Enhancing the Heterologous Expression of a Thermophilic Endoglucanase and Its Cost-Effective Production in Pichia pastoris Using Multiple Strategies
Abstract
:1. Introduction
2. Results and Discussion
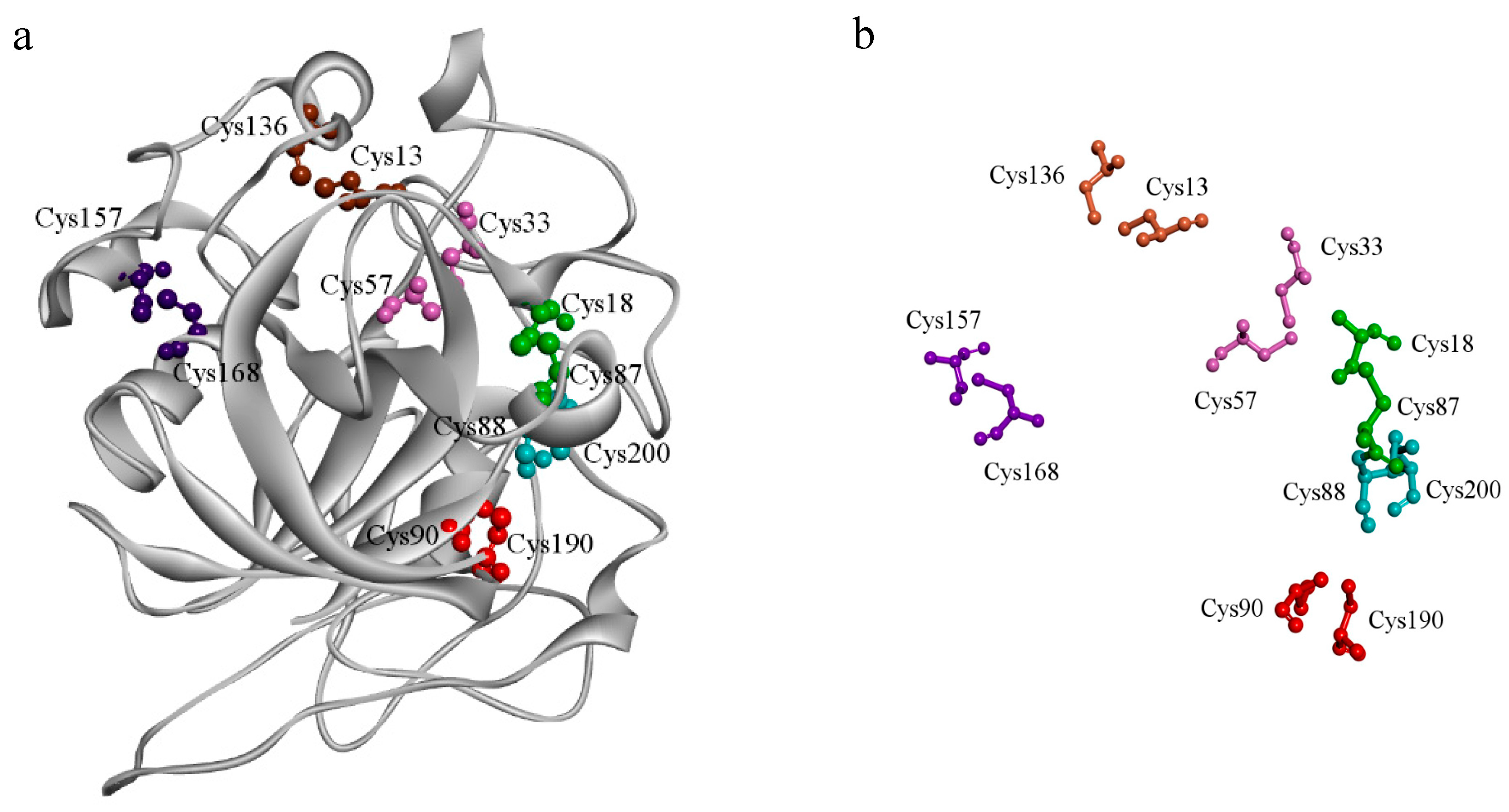
2.1. Overall Three-Dimensional Structure of Cel45A from T. terrestris
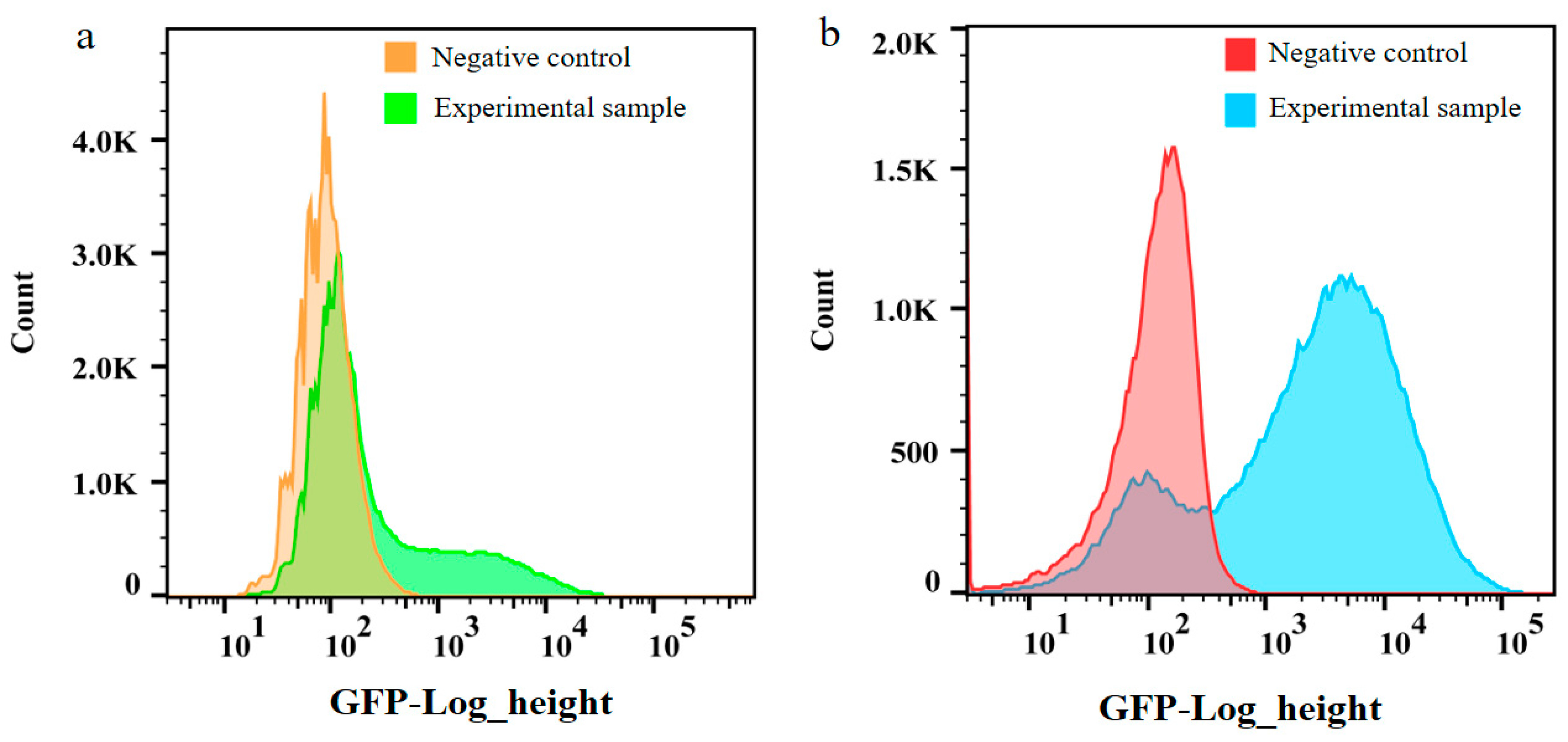
2.2. Comparison of CRISPR-Cas9 and Homologous Recombination for Chromosomal Integration
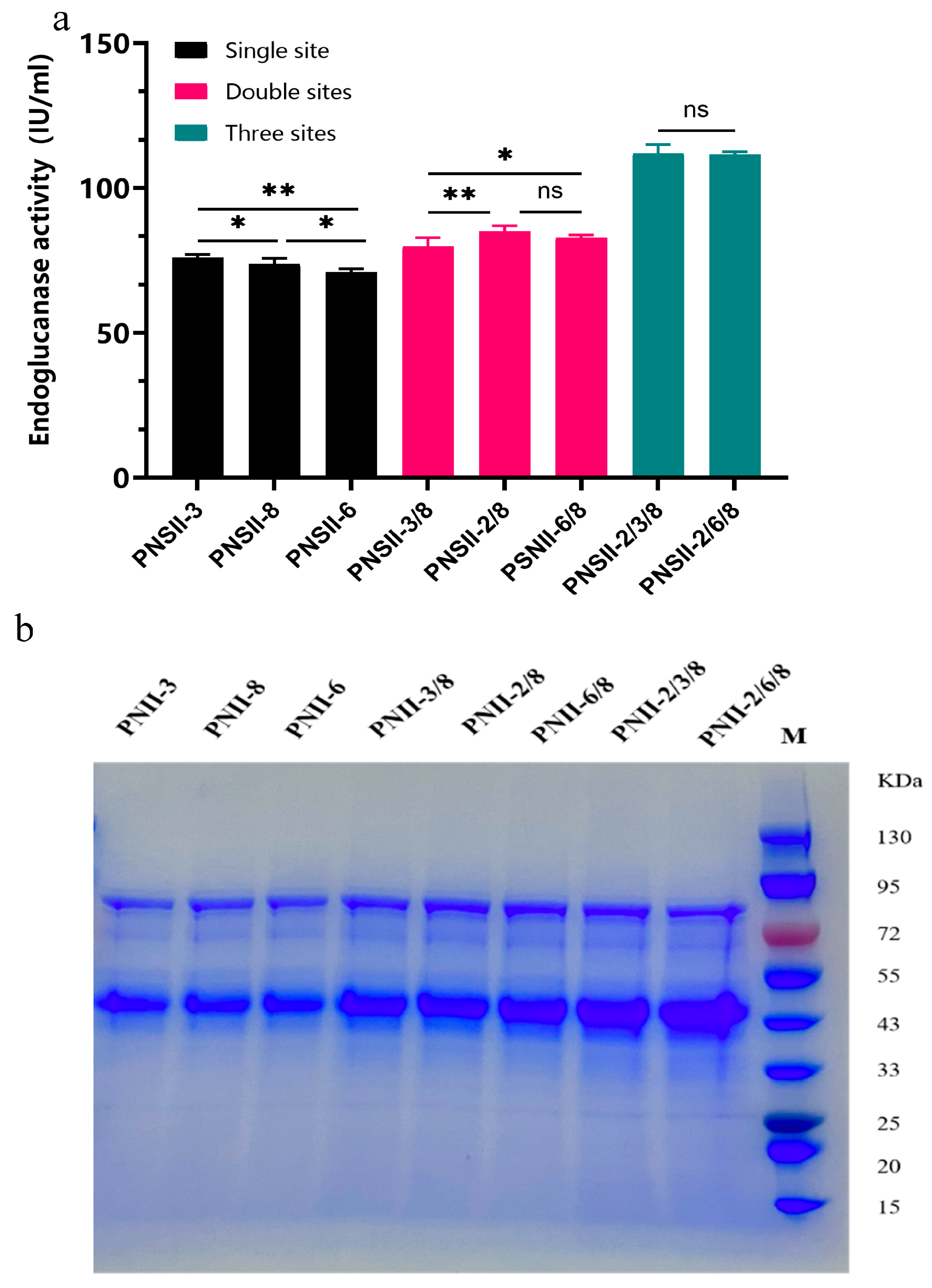
2.3. Selection of Neutral Integration Sites and Optimal Copy Numbers for Enhancing Heterologous Expression of TtCel45A in P. pastoris
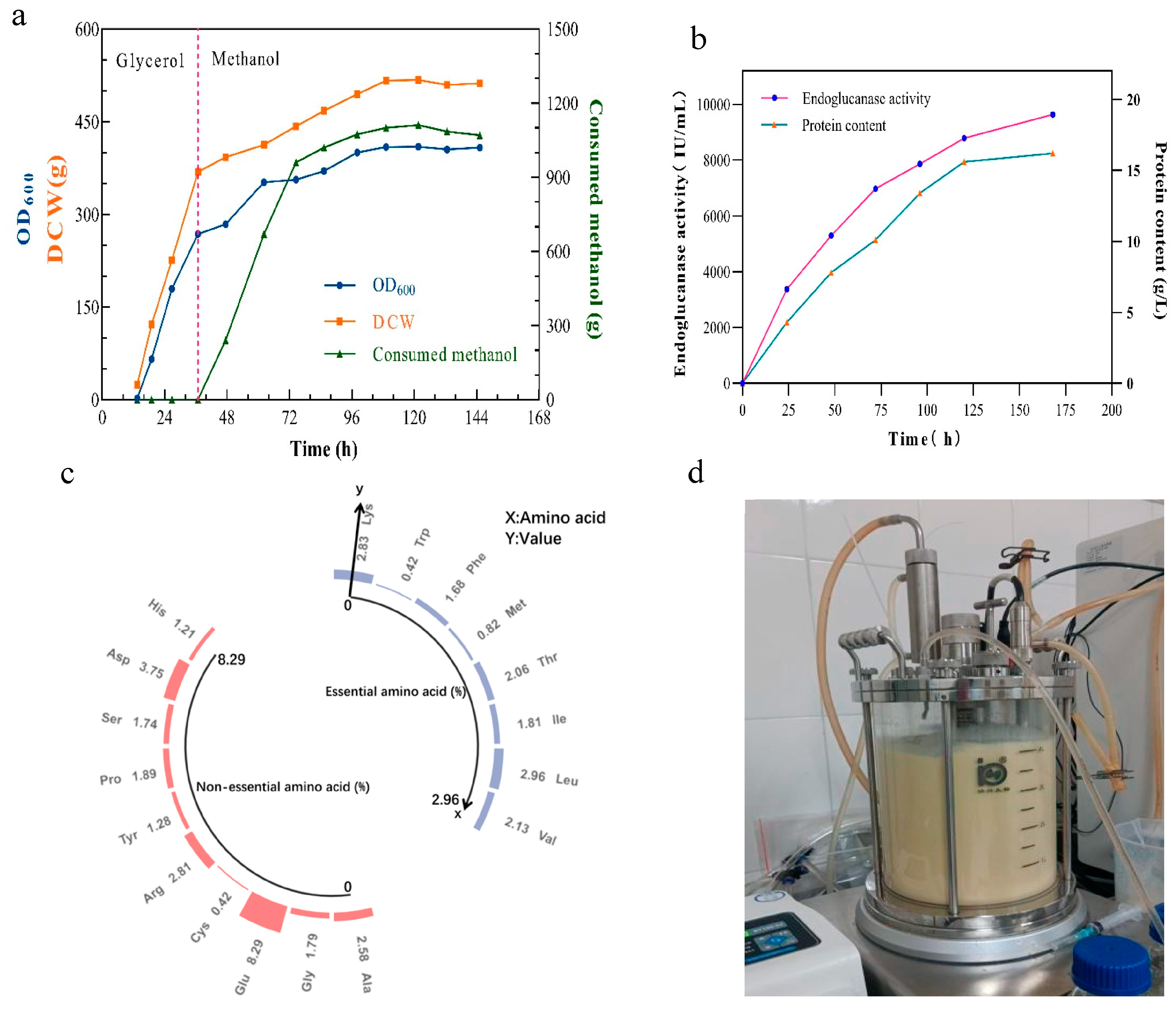
2.4. High-Cell-Density Fermentation of P. pastoris for Cost-Effective TtCel45A Production
2.5. Analyzing the Nutritional Value of Residual Single-Cell Protein to Enhance the Economic Viability of TtCel45 Fermentation
3. Materials and Methods
3.1. Strains and Culture Conditions
3.2. Plasmid and Strain Construction
3.3. Modeling of Endoglucanase Cel45A from Thermothielavioides terrestris
3.4. Enzyme Activity Measurements
3.5. Fed-Batch Fermentation
3.6. Biomass and Crude Protein Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baruah, J.; Nath, B.K.; Sharma, R.; Kumar, S.; Deka, R.C.; Baruah, D.C.; Kalita, E. Recent trends in the pretreatment of lignocellulosic biomass for value-added products. Front. Energy Res. 2018, 6, 141. [Google Scholar] [CrossRef]
- Mondal, S.; Halder, S.K.; Mondal, K.C. Tailoring in fungi for next generation cellulase production with special reference to CRISPR/CAS system. Syst. Microbiol. Biomanufacturing 2022, 2, 113–129. [Google Scholar] [CrossRef]
