; Fallini, C.; Weber, M.; Sommacal, A.; Buratti, E.; Silani, V.; Ratti, A. TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 2009, 111, 1051–1061. [Google Scholar] [CrossRef]
- Wang, C.; Duan, Y.; Duan, G.; Wang, Q.; Zhang, K.; Deng, X.; Qian, B.; Gu, J.; Ma, Z.; Zhang, S.; et al. Stress Induces Dynamic, Cytotoxicity-Antagonizing TDP-43 Nuclear Bodies via Paraspeckle LncRNA NEAT1-Mediated Liquid-Liquid Phase Separation. Mol. Cell 2020, 79, 443–458.e7. [Google Scholar] [CrossRef]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef]
- Cohen, T.J.; Hwang, A.W.; Restrepo, C.R.; Yuan, C.X.; Trojanowski, J.Q.; Lee, V.M.Y. An acetylation switch controls TDP-43 function and aggregation propensity. Nat. Commun. 2015, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Buratti, E. TDP-43 post-translational modifications in health and disease. Expert Opin. Ther. Target. 2018, 22, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Bumber, J.R.; Pilipovi, K.; Jankovi, T.; Dolenec, P.; Grzeta, N.; Kri, J.; Zupan, G. Repetitive Traumatic Brain Injury Is Associated with TDP-43 Alterations, Neurodegeneration, and Glial Activation in Mice. J. Neuropathol. Exp. Neurol. 2021, 80, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Arnold, E.S.; Ling, S.-C.; Huelga, S.C.; Lagier-Tourenne, C.; Polymenidou, M.; Ditsworth, D.; Kordasiewicz, H.B.; McAlonis-Downes, M.; Platoshyn, O.; Parone, P.A.; et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc. Natl. Acad. Sci. USA 2013, 110, E736–E745. [Google Scholar] [CrossRef] [PubMed]
- Prudencio, M.; Humphrey, J.; Pickles, S.; Brown, A.L.; Hill, S.E.; Kachergus, J.M.; Shi, J.; Heckman, M.G.; Spiegel, M.R.; Cook, C.; et al. Truncated stathmin-2 is a marker of TDP-43 pathology in frontotemporal dementia. J. Clin. Investig. 2020, 130, 6080–6092. [Google Scholar] [CrossRef]
- Brown, A.-L.; Wilkins, O.G.; Keuss, M.J.; Hill, S.E.; Zanovello, M.; Lee, W.C.; Lee, F.C.Y.; Masino, L.; Qi, Y.A.; Bryce-Smith, S.; et al. Common ALS/FTD risk variants in UNC13A exacerbate its cryptic splicing and loss upon TDP-43 mislocalization. bioRxiv 2021. bioRxiv: 2021.04.02.438170. [Google Scholar]
- Porta, S.; Xu, Y.; Restrepo, C.R.; Kwong, L.K.; Zhang, B.; Brown, H.J.; Lee, E.B.; Trojanowski, J.Q.; Lee, V.M.Y. Patient-derived frontotemporal lobar degeneration brain extracts induce formation and spreading of TDP-43 pathology in vivo. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Mishra, P.S.; Boutej, H.; Soucy, G.; Bareil, C.; Kumar, S.; Picher-Martel, V.; Dupré, N.; Kriz, J.; Julien, J.P. Transmission of ALS pathogenesis by the cerebrospinal fluid. Acta Neuropathol. Commun. 2020, 8, 1–21. [Google Scholar] [CrossRef]
- Feiler, M.S.; Strobel, B.; Freischmidt, A.; Helferich, A.M.; Kappel, J.; Brewer, B.M.; Li, D.; Thal, D.R.; Walther, P.; Ludolph, A.C.; et al. TDP-43 is intercellularly transmitted across axon terminals. J. Cell Biol. 2015, 211, 897–911. [Google Scholar] [CrossRef]
- Nehls, J.; Koppensteiner, H.; Brack-Werner, R.; Floss, T.; Schindler, M. HIV-1 replication in human immune cells is independent of TAR DNA binding protein 43 (TDP-43) expression. PLoS ONE 2014, 9, e105478. [Google Scholar] [CrossRef]
- Cabrera-Rodríguez, R.; Pérez-Yanes, S.; Montelongo, R.; Lorenzo-Salazar, J.M.; Estévez-Herrera, J.; García-Luis, J.; Íñigo-Campos, A.; Rubio-Rodríguez, L.A.; Muñoz-Barrera, A.; Trujillo-González, R.; et al. Transactive Response DNA-Binding Protein (TARDBP/TDP-43) Regulates Cell Permissivity to HIV-1 Infection by Acting on HDAC6. Int. J. Mol. Sci. 2022, 23, 6180. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.; Iketani, S.; Wang, P.; Jia, M.; Sahi, V.; Ho, D.D. CRISPR-based gene knockout screens reveal deubiquitinases involved in HIV-1 latency in two Jurkat cell models. Sci. Rep. 2020, 10, 5350. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.C.; Feuer, R.; Cashman, N.; Luo, H. Enteroviral Infection: The Forgotten Link to Amyotrophic Lateral Sclerosis? Front. Mol. Neurosci. 2018, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Ravits, J. Sporadic amyotrophic lateral sclerosis: A hypothesis of persistent (non-lytic) enteroviral infection. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2005, 6, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.C.; Liu, H.; Mohamud, Y.; Bahreyni, A.; Zhang, J.; Cashman, N.R.; Luo, H. Sublethal enteroviral infection exacerbates disease progression in an ALS mouse model. J. Neuroinflamm. 2022, 19, 1–15. [Google Scholar] [CrossRef]
- Fung, G.; Shi, J.; Deng, H.; Hou, J.; Wang, C.; Hong, A.; Zhang, J.; Jia, W.; Luo, H. Cytoplasmic translocation, aggregation, and cleavage of TDP-43 by enteroviral proteases modulate viral pathogenesis. Cell Death Differ. 2015, 22, 2087–2097. [Google Scholar] [CrossRef] [PubMed]
- Tolbert, M.; Morgan, C.E.; Pollum, M.; Crespo-Hernández, C.E.; Li, M.L.; Brewer, G.; Tolbert, B.S. HnRNP A1 Alters the Structure of a Conserved Enterovirus IRES Domain to Stimulate Viral Translation. J. Mol. Biol. 2017, 429, 2841–2858. [Google Scholar] [CrossRef]
- Lin, J.-Y.; Shih, S.-R.; Pan, M.; Li, C.; Lue, C.-F.; Stollar, V.; Li, M.-L. hnRNP A1 interacts with the 5′ untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication. J. Virol. 2009, 83, 6106–6114. [Google Scholar] [CrossRef]
