Deciphering the Effect of Different Genetic Variants on Hippocampal Subfield Volumes in the General Population
Abstract
1. Introduction
2. Results
2.1. Sample Characteristic
2.2. Direct Effects of Candidate Variants on Hippocampal Subfields
2.2.1. APOE ε4
2.2.2. 5-HTTLPR
2.2.3. BDNF
2.2.4. COMT
2.2.5. KIBRA
2.3. Direct Effects of GWAS SNPs on Hippocampal Subfields
2.4. Association between the PRS for AD and Hippocampal Subfield Volumes
2.5. Interaction Analyses between Significant Genetic Factors
2.6. Association between Memory Performance and Hippocampal Subfields
3. Discussion
4. Materials and Methods
4.1. SHIP-TREND Sample
4.1.1. Verbal Memory
4.1.2. Covariates
4.2. Genetic Data
4.2.1. Genome-Wide SNP Chip
4.2.2. APOE ε4 Carrier Status
4.2.3. Genotyping of the Serotonin Transporter
4.2.4. Polygenic Risk Score for AD
4.3. MRI Data
4.4. Statistical Analyses
4.4.1. Direct Effects of Genetic Markers on Hippocampal Subfields
4.4.2. Interaction Analyses between Significant Genetic Factors from Section 4.4.1
4.4.3. Association between Memory Performance and Hippocampal Subfields
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vilor-Tejedor, N.; Evans, T.E.; Adams, H.H.; González-de-Echávarri, J.M.; Molinuevo, J.L.; Guigo, R.; Gispert, J.D.; Operto, G. Genetic Influences on Hippocampal Subfields: An Emerging Area of Neuroscience Research. Neurol. Genet. 2021, 7, e591. [Google Scholar] [CrossRef] [PubMed]
- Hibar, D.P.; Adams, H.H.H.; Jahanshad, N.; Chauhan, G.; Stein, J.L.; Hofer, E.; Renteria, M.E.; Bis, J.C.; Arias-Vasquez, A.; Ikram, K.M.; et al. Novel genetic loci associated with hippocampal volume. Nat. Commun. 2017, 8, 13624. [Google Scholar] [CrossRef]
- Malhi, G.S.; Das, P.; Outhred, T.; Irwin, L.; Gessler, D.; Bwabi, Z.; Bryant, R.; Mannie, Z. The effects of childhood trauma on adolescent hippocampal subfields. Aust. N. Z. J. Psychiatry 2019, 53, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Zammit, A.R.; Ezzati, A.; Zimmerman, M.E.; Lipton, R.B.; Lipton, M.L.; Katz, M.J. Roles of hippocampal subfields in verbal and visual episodic memory. Behav. Brain Res. 2017, 317, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Genon, S.; Bernhardt, B.C.; La Joie, R.; Amunts, K.; Eickhoff, S.B. The many dimensions of human hippocampal organization and (dys)function. Trends Neurosci. 2021, 44, 977–989. [Google Scholar] [CrossRef]
- Shi, F.; Liu, B.; Zhou, Y.; Yu, C.; Jiang, T. Hippocampal volume and asymmetry in mild cognitive impairment and Alzheimer’s disease: Meta-analyses of MRI studies. Hippocampus 2009, 19, 1055–1064. [Google Scholar] [CrossRef]
- Li, J.-Q.; Tan, L.; Wang, H.-F.; Tan, M.-S.; Tan, L.; Xu, W.; Zhao, Q.-F.; Wang, J.; Jiang, T.; Yu, J.-T. Risk factors for predicting progression from mild cognitive impairment to Alzheimer’s disease: A systematic review and meta-analysis of cohort studies. J. Neurol. Neurosurg. Psychiatry 2016, 87, 476–484. [Google Scholar] [CrossRef]
- Feng, F.; Huang, W.; Meng, Q.; Hao, W.; Yao, H.; Zhou, B.; Guo, Y.; Zhao, C.; An, N.; Wang, L.; et al. Altered Volume and Structural Connectivity of the Hippocampus in Alzheimer’s Disease and Amnestic Mild Cognitive Impairment. Front. Aging Neurosci. 2021, 13, 705030. [Google Scholar] [CrossRef]
- Izzo, J.; Andreassen, O.A.; Westlye, L.T.; Van der Meer, D. The association between hippocampal subfield volumes in mild cognitive impairment and conversion to Alzheimer’s disease. Brain Res. 2020, 1728, 146591. [Google Scholar] [CrossRef]
- Nobis, L.; Manohar, S.G.; Smith, S.M.; Alfaro-Almagro, F.; Jenkinson, M.; Mackay, C.E.; Husain, M. Hippocampal volume across age: Nomograms derived from over 19,700 people in UK Biobank. Neuroimage Clin. 2019, 23, 101904. [Google Scholar] [CrossRef]