- Wang, B.T.; Hu, S.; Yu, X.Y.; Jin, L.; Zhu, Y.J.; Jin, F.J. Studies of cellulose and starch utilization and the regulatory mechanisms of related enzymes in fungi. Polymers 2020, 12, 530. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Soren, J.P.; Mondal, J.; Rakshit, S.; Halder, S.K.; Mondal, K.C. Contemporaneous synthesis of multiple carbohydrate debranching enzymes from newly isolated Aspergillus fumigatus SKF-2 under solid state fermentation: A unique enzyme mixture for proficient saccharification of plant bioresources. Ind. Crops. Prod. 2020, 150, 112409. [Google Scholar] [CrossRef]
- Samuel, M.S.; Govarthanan, M.; Selvarajan, E. A comprehensive review on strategic study of cellulase producing marine actinobacteria for biofuel applications. Environ. Res. 2022, 214, 114018. [Google Scholar] [CrossRef]
- Thanigaivel, S.; Priya, A.K.; Dutta, K.; Rajendran, S.; Sekar, K.; Jalil, A.A.; Soto-Moscoso, M. Role of nanotechnology for the conversion of lignocellulosic biomass into biopotent energy: A biorefinery approach for waste to value-added products. Fuel 2022, 322, 124236. [Google Scholar] [CrossRef]
- Horn, S.J.; Vaaje-Kolstad, G.; Westereng, B.; Eijsink, V. Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels. 2012, 5, 45. [Google Scholar] [CrossRef]
- Houfani, A.A.; Anders, N.; Spiess, A.C.; Baldrian, P.; Benallaoua, S. Insights from enzymatic degradation of cellulose and hemicellulose to fermentable sugars—A review. Biomass Bioenergy. 2020, 134, 105481. [Google Scholar] [CrossRef]
- Srivastava, N.; Srivastava, M.; Mishra, P.K.; Gupta, V.K.; Molina, G.; Rodriguez-Couto, S.; Manikanta, A.; Ramteke, P.W. Applications of fungal cellulases in biofuel production: Advances and limitations. Renew. Sust. Energ. Rev. 2018, 82, 2379–2386. [Google Scholar] [CrossRef]
- Singh, G.; Verma, A.K.; Kumar, V. Catalytic properties, functional attributes and industrial applications of β-glucosidases. 3 Biotech 2016, 6, 3. [Google Scholar] [CrossRef]