- Park, N.; Schweers, N.J.; Gustin, K.E. Selective Removal of FG Repeat Domains from the Nuclear Pore Complex by Enterovirus 2A(pro). J. Virol. 2015, 89, 11069–11079. [Google Scholar] [CrossRef]
- Park, N.; Katikaneni, P.; Skern, T.; Gustin, K.E. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J. Virol. 2008, 82, 1647–1655. [Google Scholar] [CrossRef]
- Feuer, R.; Ruller, C.M.; An, N.; Tabor-Godwin, J.M.; Rhoades, R.E.; Maciejewski, S.; Pagarigan, R.R.; Cornell, C.T.; Crocker, S.J.; Kiosses, W.B.; et al. Viral persistence and chronic immunopathology in the adult central nervous system following Coxsackievirus infection during the neonatal period. J. Virol. 2009, 83, 9356–9369. [Google Scholar] [CrossRef]
- Lin, T.Y.; Hsia, S.H.; Huang, Y.C.; Wu, C.T.; Chang, L.Y. Proinflammatory cytokine reactions in enterovirus 71 infections of the central nervous system. Clin. Infect. Dis. 2003, 36, 269–274. [Google Scholar] [CrossRef]
- Shi, J.; Wong, J.; Piesik, P.; Fung, G.; Zhang, J.; Jagdeo, J.; Li, L.X.; Jan, E.; Luo, H. Cleavage of sequestosome 1/p62 by an enteroviral protease results in disrupted selective autophagy and impaired NFKB signaling. Autophagy 2013, 9, 1591–1603. [Google Scholar] [CrossRef]
- Corona, A.K.; Saulsbery, H.M.; Velazquez, A.F.C.; Jackson, W.T. Enteroviruses Remodel Autophagic Trafficking through Regulation of Host SNARE Proteins to Promote Virus Replication and Cell Exit. Cell Rep. 2018, 22, 3304–3314. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.E. Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses. Virology 2015, 479–480, 457–474. [Google Scholar] [CrossRef] [PubMed]
- Yoder, J.D.; Cifuente, J.O.; Pan, J.; Bergelson, J.M.; Hafenstein, S. The Crystal Structure of a Coxsackievirus B3-RD Variant and a Refined 9-Angstrom Cryo-Electron Microscopy Reconstruction of the Virus Complexed with Decay-Accelerating Factor (DAF) Provide a New Footprint of DAF on the Virus Surface. J. Virol. 2012, 86, 12571. [Google Scholar] [CrossRef]
- Lipton, H.L.; Friedmann, A. Purification of Theiler’s murine encephalomyelitis virus and analysis of the structural virion polypeptides: Correlation of the polypeptide profile with virulence. J. Virol. 1980, 33, 1165–1172. [Google Scholar] [CrossRef]
- Luo, M.; Toth, K.S.; Zhou, L.; Pritchard, A.; Lipton, H.L. The structure of a highly virulent Theiler’s murine encephalomyelitis virus (GDVII) and implications for determinants of viral persistence. Virology 1996, 220, 246–250. [Google Scholar] [CrossRef]
- Clatch, R.J.; Melvold, R.W.; Dal Canto, M.C.; Miller, S.D.; Lipton, H.L. The Theiler’s murine encephalomyelitis virus (TMEV) model for multiple sclerosis shows a strong influence of the murine equivalents of HLA-A, B, and C. J. Neuroimmunol. 1987, 15, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Masaki, K.; Sonobe, Y.; Ghadge, G.; Pytel, P.; Roos, R.P. TDP-43 proteinopathy in Theiler’s murine encephalomyelitis virus infection. PLoS Pathog. 2019, 15, e1007574. [Google Scholar] [CrossRef]
- Wang, J.; Ho, W.Y.; Lim, K.; Feng, J.; Tucker-Kellogg, G.; Nave, K.A.; Ling, S.C. Cell-autonomous requirement of TDP-43, an ALS/FTD signature protein, for oligodendrocyte survival and myelination. Proc. Natl. Acad. Sci. USA 2018, 115, E10941–E10950. [Google Scholar] [CrossRef]
- Correia, A.S.; Patel, P.; Dutta, K.; Julien, J.P. Inflammation Induces TDP-43 Mislocalization and Aggregation. PLoS ONE 2015, 10, e0140248. [Google Scholar] [CrossRef]
- German Advisory Committee Blood (Arbeitskreis Blut). Human Immunodeficiency Virus (HIV). Transfus. Med. Hemother. 2016, 43, 203. [Google Scholar]
- Scully, C.; Cawson, R.A.; Porter, S.R. Acquired immune deficiency syndrome: Review. Br. Dent. J. 1986, 161, 53–60. [Google Scholar] [CrossRef] [PubMed]
- So-Armah, K.; Benjamin, L.A.; Bloomfield, G.S.; Feinstein, M.J.; Hsue, P.; Njuguna, B.; Freiberg, M.S. HIV and cardiovascular disease. Lancet HIV 2020, 7, e279–e293. [Google Scholar] [CrossRef]
- Hileman, C.O.; Eckard, A.R.; McComsey, G.A. Bone loss in HIV—A contemporary review. Curr. Opin. Endocrinol. Diabetes Obes. 2015, 22, 446. [Google Scholar] [CrossRef]
- Wyatt, C.M. Kidney Disease and HIV Infection. Top. Antivir. Med. 2017, 25, 13. [Google Scholar] [PubMed]
- Sherman, K.E.; Peters, M.G.; Thomas, D. Human immunodeficiency virus and liver disease: A comprehensive update. Hepatol. Commun. 2017, 1, 987. [Google Scholar] [CrossRef] [PubMed]
- Douville, R.N.; Nath, A. Human Endogenous Retrovirus-K and TDP-43 Expression Bridges ALS and HIV Neuropathology. Front. Microbiol. 2017, 8, 1986. [Google Scholar] [CrossRef]
- Manghera, M.; Ferguson-Parry, J.; Douville, R.N. TDP-43 regulates endogenous retrovirus-K viral protein accumulation. Neurobiol. Dis. 2016, 94, 226–236. [Google Scholar] [CrossRef]
- Grandi, N.; Tramontano, E. Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses. Front. Immunol. 2018, 9, 2039. [Google Scholar] [CrossRef]
- Mayer, J.; Meese, E. Human endogenous retroviruses in the primate lineage and their influence on host genomes. Cytogenet. Genome Res. 2005, 110, 448–456. [Google Scholar] [CrossRef]
