The Engineering, Expression, and Immobilization of Epimerases for D-allulose Production
Abstract
1. Introduction
2. Characterization of Newly Identified DAEases
3. Engineering of DAEases
3.1. Engineering for Improved Thermotolerance
3.2. Engineering for Improved Acid Resistance
4. Utilization and Engineering of B. subtilis for D-Allulose Biosynthesis
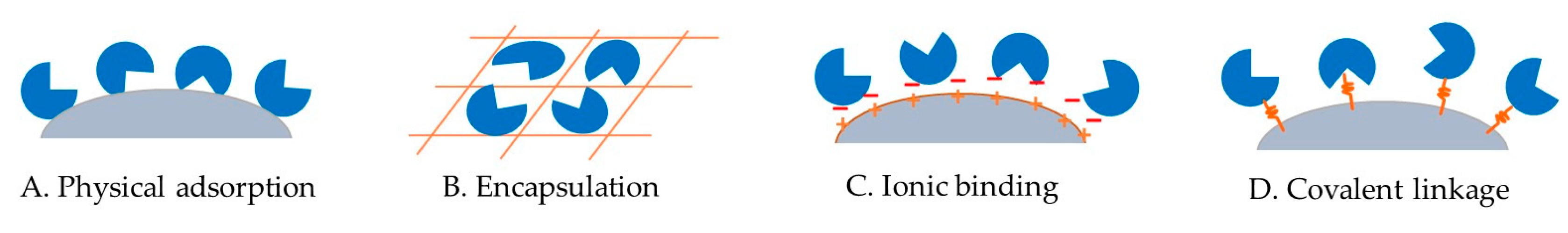
5. Immobilization
6. Summary and Future Vision
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chan, H.-C.; Zhu, Y.; Hu, Y.; Ko, T.-P.; Huang, C.-H.; Ren, F.; Chen, C.-C.; Ma, Y.; Guo, R.-T.; Sun, Y. Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars. Protein Cell 2012, 3, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, A.; Carrascosa, C.; Raheem, D.; Ramos, F.; Raposo, A. Natural Sweeteners: The Relevance of Food Naturalness for Consumers, Food Security Aspects, Sustainability and Health Impacts. Int. J. Environ. Res. Public Health 2020, 17, 6285. [Google Scholar] [CrossRef] [PubMed]
- Debras, C.; Chazelas, E.; Srour, B.; Druesne-Pecollo, N.; Esseddik, Y.; Szabo de Edelenyi, F.; Agaësse, C.; De Sa, A.; Lutchia, R.; Gigandet, S.; et al. Artificial Sweeteners and Cancer Risk: Results from the NutriNet-Santé Population-Based Cohort Study. PLoS Med. 2022, 19, e1003950. [Google Scholar] [CrossRef]
- Hossain, A.; Yamaguchi, F.; Matsuo, T.; Tsukamoto, I.; Toyoda, Y.; Ogawa, M.; Nagata, Y.; Tokuda, M. Rare sugar D-allulose: Potential role and therapeutic monitoring in maintaining obesity and type 2 diabetes mellitus. Pharmacol. Ther. 2015, 155, 49–59. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, X.-D.; Li, Z. Recent Advances Regarding the Physiological Functions and Biosynthesis of D-Allulose. Front. Microbiol. 2022, 13, 881037. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hayakawa, S.; Chuamanochan, M.; Fujimoto, M.; Innun, A.; Izumori, K. Antioxidant Effects of Maillard Reaction Products Obtained from Ovalbumin and Different D-Aldohexoses. Biosci. Biotechnol. Biochem. 2006, 70, 598–605. [Google Scholar] [CrossRef]
- Suna, S.; Yamaguchi, F.; Kimura, S.; Tokuda, M.; Jitsunari, F. Preventive effect of D-psicose, one of rare ketohexoses, on di-(2-ethylhexyl) phthalate (DEHP)-induced testicular injury in rat. Toxicol. Lett. 2007, 173, 107–117. [Google Scholar] [CrossRef]
- Kim, S.-E.; Kim, S.J.; Kim, H.-J.; Sung, M.-K. D-Psicose, a sugar substitute, suppresses body fat deposition by altering networks of inflammatory response and lipid metabolism in C57BL/6J-ob/ob mice. J. Funct. Foods 2017, 28, 265–274. [Google Scholar] [CrossRef]
- Takata, M.; Yamaguchi, F.; Nakanose, K.; Watanabe, Y.; Hatano, N.; Tsukamoto, I.; Nagata, M.; Izumori, K.; Tokuda, M. Neuroprotective effect of D-Psicose on 6-hydroxydopamine-induced apoptosis in rat pheochromocytoma (PC12) cells. J. Biosci. Bioeng 2005, 100, 511–516. [Google Scholar] [CrossRef]