- Van der Meer, D.; Rokicki, J.; Kaufmann, T.; Córdova-Palomera, A.; Moberget, T.; Alnæs, D.; Bettella, F.; Frei, O.; Doan, N.T.; Sønderby, I.E.; et al. Brain scans from 21,297 individuals reveal the genetic architecture of hippocampal subfield volumes. Mol. Psychiatry 2020, 25, 3053–3065. [Google Scholar] [CrossRef]
- El Haj, M.; Antoine, P.; Amouyel, P.; Lambert, J.-C.; Pasquier, F.; Kapogiannis, D. Apolipoprotein E (APOE) ε4 and episodic memory decline in Alzheimer’s disease: A review. Ageing Res. Rev. 2016, 27, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lin, L.; Dai, X.; Xiao, N.; Ye, Q.; Chen, X. ApoE4 increases susceptibility to stress-induced age-dependent depression-like behavior and cognitive impairment. J. Psychiatr. Res. 2021, 143, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Vervoordt, S.M.; Arnett, P.; Engeland, C.; Rabinowitz, A.R.; Hillary, F.G. Depression associated with APOE status and hippocampal volume but not cognitive decline in older adults aging with traumatic brain injury. Neuropsychology 2021, 35, 863–875. [Google Scholar] [CrossRef]
- Bonk, S.; Kirchner, K.; Ameling, S.; Garvert, L.; Völzke, H.; Nauck, M.; Völker, U.; Grabe, H.J.; Van der Auwera, S. APOE ε4 in Depression-Associated Memory Impairment-Evidence from Genetic and MicroRNA Analyses. Biomedicines 2022, 10, 1560. [Google Scholar] [CrossRef] [PubMed]
- Vilor-Tejedor, N.; Operto, G.; Evans, T.E.; Falcon, C.; Crous-Bou, M.; Minguillón, C.; Cacciaglia, R.; Milà-Alomà, M.; Grau-Rivera, O.; Suárez-Calvet, M.; et al. Effect of BDNF Val66Met on hippocampal subfields volumes and compensatory interaction with APOE-ε4 in middle-age cognitively unimpaired individuals from the ALFA study. Brain Struct. Funct. 2020, 225, 2331–2345. [Google Scholar] [CrossRef]
- Stonnington, C.M.; Velgos, S.N.; Chen, Y.; Syed, S.; Huentelman, M.; Thiyyagura, P.; Lee, W.; Richholt, R.; Caselli, R.J.; Locke, D.E.; et al. Interaction Between BDNF Val66Met and APOE4 on Biomarkers of Alzheimer’s Disease and Cognitive Decline. JAD 2020, 78, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Molendijk, M.L.; van Tol, M.-J.; Penninx, B.W.J.H.; van der Wee, N.J.A.; Aleman, A.; Veltman, D.J.; Spinhoven, P.; Elzinga, B.M. BDNF val66met affects hippocampal volume and emotion-related hippocampal memory activity. Transl. Psychiatry 2012, 2, e74. [Google Scholar] [CrossRef]
- Harrisberger, F.; Spalek, K.; Smieskova, R.; Schmidt, A.; Coynel, D.; Milnik, A.; Fastenrath, M.; Freytag, V.; Gschwind, L.; Walter, A.; et al. The association of the BDNF Val66Met polymorphism and the hippocampal volumes in healthy humans: A joint meta-analysis of published and new data. Neurosci. Biobehav. Rev. 2014, 42, 267–278. [Google Scholar] [CrossRef]
- Li, X.; Tian, P.; Hu, X. Association of Met/Val polymorphism of BDNF gene with Alzheimer’s disease in Chinese patients. Cell. Mol. Biol. 2022, 68, 46–51. [Google Scholar] [CrossRef]
- Ferreira Fratelli, C.; Willatan Siqueira, J.; Rodrigues Gontijo, B.; de Lima Santos, M.; de Souza Silva, C.M.; Da Rodrigues Silva, I.C. BDNF Genetic Variant and Its Genotypic Fluctuation in Major Depressive Disorder. Behav. Neurol. 2021, 2021, 7117613. [Google Scholar] [CrossRef]
- Toro, R.; Chupin, M.; Garnero, L.; Leonard, G.; Perron, M.; Pike, B.; Pitiot, A.; Richer, L.; Veillette, S.; Pausova, Z.; et al. Brain volumes and Val66Met polymorphism of the BDNF gene: Local or global effects? Brain Struct. Funct. 2009, 213, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Harrisberger, F.; Smieskova, R.; Schmidt, A.; Lenz, C.; Walter, A.; Wittfeld, K.; Grabe, H.J.; Lang, U.E.; Fusar-Poli, P.; Borgwardt, S. BDNF Val66Met polymorphism and hippocampal volume in neuropsychiatric disorders: A systematic review and meta-analysis. Neurosci. Biobehav. Rev. 2015, 55, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Papassotiropoulos, A.; Stephan, D.A.; Huentelman, M.J.; Hoerndli, F.J.; Craig, D.W.; Pearson, J.V.; Huynh, K.-D.; Brunner, F.; Corneveaux, J.; Osborne, D.; et al. Common Kibra alleles are associated with human memory performance. Science 2006, 314, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Vogt-Eisele, A.; Krüger, C.; Duning, K.; Weber, D.; Spoelgen, R.; Pitzer, C.; Plaas, C.; Eisenhardt, G.; Meyer, A.; Vogt, G.; et al. KIBRA (KIdney/BRAin protein) regulates learning and memory and stabilizes Protein kinase Mζ. J. Neurochem. 2014, 128, 686–700. [Google Scholar] [CrossRef]