- Perron, H.; Hamdani, N.; Faucard, R.; Lajnef, M.; Jamain, S.; Daban-Huard, C.; Sarrazin, S.; Leguen, E.; Houenou, J.; Delavest, M.; et al. Molecular characteristics of Human Endogenous Retrovirus type-W in schizophrenia and bipolar disorder. Transl. Psychiatry 2012, 2, e201. [Google Scholar] [CrossRef]
- García-Montojo, M.; de la Hera, B.; Varadé, J.; de la Encarnación, A.; Camacho, I.; Domínguez-Mozo, M.; Arias-Leal, A.; García-Martínez, Á.; Casanova, I.; Izquierdo, G.; et al. HERV-W polymorphism in chromosome X is associated with multiple sclerosis risk and with differential expression of MSRV. Retrovirology 2014, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Guo, X.; Li, J.; Han, J.; Jia, L.; Wen, H.L.; Sun, C.; Wang, X.; Zhang, B.; Li, J.; et al. Significant Upregulation of HERV-K (HML-2) Transcription Levels in Human Lung Cancer and Cancer Cells. Front. Microbiol. 2022, 13, 681. [Google Scholar] [CrossRef]
- Li, W.; Lee, M.H.; Henderson, L.; Tyagi, R.; Bachani, M.; Steiner, J.; Campanac, E.; Hoffman, D.A.; von Geldern, G.; Johnson, K.; et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med. 2015, 7, 307ra153. [Google Scholar] [CrossRef]
- Ko, E.J.; Ock, M.S.; Choi, Y.H.; Iovanna, J.L.; Mun, S.; Han, K.; Kim, H.S.; Cha, H.J. Human Endogenous Retrovirus (HERV)-K env Gene Knockout Affects Tumorigenic Characteristics of nupr1 Gene in DLD-1 Colorectal Cancer Cells. Int. J. Mol. Sci. 2021, 22, 3941. [Google Scholar] [CrossRef] [PubMed]
- Ibba, G.; Piu, C.; Uleri, E.; Serra, C.; Dolei, A. Disruption by SaCas9 Endonuclease of HERV-Kenv, a Retroviral Gene with Oncogenic and Neuropathogenic Potential, Inhibits Molecules Involved in Cancer and Amyotrophic Lateral Sclerosis. Viruses 2018, 10, 412. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.J. Hepatitis B: The Virus and Disease. Hepatology 2009, 49, S13. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.F.P.; Chan, M.; Chan, S.H.; Cheng, P.C.P.; Leung, E.H.C.; Chen, W.N.; Ren, E.C. Host heterogeneous ribonucleoprotein K (hnRNP K) as a potential target to suppress hepatitis B virus replication. PLoS Med. 2005, 2, 0673–0683. [Google Scholar] [CrossRef]
- Makokha, G.N.; Abe-Chayama, H.; Chowdhury, S.; Hayes, C.N.; Tsuge, M.; Yoshima, T.; Ishida, Y.; Zhang, Y.; Uchida, T.; Tateno, C.; et al. Regulation of the Hepatitis B virus replication and gene expression by the multi-functional protein TARDBP. Sci. Rep. 2019, 9, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Alnefeesi, Y.; Siegel, A.; Lui, L.M.W.; Teopiz, K.M.; Ho, R.C.M.; Lee, Y.; Nasri, F.; Gill, H.; Lin, K.; Cao, B.; et al. Impact of SARS-CoV-2 Infection on Cognitive Function: A Systematic Review. Front. Psychiatry 2021, 11, 1629. [Google Scholar] [CrossRef] [PubMed]
- Crunfli, F.; Carregari, V.C.; Veras, F.P.; Vendramini, P.H.; Valença, A.G.F.; Antunes, A.S.L.M.; Brandão-Teles, C.; da Silva Zuccoli, G.; Reis-de-Oliveira, G.; Silva-Costa, L.C.; et al. SARS-CoV-2 infects brain astrocytes of COVID-19 patients and impairs neuronal viability. medRxiv 2020, 18, 2020.10.09.20207464. [Google Scholar]
- Soung, A.L.; Vanderheiden, A.; Nordvig, A.S.; Sissoko, C.A.; Canoll, P.; Mariani, M.B.; Jiang, X.; Bricker, T.; Rosoklija, G.B.; Arango, V.; et al. COVID-19 induces CNS cytokine expression and loss of hippocampal neurogenesis. Brain 2022, 145, 4193–4201. [Google Scholar] [CrossRef] [PubMed]
- Idrees, D.; Kumar, V. SARS-CoV-2 spike protein interactions with amyloidogenic proteins: Potential clues to neurodegeneration. Biochem. Biophys. Res. Commun. 2021, 554, 94–98. [Google Scholar] [CrossRef]
- Paidas, M.J.; Cosio, D.S.; Ali, S.; Kenyon, N.S.; Jayakumar, A.R. Long-Term Sequelae of COVID-19 in Experimental Mice. Mol. Neurobiol. 2022, 59, 5970–5986. [Google Scholar] [CrossRef]
- Laudanski, K.; Hajj, J.; Restrepo, M.; Siddiq, K.; Okeke, T.; Rader, D.J. Dynamic changes in central and peripheral neuro-injury vs. Neuroprotective serum markers in COVID-19 are modulated by different types of anti-viral treatments but do not affect the incidence of late and early strokes. Biomedicines 2021, 9, 1791. [Google Scholar] [CrossRef]
- Kramer, L.D.; Li, J.; Shi, P.Y. West Nile virus. Lancet Neurol. 2007, 6, 171–181. [Google Scholar] [CrossRef]
- Sejvar, J.J.; Marfin, A.A. Manifestations of West Nile neuroinvasive disease. Rev. Med. Virol. 2006, 16, 209–224. [Google Scholar] [CrossRef]
- Constant, O.; Barthelemy, J.; Nagy, A.; Salinas, S.; Simonin, Y. West Nile Virus Neuroinfection in Humans: Peripheral Biomarkers of Neuroinflammation and Neuronal Damage. Viruses 2022, 14, 756. [Google Scholar] [CrossRef] [PubMed]
- Looker, K.J.; Garnett, G.P. A systematic review of the epidemiology and interaction of herpes simplex virus types 1 and 2. Sex. Transm. Infect. 2005, 81, 103. [Google Scholar] [CrossRef] [PubMed]
- AK, A.K.; Mendez, M.D. Herpes Simplex Encephalitis; StatPearls: Tampa, FL, USA, 2022. [Google Scholar]
- Berger, J.R.; Houff, S. Neurological complications of herpes simplex virus type 2 infection. Arch. Neurol. 2008, 65, 596–600. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, J.R.; Rodríguez-Izquierdo, I.; Jiménez, J.L.; Muñoz-Fernández, M.Á. Analysis of ALS-related proteins during herpes simplex virus-2 latent infection. J. Neuroinflamm. 2020, 17, 1–15. [Google Scholar] [CrossRef]