- Granström, T.; Takata, G.; Tokuda, M.; Izumori, K. Izumoring: A novel and complete strategy for bioproduction of rare sugars. J. Biosci. Bioeng. 2004, 97, 89–94. [Google Scholar] [CrossRef]
- Hu, M.; Wei, Y.; Zhang, R.; Shao, M.; Yang, T.; Xu, M.; Zhang, X.; Rao, Z. Efficient D-allulose synthesis under acidic conditions by auto-inducing expression of the tandem D-allulose 3-epimerase genes in Bacillus subtilis. Microb. Cell Factories 2022, 21, 63. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, T.; Tu, S.-J.; Mu, W. Enzymatic approaches to rare sugar production. Biotechnol. Adv. 2017, 35, 267–274. [Google Scholar] [CrossRef]
- Jiang, S.; Xiao, W.; Zhu, X.; Yang, P.; Zheng, Z.; Lu, S.; Jiang, S.; Zhang, G.; Liu, J. Review on D-Allulose: In Vivo Metabolism, Catalytic Mechanism, Engineering Strain Construction, Bio-Production Technology. Front. Bioeng. Biotechnol. 2020, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Cheng, Q.; Mu, W.; Hu, X.; Sun, Z.; Qiu, Y.; Liu, X.; Wang, Z. Research Advances of D-Allulose: An Overview of Physiological Functions, Enzymatic Biotransformation Technologies, and Production Processes. Foods 2021, 10, 2186. [Google Scholar] [CrossRef] [PubMed]
- Laksmi, F.A.; Nirwantono, R.; Nuryana, I.; Agustriana, E. Expression and characterization of thermostable D-allulose 3-epimerase from Arthrobacter psychrolactophilus (Ap DAEase) with potential catalytic activity for bioconversion of D-allulose from D-fructose. Int. J. Biol. Macromol. 2022, 214, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Feng, L.; Chen, Z.; Hu, Y.; Fei, K.; Xu, H.; Gao, X. Efficient enzymatic synthesis of D-allulose using a novel D-allulose-3-epimerase from Caballeronia insecticola. J. Sci. Food Agric. 2022, 103, 339–348. [Google Scholar] [CrossRef]
- Wulansari, S.; Heng, S.; Ketbot, P.; Baramee, S.; Waeonukul, R.; Pason, P.; Ratanakhanokchai, K.; Uke, A.; Kosugi, A.; Tachaapaikoon, C. A Novel D-Psicose 3-Epimerase from Halophilic, Anaerobic Iocasia fonsfrigidae and Its Application in Coconut Water. Int. J. Mol. Sci. 2023, 24, 6394. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.N.; Kaushal, G.; Singh, S.P. D-Allulose 3-Epimerase of Bacillus Sp. Origin Manifests Profuse Heat-Stability and Noteworthy Potential of D-Fructose Epimerization. Microb. Cell Factories 2021, 20, 60. [Google Scholar] [CrossRef]
- Mu, W.; Chu, F.; Xing, Q.; Yu, S.; Zhou, L.; Jiang, B. Cloning, Expression, and Characterization of a D-Psicose 3-Epimerase from Clostridium Cellulolyticum H10. J. Agric. Food Chem. 2011, 59, 7785–7792. [Google Scholar] [CrossRef]