- Saroj, P.P.M.; Narasimhulu, K. Biochemical characterization of thermostable carboxymethyl cellulase and β-glucosidase from Aspergillus fumigatus JCM 10253. Appl. Biochem. 2022, 194, 2503–2527. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Hirvonen, M.; Papageorgiou, A.C. Crystal structure of a family 45 endoglucanase from Melanocarpus albomyces: Mechanistic implications based on the free and cellobiose-bound forms. J. Mol. Biol. 2003, 329, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, R.; Yang, X.; Xu, Y.; Tang, Z.; Tian, W.; Shen, Q. Expression, purification and characterization of two thermostable endoglucanases cloned from a lignocellulosic decomposing fungi Aspergillus fumigatus Z5 isolated from compost. Protein Expr. Purif. 2011, 79, 176–186. [Google Scholar] [CrossRef]
- Dashtban, M.; Schraft, H.; Qin, W. Fungal bioconversion of lignocellulosic residues; opportunities & perspectives. J. Biol. Sci. 2009, 5, 578. [Google Scholar] [CrossRef]
- Zheng, F.; Zhao, H.; Wang, N.; Zhong, P.; Zhou, K.; Yu, S. Cloning and Characterization of Thermophilic Cellulase and Its Application in the Transformation of Ginsenosides; Springer: Berlin/Heidelberg, Germany, 2022. [Google Scholar] [CrossRef]
- Gibson, K.; Hansen, L. Detergent Composition Comprising Endoglucanase. Patent no. WO2004053039 A2, 24 June 2004. [Google Scholar]
- Vlasenko, E.; Schülein, M.; Cherry, J.; Xu, F. Substrate specificity of family 5, 6, 7, 9, 12, and 45 endoglucanases. Bioresour. Technol. 2010, 101, 2405–2411. [Google Scholar] [CrossRef]
- Berka, R.M.; Grigoriev, I.V.; Otillar, R.; Salamov, A.; Grimwood, J.; Reid, I.; Ishmael, N.; John, T.; Darmond, C.; Moisan, M.C.; et al. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat. Biotechnol. 2011, 29, 922–927. [Google Scholar] [CrossRef]