- Chaparro-Huerta, V.; Rivera-Cervantes, M.C.; Flores-Soto, M.E.; Gómez-Pinedo, U.; Beas-Zárate, C. Proinflammatory cytokines and apoptosis following glutamate-induced excitotoxicity mediated by p38 MAPK in the hippocampus of neonatal rats. J. Neuroimmunol. 2005, 165, 53–62. [Google Scholar] [CrossRef]
- Griffin, W.S.T.; Stanley, L.C.; Ling, C.; White, L.; MacLeod, V.; Perrot, L.J.; White, C.L.; Araoz, C. Brain interleukin 1 and S-100 immunoreactivity are elevated in Down syndrome and Alzheimer disease. Proc. Natl. Acad. Sci. USA 1989, 86, 7611–7615. [Google Scholar] [CrossRef]
- Hüll, M.; Berger, M.; Volk, B.; Bauer, J. Occurrence of interleukin-6 in cortical plaques of Alzheimer’s disease patients may precede transformation of diffuse into neuritic plaques. Ann. N. Y. Acad. Sci. 1996, 777, 205–212. [Google Scholar] [CrossRef]
- Van der Wal, E.A.; Gómez-Pinilla, F.; Cotmarv, C.W. Transforming growth factor-beta 1 is in plaques in Alzheimer and Down pathologies. Neuroreport 1993, 4, 69–72. [Google Scholar] [CrossRef]
- Dowlati, Y.; Herrmann, N.; Swardfager, W.; Liu, H.; Sham, L.; Reim, E.K.; Lanctôt, K.L. A meta-analysis of cytokines in major depression. Biol. Psychiatry 2010, 67, 446–457. [Google Scholar] [CrossRef]
- Lu, C.-H.; Allen, K.; Oei, F.; Leoni, E.; Kuhle, J.; Tree, T.; Fratta, P.; Sharma, N.; Sidle, K.; Howard, R.; et al. Systemic inflammatory response and neuromuscular involvement in amyotrophic lateral sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2016, 3, e244. [Google Scholar] [CrossRef]
- Sjögren, M.; Folkesson, S.; Blennow, K. Increased intrathecal inflammatory activity in frontotemporal dementia: Pathophysiological implications. J. Neurol. Neurosurg. Psychiatry 2004, 75, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Bossù, P.; Salani, F.; Alberici, A.; Archetti, S.; Bellelli, G.; Galimberti, D.; Scarpini, E.; Spalletta, G.; Caltagirone, C.; Padovani, A.; et al. Loss of function mutations in the progranulin gene are related to pro-inflammatory cytokine dysregulation in frontotemporal lobar degeneration patients. J. Neuroinflamm. 2011, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, L.; Rollinson, S.; Thompson, J.C.; Robinson, A.; Davidson, Y.S.; Richardson, A.; Neary, D.; Pickering-Brown, S.M.; Snowden, J.S.; Mann, D.M.A. Plasma levels of progranulin and interleukin-6 in frontotemporal lobar degeneration. Neurobiol. Aging 2015, 36, 1603.e1. [Google Scholar] [CrossRef] [PubMed]
- Wagner, R.; Myers, R.R. Endoneurial injection of TNF-alpha produces neuropathic pain behaviors. Neuroreport 1996, 7, 2897–2901. [Google Scholar] [CrossRef] [PubMed]
- Sorkin, L.S.; Doom, C.M. Epineurial application of TNF elicits an acute mechanical hyperalgesia in the awake rat. J. Peripher. Nerv. Syst. 2000, 5, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Neniskyte, U.; Vilalta, A.; Brown, G.C. Tumour necrosis factor alpha-induced neuronal loss is mediated by microglial phagocytosis. FEBS Lett. 2014, 588, 2952. [Google Scholar] [CrossRef]
- Lu, H.J.; Fu, Y.Y.; Wei, Q.Q.; Zhang, Z.J. Neuroinflammation in HIV-Related Neuropathic Pain. Front. Pharmacol. 2021, 12, 653852. [Google Scholar] [CrossRef] [PubMed]
- Duncan, D.S.; Miller, S.D. CNS Expression of B7-H1 Regulates Pro-Inflammatory Cytokine Production and Alters Severity of Theiler’s Virus-Induced Demyelinating Disease. PLoS ONE 2011, 6, 18548. [Google Scholar] [CrossRef]
- Faucard, R.; Madeira, A.; Gehin, N.; Authier, F.J.; Panaite, P.A.; Lesage, C.; Burgelin, I.; Bertel, M.; Bernard, C.; Curtin, F.; et al. Human Endogenous Retrovirus and Neuroinflammation in Chronic Inflammatory Demyelinating Polyradiculoneuropathy. EBioMedicine 2016, 6, 190. [Google Scholar] [CrossRef]
- McFarland, A.J.; Yousuf, M.S.; Shiers, S.; Price, T.J. Neurobiology of SARS-CoV-2 interactions with the peripheral nervous system: Implications for COVID-19 and pain. Pain Rep. 2021, 6, e885. [Google Scholar] [CrossRef]
- Wang, T.Y.; Chen, K.Y.; Jhan, K.Y.; Li, C.H.; Jung, S.M.; Wang, L.C. Temporal-spatial expressions of interleukin-4, interleukin-10, and interleukin-13 in the brains of C57BL/6 and BALB/c mice infected with Angiostrongylus cantonensis: An immunohistochemical study. J. Microbiol. Immunol. Infect. 2020, 53, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Szelényi, J. Cytokines and the central nervous system. Brain Res. Bull. 2001, 54, 329–338. [Google Scholar] [CrossRef]
- Sidoryk-Wegrzynowicz, M.; Wegrzynowicz, M.; Lee, E.; Bowman, A.B.; Aschner, M. Role of astrocytes in brain function and disease. Toxicol. Pathol. 2011, 39, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.S.; Garber, C.; Funk, K.E.; Salimi, H.; Soung, A.; Kanmogne, M.; Manivasagam, S.; Agner, S.; Cain, M. Neuroinflammation during RNA Viral Infections. Annu. Rev. Immunol. 2019, 37, 73. [Google Scholar] [CrossRef] [PubMed]
- de Conti, L.; Baralle, M.; Buratti, E. Neurodegeneration and RNA-binding proteins. Wiley Interdiscip. Rev. RNA 2016, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hamilton, R.; Salapa, H.E.; Levin, M.C. Pro-Inflammatory Cytokines and Antibodies Induce hnRNP A1 Dysfunction in Mouse Primary Cortical Neurons. Brain Sci. 2021, 11, 1282. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Liu, S.; Li, Y.; Yang, G.; Luo, Y.; Li, S.; Du, H.; Zhao, Y.; Wang, D.; Chen, J.; et al. The Nuclear Matrix Protein SAFA Surveils Viral RNA and Facilitates Immunity by Activating Antiviral Enhancers and Super-enhancers. Cell Host Microbe 2019, 26, 369–384.e8. [Google Scholar] [CrossRef]