- Song, L.; Tang, S.; Dong, L.; Han, X.; Cong, L.; Dong, J.; Han, X.; Zhang, Q.; Wang, Y.; Du, Y. The Neuroprotection of KIBRA in Promoting Neuron Survival and Against Amyloid β-Induced Apoptosis. Front. Cell. Neurosci. 2019, 13, 137. [Google Scholar] [CrossRef]
- Liu, J.J.; Lavebratt, C.; Lou, F.; Forsell, Y. KIBRA genetic polymorphism and cognitive dysfunction in depression. Psychiatry Res. 2015, 226, 405–406. [Google Scholar] [CrossRef] [PubMed]
- Kauppi, K.; Nilsson, L.-G.; Adolfsson, R.; Eriksson, E.; Nyberg, L. KIBRA polymorphism is related to enhanced memory and elevated hippocampal processing. J. Neurosci. 2011, 31, 14218–14222. [Google Scholar] [CrossRef]
- Palombo, D.J.; Amaral, R.S.C.; Olsen, R.K.; Müller, D.J.; Todd, R.M.; Anderson, A.K.; Levine, B. KIBRA polymorphism is associated with individual differences in hippocampal subregions: Evidence from anatomical segmentation using high-resolution MRI. J. Neurosci. 2013, 33, 13088–13093. [Google Scholar] [CrossRef]
- Witte, A.V.; Köbe, T.; Kerti, L.; Rujescu, D.; Flöel, A. Impact of KIBRA Polymorphism on Memory Function and the Hippocampus in Older Adults. Neuropsychopharmacology 2016, 41, 781–790. [Google Scholar] [CrossRef]
- Wang, J.; Wu, S.; Sun, Y.; Fang, Y.; Wu, R.; Lu, J.; Qing, Z.; Liang, X.; Wang, Z.; Zhang, W.; et al. Interaction of COMT and KIBRA modulates the association between hippocampal structure and episodic memory performance in healthy young adults. Behav. Brain Res. 2020, 384, 112550. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.P.; Logue, M.W.; Reagan, A.; Salat, D.; Wolf, E.J.; Sadeh, N.; Spielberg, J.M.; Sperbeck, E.; Hayes, S.M.; McGlinchey, R.E.; et al. COMT Val158Met polymorphism moderates the association between PTSD symptom severity and hippocampal volume. J. Psychiatry Neurosci. 2017, 42, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y.; Kakeda, S.; Sugimoto, K.; Katsuki, A.; Le Nguyen, H.; Igata, R.; Watanabe, K.; Ueda, I.; Kishi, T.; Iwata, N.; et al. COMT polymorphism regulates the hippocampal subfield volumes in first-episode, drug-naive patients with major depressive disorder. Neuropsychiatr. Dis. Treat. 2019, 15, 1537–1545. [Google Scholar] [CrossRef]
- Porter, T.; Burnham, S.C.; Milicic, L.; Savage, G.; Maruff, P.; Sohrabi, H.R.; Peretti, M.; Lim, Y.Y.; Weinborn, M.; Ames, D.; et al. COMT val158met is not associated with Aβ-amyloid and APOE ε4 related cognitive decline in cognitively normal older adults. IBRO Rep. 2019, 6, 147–152. [Google Scholar] [CrossRef]
- Haeffel, G.J.; Eastman, M.; Grigorenko, E.L. Using a cognitive endophenotype to identify risk genes for depression. Neurosci. Lett. 2012, 510, 10–13. [Google Scholar] [CrossRef]
- Taylor, W.D.; Züchner, S.; Payne, M.E.; Messer, D.F.; Doty, T.J.; MacFall, J.R.; Beyer, J.L.; Krishnan, K.R.R. The COMT Val158Met polymorphism and temporal lobe morphometry in healthy adults. Psychiatry Res. 2007, 155, 173–177. [Google Scholar] [CrossRef]
- Cerasa, A.; Gioia, M.C.; Labate, A.; Liguori, M.; Lanza, P.; Quattrone, A. Impact of catechol-O-methyltransferase Val(108/158) Met genotype on hippocampal and prefrontal gray matter volume. Neuroreport 2008, 19, 405–408. [Google Scholar] [CrossRef]