- Gao, J.; Huang, J.W.; Li, Q.; Liu, W.; Ko, T.P.; Zheng, Y.; Kuo, J.C.; Guo, R.T. Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1, 4-β-endoglucanase from Thielavia terrestris. Enzym. Microb. Tech. 2017, 99, 32–37. [Google Scholar] [CrossRef]
- Davies, G.J.; Dodson, G.G.; Hubbard, R.E.; Tolley, S.P.; Dauter, Z.; Wilson, K.S.; Rasmussen, G.; Mikkelsen, M.J.; Schülein, M. Structure and function of endoglucanase V. Nature 1993, 365, 362–364. [Google Scholar] [CrossRef]
- Pitman, D.J.; Banerjee, S.; Macari, S.J.; Castaldi, C.A.; Crone, D.E.; Bystroff, C. Exploring the folding pathway of green fluorescent protein through disulfide engineering. Protein Sci. 2015, 24, 341–353. [Google Scholar] [CrossRef]
- Le, Q.A.T.; Joo, J.C.; Yoo, Y.J.; Kim, Y.H. Development of thermostable Candida antarctica lipase B through novel in silico design of disulfide bridge. Biotechnol. Bioeng. 2012, 109, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Safder, I.; Khan, S.; Islam, I.U.; Ali, M.K.; Bibi, Z.; Waqas, M. Pichia pastoris expression system: A potential candidate to express protein in industrial and biopharmaceutical domains. Biomed. Eng. Lett. 2014, 4, 223–234. [Google Scholar] [CrossRef]
- Gasser, B.; Prielhofer, R.; Marx, H.; Maurer, M.; Nocon, J.; Steiger, M.; Puxbaum, V.; Sauer, M.; Mattanovich, D. Pichia pastoris: Protein production host and model organism for biomedical research. Future Microbiol. 2013, 8, 191–208. [Google Scholar] [CrossRef]
- Zhang, K.; Duan, X.; Cai, P.; Gao, L.; Wu, X.; Yao, L.; Zhou, Y.J. Fusing an exonuclease with Cas9 enhances homologous recombination in Pichia pastoris. Microb. Cell Factories 2022, 21, 182. [Google Scholar] [CrossRef] [PubMed]
- Patra, P.; Das, M.; Kundu, P.; Ghosh, A. Recent advances in systems and synthetic biology approaches for developing novel cell-factories in non-conventional yeasts. Biotechnol. Adv. 2021, 47, 107695. [Google Scholar] [CrossRef]