- Appocher, C.; Mohagheghi, F.; Cappelli, S.; Stuani, C.; Romano, M.; Feiguin, F.; Buratti, E. Major hnRNP proteins act as general TDP-43 functional modifiers both in Drosophila and human neuronal cells. Nucleic Acids Res. 2017, 45, 8026–8045. [Google Scholar] [CrossRef]
- Cappelli, S.; Romano, M.; Buratti, E. Systematic Analysis of Gene Expression Profiles Controlled by hnRNP Q and hnRNP R, Two Closely Related Human RNA Binding Proteins Implicated in mRNA Processing Mechanisms. Front. Mol. Biosci. 2018, 5, 1–17. [Google Scholar] [CrossRef]
- Murata, H.; Hattori, T.; Maeda, H.; Takashiba, S.; Takigawa, M.; Kido, J.; Nagata, T. Identification of transactivation-responsive DNA-binding protein 43 (TARDBP43; TDP-43) as a novel factor for TNF-α expression upon lipopolysaccharide stimulation in human monocytes. J. Periodontal Res. 2015, 50, 452–460. [Google Scholar] [CrossRef]
- Šušnjar, U.; Škrabar, N.; Brown, A.L.; Abbassi, Y.; Phatnani, H.; Phatnani, H.; Fratta, P.; Kwan, J.; Sareen, D.; Broach, J.R.; et al. Cell environment shapes TDP-43 function with implications in neuronal and muscle disease. Commun. Biol. 2022, 5, 1–17. [Google Scholar] [CrossRef]
- Lee, S.J.; Benveniste, E.N. Adhesion molecule expression and regulation on cells of the central nervous system. J. Neuroimmunol. 1999, 98, 77–88. [Google Scholar] [CrossRef]
- Dai, J.; Wang, P.; Bai, F.; Town, T.; Fikrig, E. ICAM-1 Participates in the Entry of West Nile Virus into the Central Nervous System. J. Virol. 2008, 82, 4164. [Google Scholar] [CrossRef]
- Müller, N. The role of intercellular adhesion molecule-1 in the pathogenesis of psychiatric disorders. Front. Pharmacol. 2019, 10, 1251. [Google Scholar] [CrossRef]
- Akiyama, H.; Kawamata, T.; Yamada, T.; Tooyama, I.; Ishii, T.; McGeer, P.L. Expression of intercellular adhesion molecule (ICAM)-1 by a subset of astrocytes in Alzheimer disease and some other degenerative neurological disorders. Acta Neuropathol. 1993, 85, 628–634. [Google Scholar] [CrossRef]
- Scotter, E.L.; Cao, M.C.; Jansson, D.; Rustenhoven, J.; Smyth, L.C.D.; Aalderink, M.C.; Siemens, A.; Fan, V.; Wu, J.; Mee, E.W.; et al. The amyotrophic lateral sclerosis-linked protein TDP-43 regulates interleukin-6 cytokine production by human brain pericytes. Mol. Cell Neurosci. 2022, 123, 103768. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, S.; Kang, H.Y.; Lim, H.R.; Kwon, Y.; Jo, M.; Jeon, Y.M.; Kim, S.R.; Kim, K.; Ha, C.M.; et al. The overexpression of TDP-43 in astrocytes causes neurodegeneration via a PTP1B-mediated inflammatory response. J. Neuroinflamm. 2020, 17, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, T.A.; Lee, E.; Kang, S.; Park, A.; Kim, S.W.; Park, H.J.; Yoon, J.H.; Ha, S.J.; Park, T.; et al. Identification of a subnuclear body involved in sequence-specific cytokine RNA processing. Nat. Commun. 2015, 6, 5791. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in Inflammation, Immunity, and Disease. Cold Spring Harb. Perspect. Biol. 2014, 6, 16295–16296. [Google Scholar] [CrossRef]
- Bouezzedine, F.; Fardel, O.; Gripon, P. Interleukin 6 inhibits HBV entry through NTCP down regulation. Virology 2015, 481, 34–42. [Google Scholar] [CrossRef]
- Palumbo, G.A.; Scisciani, C.; Pediconi, N.; Lupacchini, L.; Alfalate, D.; Guerrieri, F.; Calvo, L.; Salerno, D.; di Cocco, S.; Levrero, M.; et al. IL6 Inhibits HBV Transcription by Targeting the Epigenetic Control of the Nuclear cccDNA Minichromosome. PLoS ONE 2015, 10, e0142599. [Google Scholar] [CrossRef] [PubMed]
- Hösel, M.; Quasdorff, M.; Wiegmann, K.; Webb, D.; Zedler, U.; Broxtermann, M.; Tedjokusumo, R.; Esser, K.; Arzberger, S.; Kirschning, C.J.; et al. Not interferon, but interleukin-6 controls early gene expression in hepatitis B virus infection. Hepatology 2009, 50, 1773–1782. [Google Scholar] [CrossRef] [PubMed]
- Ulhaq, Z.S.; Soraya, G.V. Interleukin-6 as a potential biomarker of COVID-19 progression. Med. Mal. Infect. 2020, 50, 382–383. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Xie, J.; Yin, M.; Yang, Y.; Ding, C.; Gao, Y.; Ma, X. Interleukin-6 and severity of COVID-19 patients in Hefei, China. Med. Mal. Infect. 2020, 50, 629–631. [Google Scholar] [CrossRef]
- Trovato, M.; Ruggeri, R.M.; Sciacchitano, S.; Vicchio, T.M.; Picerno, I.; Pellicanò, G.; Valenti, A.; Visalli, G. Serum interleukin-6 levels are increased in HIV-infected patients that develop autoimmune disease during long-term follow-up. Immunobiology 2018, 223, 264–268. [Google Scholar] [CrossRef]
- Chen, Y.; Xia, K.; Chen, L.; Fan, D. Increased interleukin-6 levels in the astrocyte-derived exosomes of sporadic amyotrophic lateral sclerosis patients. Front. Neurosci. 2019, 13, 574. [Google Scholar] [CrossRef]
- Hofmann, K.W.; Schuh, A.F.S.; Saute, J.; Townsend, R.; Fricke, D.; Leke, R.; Souza, D.O.; Portela, L.V.; Chaves, M.L.F.; Rieder, C.R.M. Interleukin-6 serum levels in patients with Parkinson’s disease. Neurochem. Res. 2009, 34, 1401–1404. [Google Scholar] [CrossRef]