- Honea, R.; Verchinski, B.A.; Pezawas, L.; Kolachana, B.S.; Callicott, J.H.; Mattay, V.S.; Weinberger, D.R.; Meyer-Lindenberg, A. Impact of interacting functional variants in COMT on regional gray matter volume in human brain. Neuroimage 2009, 45, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Border, R.; Johnson, E.C.; Evans, L.M.; Smolen, A.; Berley, N.; Sullivan, P.F.; Keller, M.C. No Support for Historical Candidate Gene or Candidate Gene-by-Interaction Hypotheses for Major Depression Across Multiple Large Samples. Am. J. Psychiatry 2019, 176, 376–387. [Google Scholar] [CrossRef]
- Eker, M.C.; Kitis, O.; Okur, H.; Eker, O.D.; Ozan, E.; Isikli, S.; Akarsu, N.; Gonul, A.S. Smaller hippocampus volume is associated with short variant of 5-HTTLPR polymorphism in medication-free major depressive disorder patients. Neuropsychobiology 2011, 63, 22–28. [Google Scholar] [CrossRef]
- Frodl, T.; Koutsouleris, N.; Bottlender, R.; Born, C.; Jäger, M.; Mörgenthaler, M.; Scheuerecker, J.; Zill, P.; Baghai, T.; Schüle, C.; et al. Reduced gray matter brain volumes are associated with variants of the serotonin transporter gene in major depression. Mol. Psychiatry 2008, 13, 1093–1101. [Google Scholar] [CrossRef]
- Rabl, U.; Meyer, B.M.; Diers, K.; Bartova, L.; Berger, A.; Mandorfer, D.; Popovic, A.; Scharinger, C.; Huemer, J.; Kalcher, K.; et al. Additive gene-environment effects on hippocampal structure in healthy humans. J. Neurosci. 2014, 34, 9917–9926. [Google Scholar] [CrossRef] [PubMed]
- Little, K.; Olsson, C.A.; Whittle, S.; Youssef, G.J.; Byrne, M.L.; Simmons, J.G.; Yücel, M.; Foley, D.L.; Allen, N.B. Association between serotonin transporter genotype, brain structure and adolescent-onset major depressive disorder: A longitudinal prospective study. Transl. Psychiatry 2014, 4, e445. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.L.; Batten, L.A.; Tremblay, P.; Aldosary, F.; Du, L.; Blier, P. Impact of monoamine-related gene polymorphisms on hippocampal volume in treatment-resistant depression. Acta Neuropsychiatr. 2015, 27, 353–361. [Google Scholar] [CrossRef]
- Ahdidan, J.; Foldager, L.; Rosenberg, R.; Rodell, A.; Videbech, P.; Mors, O. Hippocampal volume and serotonin transporter polymorphism in major depressive disorder. Acta Neuropsychiatr. 2013, 25, 206–214. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, D.; Cui, H.; Zhao, T.; Chu, C.; Wang, J. Group-guided individual functional parcellation of the hippocampus and application to normal aging. Hum. Brain Mapp. 2021, 42, 5973–5984. [Google Scholar] [CrossRef] [PubMed]
- Tzioras, M.; Davies, C.; Newman, A.; Jackson, R.; Spires-Jones, T. Invited Review: APOE at the interface of inflammation, neurodegeneration and pathological protein spread in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 2019, 45, 327–346. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.-J. Critical Issues in BDNF Val66Met Genetic Studies of Neuropsychiatric Disorders. Front. Mol. Neurosci. 2018, 11, 156. [Google Scholar] [CrossRef]
- Popova, N.K.; Naumenko, V.S. Neuronal and behavioral plasticity: The role of serotonin and BDNF systems tandem. Expert Opin. Ther. Targets 2019, 23, 227–239. [Google Scholar] [CrossRef]
- Weerasinghe-Mudiyanselage, P.D.E.; Ang, M.J.; Kang, S.; Kim, J.-S.; Moon, C. Structural Plasticity of the Hippocampus in Neurodegenerative Diseases. Int. J. Mol. Sci. 2022, 23, 3349. [Google Scholar] [CrossRef]
- Stepan, J.; Dine, J.; Eder, M. Functional optical probing of the hippocampal trisynaptic circuit in vitro: Network dynamics, filter properties, and polysynaptic induction of CA1 LTP. Front. Neurosci. 2015, 9, 160. [Google Scholar] [CrossRef] [PubMed]
- Lazarov, O.; Hollands, C. Hippocampal neurogenesis: Learning to remember. Prog. Neurobiol. 2016, 138–140, 1–18. [Google Scholar] [CrossRef]