- Liu, S.; Dong, H.; Hong, K.; Meng, J.; Lin, L.; Wu, X. Improving Methanol Utilization by Reducing Alcohol Oxidase Activity and Adding Co-Substrate of Sodium Citrate in Pichia pastoris. J. Fungi. 2023, 9, 422. [Google Scholar] [CrossRef] [PubMed]
- Cai, P.; Duan, X.; Wu, X.; Gao, L.; Ye, M.; Zhou, Y.J. Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris. Nucleic Acids Res. 2021, 49, 7791–7805. [Google Scholar] [CrossRef]
- Liu, Q.; Shi, X.; Song, L.; Liu, H.; Zhou, X.; Wang, Q.; Zhang, X.Y.; Cai, M. CRISPR–Cas9-mediated genomic multiloci integration in Pichia pastoris. Microb. Cell Factories. 2019, 18, 144. [Google Scholar] [CrossRef]
- Arnone, J.T. Genomic considerations for the modification of Saccharomyces cerevisiae for biofuel and metabolite biosynthesis. Microorganisms 2020, 8, 321. [Google Scholar] [CrossRef]
- Kong, S.; Yu, W.; Gao, N.; Zhai, X.; Zhou, Y.J. Expanding the neutral sites for integrated gene expression in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2022, 369, fnac081. [Google Scholar] [CrossRef]
- Yu, W.; Gao, J.; Zhai, X.; Zhou, Y.J. Screening neutral sites for metabolic engineering of methylotrophic yeast Ogataea polymorpha. Synth. Syst. Biotechnol. 2021, 6, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Meng, Q.; Dong, G.; Qi, N.; Zhao, H.; Shi, S. Metabolic engineering of threonine catabolism enables Saccharomyces cerevisiae to produce propionate under aerobic conditions. Biotechnol. J. 2022, 17, 2100579. [Google Scholar] [CrossRef] [PubMed]
- Boël, G.; Letso, R.; Neely, H.; Price, W.N.; Wong, K.H.; Su, M.; Acton, B.T.; Xiao, R.; Montelione, T.G.; Aalberts, P.D.; et al. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature 2016, 529, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Cittadino, G.M.; Andrews, J.; Purewal, H.; Estanislao Acuña Avila, P.; Arnone, J.T. Functional Clustering of Metabolically Related Genes Is Conserved across Dikarya. J. Fungi. 2023, 9, 523. [Google Scholar] [CrossRef] [PubMed]