- Bauer, J.; Strauss, S.; Schreiter-Gasser, U.; Ganter, U.; Schlegel, P.; Witt, I.; Yolk, B.; Berger, M. Interleukin-6 and α-2-macroglobulin indicate an acute-phase state in Alzheimer’s disease cortices. FEBS Lett. 1991, 285, 111–114. [Google Scholar] [CrossRef]
- Wertz, M.H.; Pineda, S.S.; Lee, H.; Kulicke, R.; Kellis, M.; Heiman, M. Interleukin-6 deficiency exacerbates Huntington’s disease model phenotypes. Mol. Neurodegener. 2020, 15, 1–8. [Google Scholar] [CrossRef]
- Lopez-Castejon, G.; Brough, D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 2011, 22, 189. [Google Scholar] [CrossRef]
- Mendiola, A.S.; Cardona, A.E. The IL-1β phenomena in neuroinflammatory diseases. J. Neural Transm. 2018, 125, 781. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 1–9. [Google Scholar] [CrossRef]
- Meissner, F.; Molawi, K.; Zychlinsky, A. Mutant superoxide dismutase 1-induced IL-1β accelerates ALS pathogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 13046–13050. [Google Scholar] [CrossRef] [PubMed]
- Cioccarelli, C.; Sánchez-Rodríguez, R.; Angioni, R.; Venegas, F.C.; Bertoldi, N.; Munari, F.; Cattelan, A.; Molon, B.; Viola, A. IL1β Promotes TMPRSS2 Expression and SARS-CoV-2 Cell Entry through the p38 MAPK-GATA2 Axis. Front. Immunol. 2021, 12, 5173. [Google Scholar] [CrossRef] [PubMed]
- Frakes, A.E.; Ferraiuolo, L.; Haidet-Phillips, A.M.; Schmelzer, L.; Braun, L.; Miranda, C.J.; Ladner, K.J.; Bevan, A.K.; Foust, K.D.; Godbout, J.P.; et al. Microglia induce motor neuron death via the classical NF-κB pathway in amyotrophic lateral sclerosis. Neuron 2014, 81, 1009–1023. [Google Scholar] [CrossRef]
- Crosio, C.; Valle, C.; Casciati, A.; Iaccarino, C.; Carrì, M.T. Astroglial inhibition of NF-κB does not ameliorate disease onset and progression in a mouse model for amyotrophic lateral sclerosis (ALS). PLoS ONE 2011, 6, e17187. [Google Scholar] [CrossRef] [PubMed]
- Krabbe, G.; Minami, S.S.; Etchegaray, J.I.; Taneja, P.; Djukic, B.; Davalos, D.; Le, D.; Lo, I.; Zhan, L.; Reichert, M.C.; et al. Microglial NFκB-TNFα hyperactivation induces obsessive-compulsive behavior in mouse models of progranulin-deficient frontotemporal dementia. Proc. Natl. Acad. Sci. USA 2017, 114, 5029–5034. [Google Scholar] [CrossRef] [PubMed]
- Hunot, S.; Brugg, B.; Ricard, D.; Michel, P.P.; Muriel, M.P.; Ruberg, M.; Faucheux, B.A.; Agid, Y.; Hirsch, E.C. Nuclear translocation of NF-κB is increased in dopaminergic neurons of patients with Parkinson disease. Proc. Natl. Acad. Sci. USA 1997, 94, 7531. [Google Scholar] [CrossRef]
- Hsiao, H.Y.; Chen, Y.C.; Chen, H.M.; Tu, P.H.; Chern, Y. A critical role of astrocyte-mediated nuclear factor-κB-dependent inflammation in Huntington’s disease. Hum. Mol. Genet. 2013, 22, 1826–1842. [Google Scholar] [CrossRef]
- Kircheis, R.; Haasbach, E.; Lueftenegger, D.; Heyken, W.T.; Ocker, M.; Planz, O. NF-κB Pathway as a Potential Target for Treatment of Critical Stage COVID-19 Patients. Front. Immunol. 2020, 11, 3446. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, H.; Wang, F.; Xia, X.; Sun, Q.; Wang, R.; Cheng, B. The hepatitis B virus X protein downregulates NF-κB signaling pathways through decreasing the Notch signaling pathway in HBx-transformed L02 cells. Int. J. Oncol. 2013, 42, 1636–1643. [Google Scholar] [CrossRef]
- Fiume, G.; Vecchio, E.; de Laurentiis, A.; Trimboli, F.; Palmieri, C.; Pisano, A.; Falcone, C.; Pontoriero, M.; Rossi, A.; Scialdone, A.; et al. Human immunodeficiency virus-1 Tat activates NF-κB via physical interaction with IκB-α and p65. Nucleic Acids Res. 2012, 40, 3548–3562. [Google Scholar] [CrossRef]
- Fink, D.L.; Cai, J.; Whelan, M.V.X.; Monit, C.; de Motes, C.M.; Towers, G.J.; Sumner, R.P. HIV-2/SIV Vpx antagonises NF-κB activation by targeting p65. Retrovirology 2022, 19, 1–14. [Google Scholar] [CrossRef]
- Saura, M.; Lizarbe, T.R.; Rama-Pacheco, C.; Lowenstein, C.J.; Zaragoza, C. Cell Cycle Inhibitor of NFκB Alpha is a Host Sensor of Coxsackievirus Infection. Cell Cycle 2007, 6, 503–506. [Google Scholar] [CrossRef]
- Amici, C.; Rossi, A.; Costanzo, A.; Ciafrè, S.; Marinari, B.; Balsamo, M.; Levrero, M.; Santoro, M.G. Herpes simplex virus disrupts NF-κB regulation by blocking its recruitment on the IκBα promoter and directing the factor on viral genes. J. Biol. Chem. 2006, 281, 7110–7117. [Google Scholar] [CrossRef] [PubMed]
- Enguita, F.J.; Leitão, A.L.; McDonald, J.T.; Zaksas, V.; Das, S.; Galeano, D.; Taylor, D.; Wurtele, E.S.; Saravia-Butler, A.; Baylin, S.B.; et al. The interplay between lncRNAs, RNA-binding proteins and viral genome during SARS-CoV-2 infection reveals strong connections with regulatory events involved in RNA metabolism and immune response. Theranostics 2022, 12, 3946. [Google Scholar] [CrossRef] [PubMed]
- Ginn, L.; la Montagna, M.; Wu, Q.; Shi, L. Diverse roles of long non-coding RNAs in viral diseases. Rev. Med. Virol. 2021, 31, e2198. [Google Scholar] [CrossRef]
- Matsukawa, K.; Kukharsky, M.S.; Park, S.K.; Park, S.; Watanabe, N.; Iwatsubo, T.; Hashimoto, T.; Liebman, S.W.; Shelkovnikova, T.A. Long non-coding RNA NEAT1_1 ameliorates TDP-43 toxicity in in vivo models of TDP-43 proteinopathy. RNA Biol. 2021, 18, 1546. [Google Scholar] [CrossRef]