- Tahami Monfared, A.A.; Byrnes, M.J.; White, L.A.; Zhang, Q. Alzheimer’s Disease: Epidemiology and Clinical Progression. Neurol. Ther. 2022, 11, 553–569. [Google Scholar] [CrossRef] [PubMed]
- Kayed, R.; Lasagna-Reeves, C.A. Molecular Mechanisms of Amyloid Oligomers Toxicity. J. Alzheimer’s Dis. 2013, 33 (Suppl. 1), 67–78. [Google Scholar] [CrossRef]
- Sengupta, U.; Nilson, A.N.; Kayed, R. The Role of Amyloid-β Oligomers in Toxicity, Propagation, and Immunotherapy. EBioMedicine 2016, 6, 42–49. [Google Scholar] [CrossRef]
- Grabe, H.J.; Schwahn, C.; Mahler, J.; Appel, K.; Schulz, A.; Spitzer, C.; Fenske, K.; Barnow, S.; Freyberger, H.J.; Teumer, A.; et al. Genetic epistasis between the brain-derived neurotrophic factor Val66Met polymorphism and the 5-HTT promoter polymorphism moderates the susceptibility to depressive disorders after childhood abuse. Prog. Neuropsychopharmacol. Biol. Psychiatry 2012, 36, 264–270. [Google Scholar] [CrossRef]
- Li, S.; Weinstein, G.; Zare, H.; Teumer, A.; Völker, U.; Friedrich, N.; Knol, M.J.; Satizabal, C.L.; Petyuk, V.A.; Adams, H.H.H.; et al. The genetics of circulating BDNF: Towards understanding the role of BDNF in brain structure and function in middle and old ages. Brain Commun. 2020, 2, fcaa176. [Google Scholar] [CrossRef] [PubMed]
- Teng, H.K.; Teng, K.K.; Lee, R.; Wright, S.; Tevar, S.; Almeida, R.D.; Kermani, P.; Torkin, R.; Chen, Z.-Y.; Lee, F.S.; et al. ProBDNF induces neuronal apoptosis via activation of a receptor complex of p75NTR and sortilin. J. Neurosci. 2005, 25, 5455–5463. [Google Scholar] [CrossRef]
- Fleitas, C.; Piñol-Ripoll, G.; Marfull, P.; Rocandio, D.; Ferrer, I.; Rampon, C.; Egea, J.; Espinet, C. proBDNF is modified by advanced glycation end products in Alzheimer’s disease and causes neuronal apoptosis by inducing p75 neurotrophin receptor processing. Mol. Brain 2018, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Wang, L.; Nie, Y.; Wei, W.; Xiong, W. proBDNF expression induces apoptosis and inhibits synaptic regeneration by regulating the RhoA-JNK pathway in an in vitro post-stroke depression model. Transl. Psychiatry 2021, 11, 578. [Google Scholar] [CrossRef]
- Smith, G.S.; Barret, F.S.; Joo, J.H.; Nassery, N.; Savonenko, A.; Sodums, D.J.; Marano, C.M.; Munro, C.A.; Brandt, J.; Kraut, M.A.; et al. Molecular imaging of serotonin degeneration in mild cognitive impairment. Neurobiol. Dis. 2017, 105, 33–41. [Google Scholar] [CrossRef]
- Mattson, M.P.; Maudsley, S.; Martin, B. BDNF and 5-HT: A dynamic duo in age-related neuronal plasticity and neurodegenerative disorders. Trends Neurosci. 2004, 27, 589–594. [Google Scholar] [CrossRef]
- Haase, J.; Brown, E. Integrating the monoamine, neurotrophin and cytokine hypotheses of depression--a central role for the serotonin transporter? Pharmacol. Ther. 2015, 147, 1–11. [Google Scholar] [CrossRef]
- Levy, M.J.F.; Boulle, F.; Steinbusch, H.W.; van den Hove, D.L.A.; Kenis, G.; Lanfumey, L. Neurotrophic factors and neuroplasticity pathways in the pathophysiology and treatment of depression. Psychopharmacology 2018, 235, 2195–2220. [Google Scholar] [CrossRef] [PubMed]
- Porcelli, S.; Fabbri, C.; Serretti, A. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with antidepressant efficacy. Eur. Neuropsychopharmacol. 2012, 22, 239–258. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, E.M.; Gupta, N.K.; Syed, S.A.; Rozenboym, A.V.; Fulton, S.L.; Jackowski, A.P.; Perera, T.D.; Coplan, J.D. Developmental Antecedents of Adult Macaque Neurogenesis: Early-Life Adversity, 5-HTTLPR Polymorphisms, and Adolescent Hippocampal Volume. J. Affect. Disord. 2021, 286, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Kwak, G.-H.; Kamble, K.; Kim, H.-Y. Methionine sulfoxide reductase B3 deficiency inhibits cell growth through the activation of p53-p21 and p27 pathways. Arch. Biochem. Biophys. 2014, 547, 1–5. [Google Scholar] [CrossRef]