- Hagee, D.; Hardan, A.A.; Botero, J.; Arnone, J.T. Genomic clustering within functionally related gene families in Ascomycota fungi. Comput. Struct. Biotechnol. J. 2020, 18, 3267–3277. [Google Scholar] [CrossRef]
- Quintero-Cadena, P.; Sternberg, P.W. Enhancer sharing promotes neighborhoods of transcriptional regulation across eukaryotes. G3-Genes Genom Genet. 2016, 6, 4167–4174. [Google Scholar] [CrossRef]
- Riesenberg, D.; Guthke, R. High-cell-density cultivation of microorganisms. Appl. Microbiol. Biot. 1999, 51, 422–430. [Google Scholar] [CrossRef]
- Lu, Y.; Fang, C.; Wang, Q.; Zhou, Y.; Zhang, G.; Ma, Y. High-level expression of improved thermo-stable alkaline xylanase variant in Pichia Pastoris through codon optimization, multiple gene insertion and high-density fermentation. Sci. Rep. 2016, 6, 37869. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, Y.; Shi, X.; Zhou, M.; Bao, L.; Hatanpaa, K.J.; Patel, T.; DeBerardinis, J.R.; Wang, F.Y.; Luo, W. Regulation of branched-chain amino acid metabolism by hypoxia-inducible factor in glioblastoma. Cell. Mol. Life Sci. 2021, 78, 195–206. [Google Scholar] [CrossRef]
- Pires, C.M.; Ribeiro, A.B.; Mateus, E.P.; Ponte, H.A.; Ponte, M.J.J. Extraction of rare earth elements via electric field assisted mining applying deep eutectic solvents. Sustain. Chem. Pharm. 2022, 26, 100638. [Google Scholar] [CrossRef]
- Yap, M.; Ercolini, D.; Álvarez-Ordóñez, A.; O’Toole, P.W.; O’Sullivan, O.; Cotter, P.D. Next-generation food research: Use of meta-omic approaches for characterizing microbial communities along the food chain. Annu. Rev. Food. 2022, 13, 361–384. [Google Scholar] [CrossRef] [PubMed]