- An, H.; Williams, N.G.; Shelkovnikova, T.A. NEAT1 and paraspeckles in neurodegenerative diseases: A missing lnc found? Noncoding RNA Res. 2018, 3, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Vrati, S. The Malat1 long non-coding RNA is upregulated by signalling through the PERK axis of unfolded protein response during flavivirus infection. Sci. Rep. 2015, 5, 1–8. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, Y.; Fang, S.; Jiang, B.; Qin, C.; Xie, P.; Zhou, G.; Li, G. The role of MALAT1 correlates with HPV in cervical cancer. Oncol. Lett. 2014, 7, 2135–2141. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Z.; Liu, L.; Yang, Z.; Liu, S.; Ma, Z.; Liu, Y.; Ma, Y.; Zhang, L.; Zhang, X.; et al. LncRNA Malat1 inhibition of TDP43 cleavage suppresses IRF3-initiated antiviral innate immunity. Proc. Natl. Acad. Sci. USA 2020, 117, 23695–23706. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, C.Y.; Yedavalli, V.S.R.K.; Jeang, K.T. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. mBio 2013, 4, e00596-12. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fan, P.; Zhao, Y.; Zhang, S.; Lu, J.; Xie, W.; Jiang, Y.; Lei, F.; Xu, N.; Zhang, Y. NEAT1 modulates herpes simplex virus-1 replication by regulating viral gene transcription. Cell. Mol. Life Sci. 2017, 74, 1117. [Google Scholar] [CrossRef]
- Buratti, E.; Baralle, F.E. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front. Biosci. 2008, 13, 867–878. [Google Scholar] [CrossRef]
- Bellmann, J.; Monette, A.; Tripathy, V.; Sójka, A.; Abo-Rady, M.; Janosh, A.; Bhatnagar, R.; Bickle, M.; Mouland, A.J.; Sterneckert, J. Viral Infections Exacerbate FUS-ALS Phenotypes in iPSC-Derived Spinal Neurons in a Virus Species-Specific Manner. Front. Cell Neurosci. 2019, 13, 480. [Google Scholar] [CrossRef] [PubMed]
- Buratti, E. Targeting TDP-43 proteinopathy with drugs and drug-like small molecules. Br. J. Pharmacol. 2021, 178, 1298–1315. [Google Scholar] [CrossRef]
- Mollasalehi, N.; Francois-Moutal, L.; Scott, D.D.; Tello, J.A.; Williams, H.; Mahoney, B.; Carlson, J.M.; Dong, Y.; Li, X.; Miranda, V.G.; et al. An Allosteric Modulator of RNA Binding Targeting the N-Terminal Domain of TDP-43 Yields Neuroprotective Properties. ACS Chem. Biol. 2020, 15, 2854–2859. [Google Scholar] [CrossRef]
- Bhandare, V.V.; Ramaswamy, A. Identification of possible siRNA molecules for TDP43 mutants causing amyotrophic lateral sclerosis: In silico design and molecular dynamics study. Comput. Biol. Chem. 2016, 61, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Sckaff, M.; Gill, K.; Sachdev, A.; Birk, A.; Arogundade, O.A.; Watry, H.; Keough, K.; Tsai, Y.-C.; Ziegle, J.; Conklin, B.; et al. Two therapeutic CRISPR/Cas9 gene editing approaches revert FTD/ALS cellular pathology caused by a C9orf72 repeat expansion mutation in patient derived cells. bioRxiv 2022. bioRxiv: 2022.05.21.492887. [Google Scholar]
- Becker, L.A.; Huang, B.; Bieri, G.; Ma, R.; Knowles, D.A.; Jafar-Nejad, P.; Messing, J.; Kim, H.J.; Soriano, A.; Auburger, G.; et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 2017, 544, 367–371. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Feuer, R.; Mena, I.; Pagarigan, R.R.; Harkins, S.; Hassett, D.E.; Whitton, J.L. Coxsackievirus B3 and the Neonatal CNS: The Roles of Stem Cells, Developing Neurons, and Apoptosis in Infection, Viral Dissemination, and Disease. Am. J. Pathol. 2003, 163, 1379. [Google Scholar] [CrossRef]
- Wong, A.H.; Lau, C.S.; Cheng, P.K.C.; Ng, A.Y.Y.; Lim, W.W.L. Coxsackievirus B3-associated aseptic meningitis: An emerging infection in Hong Kong. J. Med. Virol. 2011, 83, 483–489. [Google Scholar] [CrossRef]
- Omura, S.; Kawai, E.; Sato, F.; Martinez, N.E.; Minagar, A.; Al-Kofahi, M.; Yun, J.W.; Cvek, U.; Trutschl, M.; Alexander, J.S.; et al. Theiler’s Virus-Mediated Immunopathology in the CNS and Heart: Roles of Organ-Specific Cytokine and Lymphatic Responses. Front. Immunol. 2018, 9, 2870. [Google Scholar] [CrossRef]
- Zink, W.E.; Zheng, J.; Persidsky, Y.; Poluektova, L.; Gendelman, H.E. The neuropathogenesis of HIV-1 infection. FEMS Immunol. Med. Microbiol. 1999, 26, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Bowen, L.N.; Smith, B.; Reich, D.; Quezado, M.; Nath, A. HIV-associated opportunistic CNS infections: Pathophysiology, diagnosis and treatment. Nat. Rev. Neurol. 2016, 12, 662–674. [Google Scholar] [CrossRef]
- Gröger, V.; Emmer, A.; Staege, M.S.; Cynis, H. Endogenous Retroviruses in Nervous System Disorders. Pharmaceuticals 2021, 14, 70. [Google Scholar] [CrossRef]
- Lin, S.; Guo, Z.; Chen, S.; Lin, X.; Ye, M.; Qiu, Y. Progressive Brain Structural Impairment Assessed via Network and Causal Analysis in Patients with Hepatitis B Virus-Related Cirrhosis. Front. Neurol. 2022, 13, 849571. [Google Scholar] [CrossRef]
- Crunfli, F.; Carregari, V.C.; Veras, F.P.; Silva, L.S.; Nogueira, M.H.; Antunes, A.S.L.M.; Vendramini, P.H.; Valença, A.G.F.; Brandão-Teles, C.; da Silva Zuccoli, G.; et al. Morphological, cellular, and molecular basis of brain infection in COVID-19 patients. Proc. Natl. Acad. Sci. USA 2022, 119, e2200960119. [Google Scholar] [CrossRef]