- Adams, S.L.; Benayoun, L.; Tilton, K.; Chavez, O.R.; Himali, J.J.; Blusztajn, J.K.; Seshadri, S.; Delalle, I. Methionine Sulfoxide Reductase-B3 (MsrB3) Protein Associates with Synaptic Vesicles and its Expression Changes in the Hippocampi of Alzheimer’s Disease Patients. JAD 2017, 60, 43–56. [Google Scholar] [CrossRef]
- Zheng, T.; Ge, B.; Qin, L.; Chen, B.; Tian, L.; Gao, Y.; Xiao, L.; Hu, X.; Pan, H.; Chen, Y. Association of Plasma DPP4 Activity and Brain-Derived Neurotrophic Factor With Moderate to Severe Depressive Symptoms in Patients With Type 2 Diabetes: Results From a Cross-Sectional Study. Psychosom. Med. 2020, 82, 350–358. [Google Scholar] [CrossRef]
- Behesti, H.; Fore, T.R.; Wu, P.; Horn, Z.; Leppert, M.; Hull, C.; Hatten, M.E. ASTN2 modulates synaptic strength by trafficking and degradation of surface proteins. Proc. Natl. Acad. Sci. USA. 2018, 115, E9717–E9726. [Google Scholar] [CrossRef]
- Bauleo, A.; Montesanto, A.; Pace, V.; Brando, R.; de Stefano, L.; Puntorieri, D.; Cento, L.; Loddo, S.; Calacci, C.; Novelli, A.; et al. Rare copy number variants in ASTN2 gene in patients with neurodevelopmental disorders. Psychiatr. Genet. 2021, 31, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Völzke, H.; Schössow, J.; Schmidt, C.O.; Jürgens, C.; Richter, A.; Werner, A.; Werner, N.; Radke, D.; Teumer, A.; Ittermann, T.; et al. Cohort Profile Update: The Study of Health in Pomerania (SHIP). Int. J. Epidemiol. 2022, 51, e372–e383. [Google Scholar] [CrossRef] [PubMed]
- Oswald, W.D.; Fleischmann, U.M. (Eds.) Nürnberger-Alters-Inventar: (NAI). In NAI-Testmanual und Textband; Hogrefe: Boston, MA, USA, 1999. [Google Scholar]
- Van der Auwera, S.; Terock, J.; Teumer, A.; Schomerus, G.; Homuth, G.; Grabe, H.J. Sex effects for the interaction of dopamine related genetic variants for COMT and BDNF on declarative memory performance. Genes Brain Behav. 2021, 20, e12737. [Google Scholar] [CrossRef]
- Wittchen, H.U. Reliability and Validity Studies of the WHO-Composite International Diagnostic Interview (CIDI: A Critical Review. J. Psychiatr. Res. 1994, 28, 57–84. [Google Scholar] [CrossRef]
- Wittchen, H.U.; Lachner, G.; Wunderlich, U.; Pfister, H. Test-retest reliability of the computerized DSM-IV version of the Munich-Composite International Diagnostic Interview (M-CIDI). Soc. Psychiatry Psychiatr. Epidemiol. 1998, 33, 568–578. [Google Scholar] [CrossRef]
- Kroenke, K.; Spitzer, R.L.; Williams, J.W. The PHQ-9: Validity of a Brief Depression Severity Measure. J. Gen. Intern. Med. 2001, 16, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Habes, M.; Toledo, J.B.; Resnick, S.M.; Doshi, J.; van der Auwera, S.; Erus, G.; Janowitz, D.; Hegenscheid, K.; Homuth, G.; Völzke, H.; et al. Relationship between APOE Genotype and Structural MRI Measures throughout Adulthood in the Study of Health in Pomerania Population-Based Cohort. AJNR. Am. J. Neuroradiol. 2016, 37, 1636–1642. [Google Scholar] [CrossRef]
- Ge, T.; Chen, C.-Y.; Ni, Y.; Feng, Y.-C.A.; Smoller, J.W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 2019, 10, 1776. [Google Scholar] [CrossRef]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; van der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef]
- Grabe, H.J.; Wittfeld, K.; Hegenscheid, K.; Hosten, N.; Lotze, M.; Janowitz, D.; Völzke, H.; John, U.; Barnow, S.; Freyberger, H.J. Alexithymia and brain gray matter volumes in a general population sample. Hum. Brain Mapp. 2014, 35, 5932–5945. [Google Scholar] [CrossRef] [PubMed]