- Iwai, S.; Hasegawa, T.; Ikeda, H.O.; Tsujikawa, A. Branched Chain Amino Acids Promote ATP Production via Translocation of Glucose Transporters. Invest. Ophth. Vis. Sci. 2022, 63, 7. [Google Scholar] [CrossRef]
- Mahalingam, R.; Ahmad Ahanger, W. New technologies-BlockChain as a Service (BCaaS) for healthcare. J. Emerg. Med. 2021, 2020, 15. [Google Scholar] [CrossRef]
- Rachamontree, P.; Phusantisampan, T.; Woravutthikul, N.; Pornwongthong, P.; Sriariyanun, M. Selection of Pichia kudriavzevii strain for the production of single-cell protein from cassava processing waste. Int. J. Bioeng. Life Sci. 2015, 9, 517–521. [Google Scholar] [CrossRef]
- Ritala, A.; Häkkinen, S.T.; Toivari, M.; Wiebe, M.G. Single cell protein—State-of-the-art, industrial landscape and patents 2001–2016. Front Microbiol. 2017, 8, 2009. [Google Scholar] [CrossRef] [PubMed]
- Anupama, R.P.; Ravindra, P. Value-added food: Single cell protein. Biotechnol. Adv. 2000, 18, 459–479. [Google Scholar] [CrossRef]
- Olstorpe, M.; Passoth, V. Pichia anomala in grain biopreservation. Antonie Van Leeuwenhoek 2011, 99, 57–62. [Google Scholar] [CrossRef]
- Kuhad, R.C.; Singh, A.; Tripathi, K.K.; Saxena, R.K.; Eriksson, K.E.L. Microorganisms as an alternative source of protein. Nutr. Rev. 1997, 55, 65–75. [Google Scholar] [CrossRef]
- Bydlinski, N.; Maresch, D.; Schmieder, V.; Klanert, G.; Strasser, R.; Borth, N. The contributions of individual galactosyltransferases to protein specific N-glycan processing in Chinese Hamster Ovary cells. J. Biotechnol. 2018, 282, 101–110. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Yan, J.; Liu, W.; Li, Y.; Lai, H.L.; Zheng, Y.; Huang, J.W.; Li, Z.H.; Guo, R.T. Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-β-endoglucanase. Biochem. Biophys. Res. Commun. 