- Zhang, W.; Li, H.; Zhang, T.; Tu, S.-J.; Zhou, L.; Mu, W. Characterization of a D-Psicose 3-Epimerase from Dorea Sp. CAG317 with an Acidic PH Optimum and a High Specific Activity. J. Mol. Catal. B Enzym. 2015, 120, 68–74. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Liu, S.; Guo, X.; Zhao, T.; Wu, J.; Chen, S. Boosting the Heterologous Expression of D-Allulose 3-Epimerase in Bacillus subtilis through Protein Engineering and Catabolite-Responsive Element Box Engineering. J. Agric. Food Chem. 2022, 70, 12128–12134. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Pu, Z.; Zhu, L.; Wu, M.; Yang, L.; Yu, H.; Lin, J. Enhancing the thermostability of D-allulose 3-epimerase from Clostridium cellulolyticum H10 via a dual-enzyme screening system. Enzym. Microb. Technol. 2022, 159, 110054. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, D.-S.; Chen, Q.; Xu, W.; Zhang, W.; Mu, W. Computer-Aided Targeted Mutagenesis of Thermoclostridium caenicola D-Allulose 3-Epimerase for Improved Thermostability. J. Agric. Food Chem. 2022, 70, 1943–1951. [Google Scholar] [CrossRef] [PubMed]
- Tseng, W.-C.; Hsu, C.-T.; Chang, H.-C.; Wang, M.-J.; Fang, T.-Y. Fusion of the peptide derived from the acidic tail of alpha-synuclein improves the thermostability and soluble expression of recombinant Agrobacterium sp. D-allulose 3-epimerase. Biochem. Eng. J. 2021, 165, 107828. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.M.; Zhao, J.; Li, H.; Wei, X.; Zhou, J. Improved thermostability of D-allulose 3-epimerase from Clostridium bolteae ATCC BAA-613 by proline residue substitution. Protein Expr. Purif. 2022, 199, 106145. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Wang, T.; Qi, H.R.; Wei, M.; Lu, F.; Guan, L.; Mao, S.; Qin, H.-M. Engineering of Acid-Resistant D-Allulose 3-Epimerase for Functional Juice Production. J. Agric. Food Chem. 2022, 70, 16298–16306. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Tseng, W.-C.; Chen, C.-N.; Hsu, C.-T.; Lee, H.-C.; Fang, H.; Zhang, D.; Wu, Y.-H.; Fang, T.-Y. Characterization of a recombinant D-allulose 3-epimerase from Agrobacterium sp. ATCC 31749 and identification of an important interfacial residue. Int. J. Biol. Macromol. 2018, 112, 767–774. [Google Scholar] [CrossRef]
- Zhang, W.; Wei, M.; Sun, X.; Lu, F.; Guan, L.-J.; Mao, S.; Qin, H.-M. Fine-Tuning of Carbon Flux and Artificial Promoters in Bacillus subtilis Enables High-Level Biosynthesis of D-Allulose. J. Agric. Food Chem. 2022, 70, 13935–13944. [Google Scholar] [CrossRef]
- Weickert, M.J.; Chambliss, G.H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 1990, 87, 6238–6242. [Google Scholar] [CrossRef]
- Su, L.; Sun, F.; Liu, Z.; Zhang, K.; Wu, J. Highly efficient production of Clostridium cellulolyticum H10 D-psicose 3-epimerase in Bacillus subtilis and use of these cells to produce D-psicose. Microb. Cell Factories 2018, 17, 188. [Google Scholar] [CrossRef]
- Takeshita, K.; Suga, A.; Takada, G.; Izumori, K. Mass production of D-psicose from d-fructose by a continuous bioreactor system using immobilized D-tagatose 3-epimerase. J. Biosci. Bioeng. 2000, 90, 453–455. [Google Scholar] [CrossRef]