- Hosten, N.; Bülow, R.; Völzke, H.; Domin, M.; Schmidt, C.O.; Teumer, A.; Ittermann, T.; Nauck, M.; Felix, S.B.; Dörr, M.; et al. SHIP-MR and Radiology: 12 Years of Whole-Body Magnetic Resonance Imaging in a Single Center. Healthcare 2022, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, J.E.; Augustinack, J.C.; Nguyen, K.; Player, C.M.; Player, A.; Wright, M.; Roy, N.; Frosch, M.P.; McKee, A.C.; Wald, L.L.; et al. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. Neuroimage 2015, 115, 117–137. [Google Scholar] [CrossRef] [PubMed]
- von Bohlen und Halbach, O. Involvement of BDNF in age-dependent alterations in the hippocampus. Front. Aging Neurosci. 2010, 2, 36. [Google Scholar] [CrossRef]
- StataCorp LLC. STATA 14; StataCorp: College Station, TX, USA, 2016. [Google Scholar]
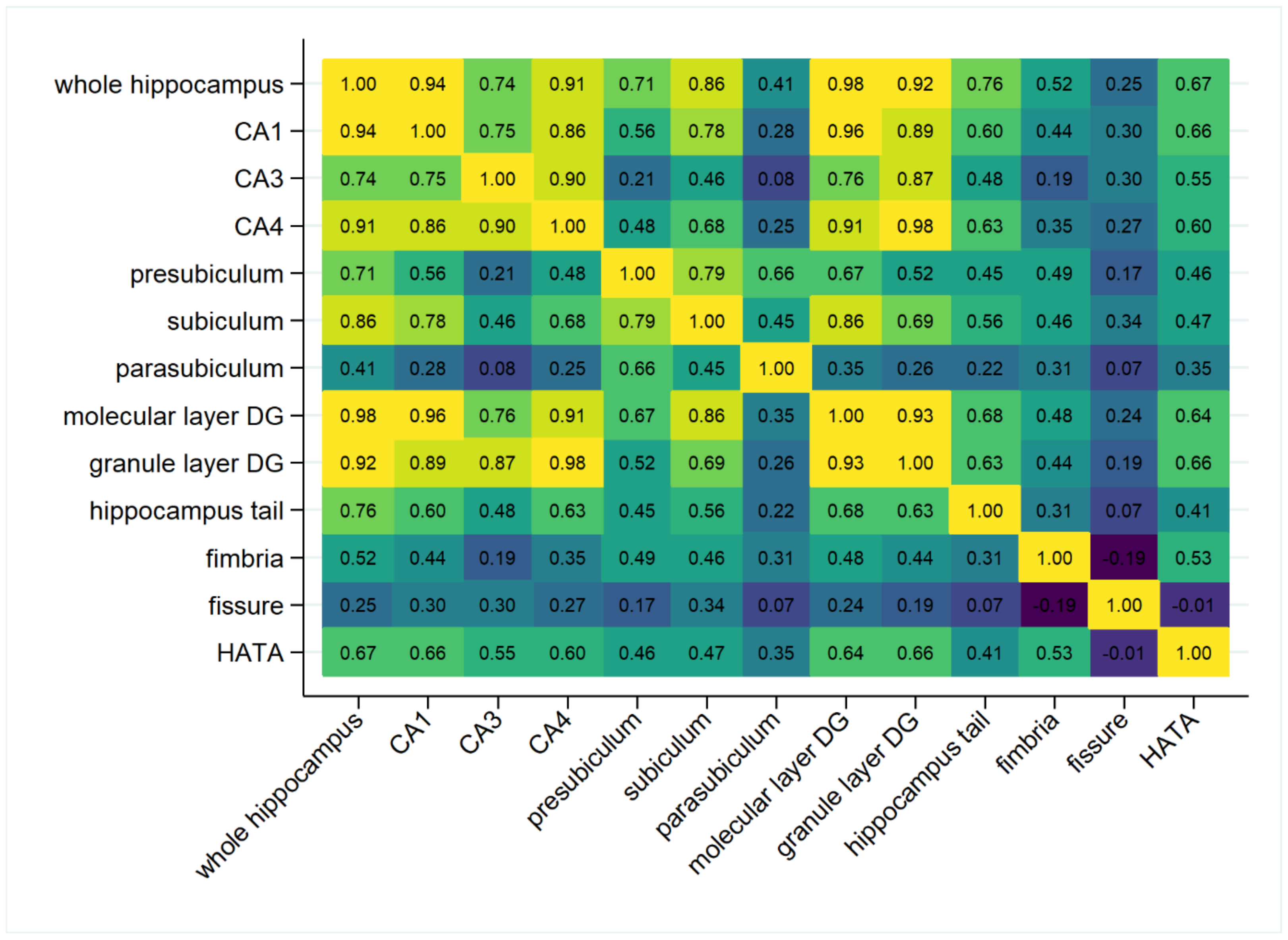
| Females (n = 955) | Males (n = 851) | Comparison | |
|---|---|---|---|
| Age | 50.6 (13.5), [21–81] | 50.4 (14.3), [21–81] | T = −0.23, p = 0.82 |
| Education | χ2 = 23.5, p < 0.001 | ||
| <10 years | 139 (14.6%) | 115 (13.5%) | |
| =10 years | 567 (59.4%) | 425 (49.9%) | |
| >10 years | 249 (26.0%) | 311 (36.6%) | |
| Hippocampal volume in cm3 | 6.6 (0.57), [4.5–8.3] | 7.2 (0.67), [5.0–9.7] | T = 20.7, p < 0.001 |
| ICV in cm3 | 1496 (116), [1054–1914] | 1693 (131), [1314–2135] | T = 33.8, p < 0.001 |
| MDD lifetime (yes) | 219 (22.9%) | 106 (12.5%) | χ2 = 33.9, p < 0.001 |
| Missing | 8 (0.8%) | 4 (0.5%) | |
| PHQ-9 sum score | 4.3 (3.68), [0–25] | 3.2 (3.36), [0–26] | T = −6.2, p < 0.001 |
| Missing | 66 (6.9%) | 53 (6.2%) | |
| Immediate verbal memory score | 5.54 (1.26), [0–8] | 5.25 (1.22), [0–8] | T = −5.07, p < 0.001 |
| Delayed verbal memory score | 6.03 (1.59), [−3–8] | 5.51 (1.68), [–1, 8] | T = −6.82, p < 0.001 |
| Missing | 6 (0.6%) | 9 (1.1%) | |
| APOE ε4 status | χ2 = 0.48, p = 0.49 | ||
| ε4 allele carrier | 217 (22.7) | 204 (24.0%) | |
| missing | 22 (2.3%) | 24 (2.8%) | |
| 5-HTTLPR | χ2 = 1.99, p = 0.37 | ||
| SS | 202 (21.2%) | 159 (18.7%) | |
| SL | 439 (46.0%) | 395 (46.4%) | |
| LL | 224 (23.5%) | 215 (25.3%) | |
| Missing | 90 (9.4%) | 82 (9.6%) | |
| COMT Val158Met | χ2 = 2.83, p = 0.24 | ||
| GG (Val/Val) | 254 (26.6%) | 252 (29.6%) | |
| GA (Val/Met) | 505 (52.9%) | 418 (49.1%) | |
| AA (Met/Met) | 196 (20.5%) | 181 (21.3%) | |
| BDNF Val66Met | χ2 = 3.68, p = 0.16 | ||
| AA (Met/Met) | 36 (3.8%) | 41 (4.8%) | |
| GA (Val/Met) | 274 (28.7%) | 270 (31.7%) | |
| GG (Val/Val) | 645 (67.5%) | 540 (63.5%) | |
| KIBRA rs17070145 | χ2 = 0.60, p = 0.74 | ||
| TT | 105 (11.0%) | 92 (10.8%) | |
| CT | 416 (43.6%) | 357 (42.0%) | |
| CC | 434 (45.4%) | 402 (47.2%) |
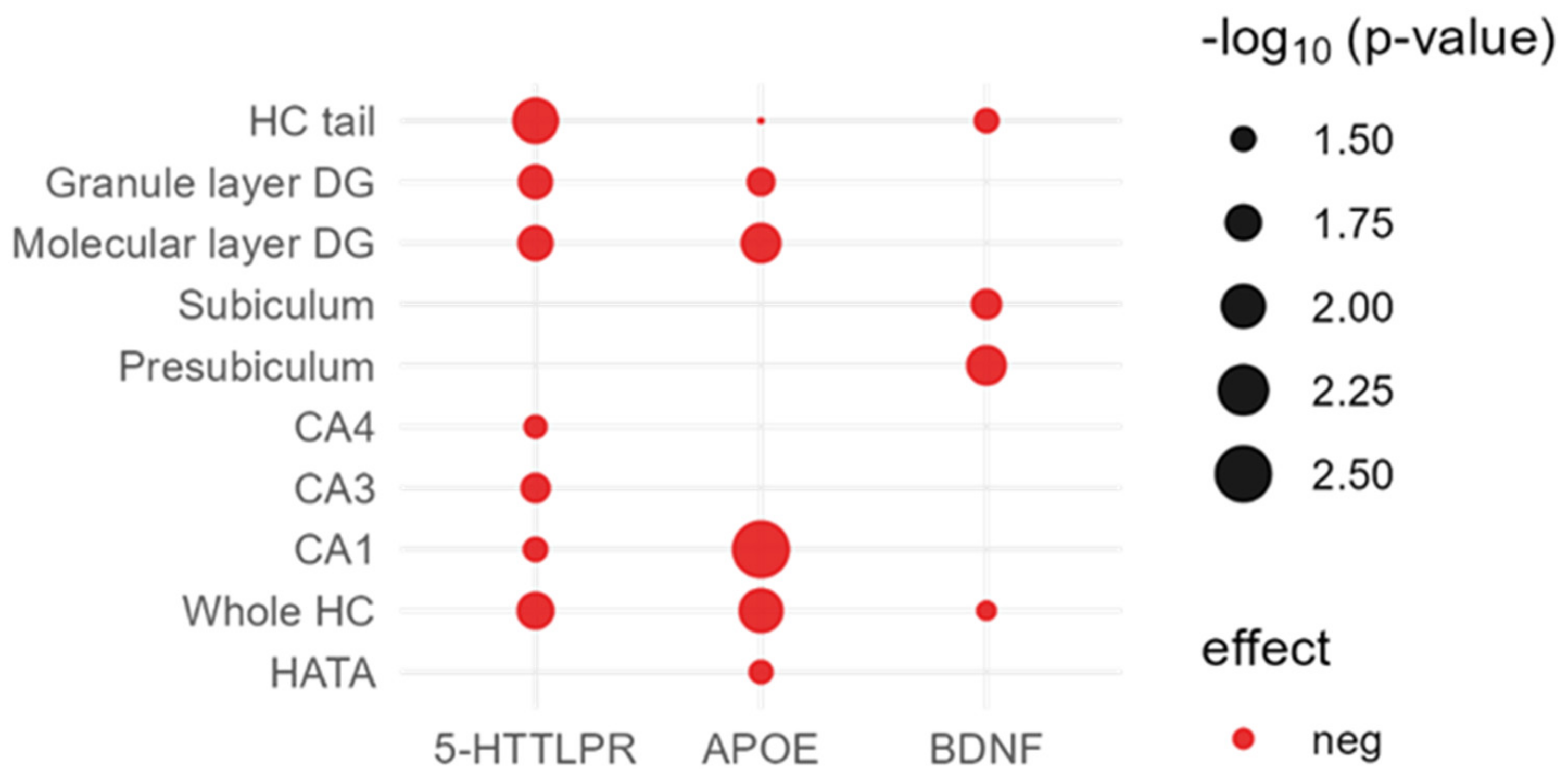
| HC Volume | APOE | 5-HTTLPR | BDNF | COMT | KIBRA | PRS AD |
|---|---|---|---|---|---|---|
| Whole HC | 0.010(−) | 0.017(−) | 0.038(−) | 0.180 (−) | 0.370 (−) | 0.564 (−) |
| CA1 | 0.003(−) | 0.032(−) | 0.310 (−) | 0.200 (−) | 0.540 (−) | 0.450 (−) |
| CA3 | 0.110 (−) | 0.026(−) | 0.640 (+) | 0.440 (−) | 0.680 (+) | 0.346 (+) |
| CA4 | 0.071 (−) | 0.034(−) | 0.210 (−) | 0.410 (−) | 0.390 (−) | 0.800 (+) |
| Presubiculum | 0.450 (−) | 0.740 (−) | 0.014(−) | 0.480 (−) | 0.580 (−) | 0.208 (−) |
| Subiculum | 0.190 (−) | 0.400 (−) | 0.025(−) | 0.500 (−) | 0.230 (−) | 0.347 (−) |
| Parasubiculum | 0.770 (+) | 0.740 (+) | 0.170 (−) | 0.910 (−) | 0.560 (−) | 0.742 (+) |
| Molecular layer DG | 0.014(−) | 0.019(−) | 0.084 (−) | 0.210 (−) | 0.500 (−) | 0.555 (−) |
| Granule layer DG | 0.028(−) | 0.020(−) | 0.140 (−) | 0.320 (−) | 0.500 (−) | 0.911 (−) |
| HC tail | 0.046(−) | 0.009(−) | 0.032(−) | 0.210 (−) | 0.170 (−) | 0.926 (−) |
| Fimbria | 0.350 (−) | 0.670 (−) | 0.910 (−) | 0.960 (+) | 0.180 (+) | 0.327 (−) |
| Fissure | 0.790 (+) | 0.810 (+) | 0.400 (+) | 0.580 (+) | 0.520 (−) | 0.351 (+) |
| HATA | 0.033(−) | 0.510 (−) | 0.840 (−) | 0.500 (−) | 0.640 (+) | 0.539 (−) |
| Lead SNP (Effect Allele) | Mapped Genes | Sig. Subfields GWAS | Sig. Subfields TREND-0 |
|---|---|---|---|
| rs12218858 (C) | FAM175B, FAM53B, METTL10 | Whole HC (+) | HC tail (+) |