2016, 475, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Chen, M.; Zeng, X.; Li, W.; Liang, S.; Lin, Y. Improving the catalytic performance of Pichia pastoris whole-cell biocatalysts by fermentation process. RSC Adv. 2021, 11, 36329–36339. [Google Scholar] [CrossRef] [PubMed]
- Marcó, A.; Rubio, R.; Compañó, R.; Casals, I. Comparison of the Kjeldahl method and a combustion method for total nitrogen determination in animal feed. Talanta 2002, 57, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, W.; Dong, H.; Zhang, Z.; Wu, X.; Bao, T.; Gao, L.; Chen, X. Enhancing the Heterologous Expression of a Thermophilic Endoglucanase and Its Cost-Effective Production in Pichia pastoris Using Multiple Strategies. Int. J. Mol. Sci. 2023, 24, 15017. https://doi.org/10.3390/ijms241915017
Dai W, Dong H, Zhang Z, Wu X, Bao T, Gao L, Chen X. Enhancing the Heterologous Expression of a Thermophilic Endoglucanase and Its Cost-Effective Production in Pichia pastoris Using Multiple Strategies. International Journal of Molecular Sciences. 2023; 24(19):15017. https://doi.org/10.3390/ijms241915017
Chicago/Turabian StyleDai, Wuling, Haofan Dong, Zhaokun Zhang, Xin Wu, Tongtong Bao, Le Gao, and Xiaoyi Chen. 2023. "Enhancing the Heterologous Expression of a Thermophilic Endoglucanase and Its Cost-Effective Production in Pichia pastoris Using Multiple Strategies" International Journal of Molecular Sciences 24, no. 19: 15017. https://doi.org/10.3390/ijms241915017
APA StyleDai, W., Dong, H., Zhang, Z., Wu, X., Bao, T., Gao, L., & Chen, X. (2023). Enhancing the Heterologous Expression of a Thermophilic Endoglucanase and Its Cost-Effective Production in Pichia pastoris Using Multiple Strategies. International Journal of Molecular Sciences, 24(19), 15017. https://doi.org/10.3390/ijms241915017