- Strong, M.J. SARS-CoV-2, aging, and Post-COVID-19 neurodegeneration. J. Neurochem. 2022; ahead of print. [Google Scholar] [CrossRef]
- Hart, J.; Tillman, G.; Kraut, M.A.; Chiang, H.S.; Strain, J.F.; Li, Y.; Agrawal, A.G.; Jester, P.; Gnann, J.W.; Whitley, R.J. West Nile virus neuroinvasive disease: Neurological manifestations and prospective longitudinal outcomes. BMC Infect. Dis. 2014, 14, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Orba, Y.; Yamaguchi, H.; Kimura, T.; Sawa, H. Accumulation of ubiquitinated proteins is related to West Nile virus-induced neuronal apoptosis. Neuropathology 2012, 32, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Garber, C.; Soung, A.; Vollmer, L.L.; Kanmogne, M.; Last, A.; Brown, J.; Klein, R.S. T cells promote microglia-mediated synaptic elimination and cognitive dysfunction during recovery from neuropathogenic flaviviruses. Nat. Neurosci. 2019, 22, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Garber, C.; Vasek, M.J.; Vollmer, L.L.; Sun, T.; Jiang, X.; Klein, R.S. Astrocytes decrease adult neurogenesis during virus-induced memory dysfunction via IL-1. Nat. Immunol. 2018, 19, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Zukor, K.; Wang, H.; Hurst, B.L.; Siddharthan, V.; van Wettere, A.; Pilowsky, P.M.; Morrey, J.D. Phrenic nerve deficits and neurological immunopathology associated with acute West Nile virus infection in mice and hamsters. J. Neurovirol. 2017, 23, 186–204. [Google Scholar] [CrossRef]
- Margolis, T.P.; Imai, Y.; Yang, L.; Vallas, V.; Krause, P.R. Herpes simplex virus type 2 (HSV-2) establishes latent infection in a different population of ganglionic neurons than HSV-1: Role of latency-associated transcripts. J. Virol. 2007, 81, 1872–1878. [Google Scholar] [CrossRef]
| Virus | Brain Involvement |
|---|---|
| Coxsackievirus B3 (CVB3) | CNS of neonatal mice were found to be susceptible to CVB illness likely through the infection of progenitor cells [155]. CVB3 was found to be associated with aseptic meningitis in a Hong Kong population [156]. |
| Theiler’s murine encephalomyelitis virus (TMEV) | TMEV is widely used as a model to study multiple sclerosis [39]. It can induce apoptosis, neuronophagia and inflammation in infected neurons [157]. |
| Human immunodeficiency virus (HIV) | HIV is the etiological agent of HIV encephalitis. HIV can be carried into CNS through infected CD4+ T cells and/or monocytes and, as a result, brain macrophages and microglia are widely considered the reservoirs for persistent viral infection. When these cells are activated, they can trigger an immunological response leading to neuronal death and the consequent establishment of HIV-associated dementia (HAD) [158]. HIV is responsible for opportunistic infection in CNS of HIV-positive individuals, including cerebral toxoplasmosis, progressive multifocal leukoencephalopathy (PML), tuberculous meningitis, cryptococcal meningitis and cytomegalovirus infection [159]. |
| Human endogenous retroviruses (HERV) | HERV endogenous expression can be induced by viral RNAs and proteins, following infection of HIV-1, HBV and influenza A viruses [160,56]. Interestingly, increased HERV-K expression has been detected in brain of ALS patients, and it was found to induce neuronal injury in a model of transgenic animals [56]. |
| Hepatitis B virus (HBV) | Impairment of neuronal structures has been observed to occur in HBV-positive individuals with associated chronic liver dysfunction [161]. |
| Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) | Cognitive dysfunctions and encephalitis were reported in patients suffering from SAR-CoV-2 infection [162,163]. SARS-CoV-2 has been proposed to induce morphological and cellular alteration of brain structures [162]. |
| West Nile virus (WNV) | WNV causes severe neurological illness, generally referred to West Nile neuroinvasive disease (WNND) and including West Nile encephalitis (WNE), West Nile meningitis (WNM), and West Nile paralysis (WNP) [164]. WNV infection has been linked to neuronal dysfunction, loss of synapses, and astrocytic gliosis, both in human patients and animal models [165,166,167,168]. |
| Herpes simplex virus-2 (HSV-2) | HSV-2 has been observed to cause neurological complication and establishes latent infection in neurons of ganglia [74,169]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahic, Z.; Buratti, E.; Cappelli, S. Reviewing the Potential Links between Viral Infections and TDP-43 Proteinopathies. Int. J. Mol. Sci. 2023, 24, 1581. https://doi.org/10.3390/ijms24021581
Rahic Z, Buratti E, Cappelli S. Reviewing the Potential Links between Viral Infections and TDP-43 Proteinopathies. International Journal of Molecular Sciences. 2023; 24(2):1581. https://doi.org/10.3390/ijms24021581
Chicago/Turabian StyleRahic, Zerina, Emanuele Buratti, and Sara Cappelli. 2023. "Reviewing the Potential Links between Viral Infections and TDP-43 Proteinopathies" International Journal of Molecular Sciences 24, no. 2: 1581. https://doi.org/10.3390/ijms24021581
APA StyleRahic, Z., Buratti, E., & Cappelli, S. (2023). Reviewing the Potential Links between Viral Infections and TDP-43 Proteinopathies. International Journal of Molecular Sciences, 24(2), 1581. https://doi.org/10.3390/ijms24021581