| rs1419859 (C) | PARP11 | Whole HC (+) | Subiculum (−) |
| rs17178139 (G) | MSRB3 | Whole HC (+) | Whole HC, CA1, CA3, CA4, Molecular layer DG, Granule layer DG (all +) |
| rs160459 (A) | DACT1 | CA1 (−), Granule layer DG (−), HC tail (+) | CA3, CA4, Granule layer DG (all −) |
| rs6675690 (T) | / | HC tail (−) | None |
| rs10888696 (G) | DMRTA2, FAF1, CDKN2C | HC tail (−) | CA1, CA3, Fissure (all −) |
| rs1861979 (T) | DPP4 | Whole HC (+) | Whole HC, CA4, Granule layer DG, HC tail (all +) |
| rs7630893 (C) | ATP1B3, TFDP2 | Whole HC (+) | Fimbria (−) |
| rs57246240 (G) | MAST4 | Whole HC (−) | Whole HC, CA1, CA3, CA4, Presubiculum, Subiculum, Molecular layer DG, Granule layer DG, HC tail, Fissure (all −) |
| rs13188633 (C) | / | HC tail (+) | CA3, HATA (all −) |
| rs10474356 (A) | / | HC tail (+) | None |
| rs55736786 (C) | FAM172A, POU5F2 | HC tail (+) | None |
| rs9399619 (G) | SAMD5 | Subiculum (+) | None |
| rs7873551 (T) | ASTN2 | Whole HC (+) | Whole HC, CA1, CA4, Subiculum, Molecular layer DG, Granule layer DG, HC tail, HATA (all +) |
| rs4962694 (G) | FAM175B, FAM53B, METTL10 | Molecular layer DG (−) | HC tail (+) |
| rs17178006 (T) | WIF1, LEMD3, MSRB3 | CA1 (+), Presubiculum (−) | Whole HC, CA1, Molecular layer DG, HC tail (all +) |
| rs2909443 (G) | SLC4A10, DPP4 | HC tail (+) | Whole HC, CA4, Granule layer DG, HC tail (all +) |
| Volume | Short-Term Retrieval | Long-Term Retrieval | Long-Term Retrieval (Young) | Long-Term Retrieval (Old) |
|---|---|---|---|---|
| Whole HC | 0.550 (+) | 0.100 (+) | 0.740 (− | 0.017 (+) |
| CA1 | 0.930 (+) | 0.160 (+) | 0.540 (−) | 0.015 (+) |
| CA3 | 0.320 (+) | 0.210 (+) | 0.940 (+) | 0.120 (+) |
| CA4 | 0.340 (+) | 0.071 (+) | 10.000 (+) | 0.027 (+) |
| Presubiculum | 0.650 (+) | 0.120 (+) | 0.550 (+) | 0.100 (+) |
| Subiculum | 0.620 (+) | 0.170 (+) | 0.400 (−) | 0.018 (+) |
| Parasubiculum | 0.660 (+) | 0.350 (+) | 0.510 (+) | 0.430 (+) |
| Molecular layer DG | 0.690 (+) | 0.080 (+) | 0.710 (+) | 0.009 (+) |
| Granule layer DG | 0.360 (+) | 0.060 (+) | 0.980 (−) | 0.019 (+) |
| HC tail | 0.980 (+) | 0.970 (+) | 0.760 (−) | 0.760 (+) |
| Fimbria | 0.490 (+) | 0.330 (+) | 0.610 (+) | 0.310 (+) |
| Fissure | 0.330 (−) | 0.530 (+) | 0.510 (−) | 0.280 (+) |
| HATA | 0.350 (+) | 0.870 (+) | 0.410 (−) | 0.310 (+) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kirchner, K.; Garvert, L.; Wittfeld, K.; Ameling, S.; Bülow, R.; Meyer zu Schwabedissen, H.; Nauck, M.; Völzke, H.; Grabe, H.J.; Van der Auwera, S. Deciphering the Effect of Different Genetic Variants on Hippocampal Subfield Volumes in the General Population. Int. J. Mol. Sci. 2023, 24, 1120. https://doi.org/10.3390/ijms24021120
Kirchner K, Garvert L, Wittfeld K, Ameling S, Bülow R, Meyer zu Schwabedissen H, Nauck M, Völzke H, Grabe HJ, Van der Auwera S. Deciphering the Effect of Different Genetic Variants on Hippocampal Subfield Volumes in the General Population. International Journal of Molecular Sciences. 2023; 24(2):1120. https://doi.org/10.3390/ijms24021120
Chicago/Turabian StyleKirchner, Kevin, Linda Garvert, Katharina Wittfeld, Sabine Ameling, Robin Bülow, Henriette Meyer zu Schwabedissen, Matthias Nauck, Henry Völzke, Hans J. Grabe, and Sandra Van der Auwera. 2023. "Deciphering the Effect of Different Genetic Variants on Hippocampal Subfield Volumes in the General Population" International Journal of Molecular Sciences 24, no. 2: 1120. https://doi.org/10.3390/ijms24021120
APA StyleKirchner, K., Garvert, L., Wittfeld, K., Ameling, S., Bülow, R., Meyer zu Schwabedissen, H., Nauck, M., Völzke, H., Grabe, H. J., & Van der Auwera, S. (2023). Deciphering the Effect of Different Genetic Variants on Hippocampal Subfield Volumes in the General Population. International Journal of Molecular Sciences, 24(2), 1120. https://doi.org/10.3390/ijms24021120