- Hu, M.; Li, M.; Jiang, B.; Zhang, T. Bioproduction of D-allulose: Properties, applications, purification, and future perspectives. Compr. Rev. Food Sci. Food Saf. 2021, 20, 6012–6026. [Google Scholar] [CrossRef]
- Dedania, S.R.; Patel, V.K.; Soni, S.S.; Patel, D.H. Immobilization of Agrobacterium tumefaciens D-psicose 3-epimerase onto titanium dioxide for bioconversion of rare sugar. Enzym. Microb. Technol. 2020, 140, 109605. [Google Scholar] [CrossRef] [PubMed]
- Xue, K.; Liu, C.-L.; Yang, Y.; Liu, X.; Zhan, J.; Bai, Z. Immobilization of D-allulose 3-epimerase into magnetic metal-organic framework nanoparticles for efficient biocatalysis. World J. Microbiol. Biotechnol. 2022, 38, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Fan, D.; Zhao, F.; Lin, Y.; Zheng, S.; Han, S. Characterization of D-allulose-3-epimerase from Ruminiclostridium papyrosolvens and immobilization within metal-organic frameworks. Front. Bioeng. Biotechnol. 2022, 10, 869536. [Google Scholar] [CrossRef]
- Gao, X.; Wei, C.; Qi, H.; Li, C.; Lu, F.; Qin, H.-M. Directional immobilization of D-allulose 3-epimerase using SpyTag/SpyCatcher strategy as a robust biocatalyst for synthesizing D-allulose. Food Chem. 2023, 401, 134199. [Google Scholar] [CrossRef]
- Zhong, G.; Liu, D.; Zhang, J. The application of ZIF-67 and its derivatives: Adsorption, separation, electrochemistry and catalysts. J. Mater. Chem. A 2018, 6, 1887–1899. [Google Scholar] [CrossRef]
- Hatlem, D.; Trunk, T.; Linke, D.; Leo, J.C. Catching a SPY: Using the SpyCatcher-SpyTag and Related Systems for Labeling and Localizing Bacterial Proteins. Int. J. Mol. Sci. 2019, 20, 2129. [Google Scholar] [CrossRef]
- Qiu, Y.; Wei, G.-W. Artificial Intelligence-Aided Protein Engineering: From Topological Data Analysis to Deep Protein Language Models. arXiv 2023. [Google Scholar] [CrossRef]
- Frisby, T.S.; Langmead, C.J. Identifying Promising Sequences for Protein Engineering Using a Deep Transformer Protein Language Model. Proteins Struct. Funct. Genet. 2023, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Repecka, D.; Jauniskis, V.; Karpus, L.; Rembeza, E.; Rokaitis, I.; Zrimec, J.; Povilonienė, S.; Laurynėnas, A.; Viknander, S.; Abuajwa, W.; et al. Expanding Functional Protein Sequence Spaces Using Generative Adversarial Networks. Nat. Mach. Intell. 2021, 3, 324–333. [Google Scholar] [CrossRef]

| Enzyme Source | Expression Strain | Optimal Reaction Conditions | Metal Ion | Conversion (%) | kcat (s−1) | Km (mM) | kcat/Km (mM−1 s−1) | t1/2 (mins) | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|
| pH | Temperature (°C) | |||||||||
| DAEase from Anthrobacter psychrolactophilus | E. coli BL21(DE3) | 8.5 | 70 | Mg2+ | 27% | 2920 | 738.7 | 3.953 | 1386.3 (55 °C + Mg2+) 128.4 (70 °C) | [15] |
| DAEase from Caballeronia insecticola | E. coli BL21(DE3) | 9 | 65 | Mn2+ | 31% | 204.05 | 137.7 | 1.482 | Not reported | [16] |
| DPEase from Iocasia fonsfrigidae | E. coli BL21(DE3) | 7.5 | 50 | Mn2+ | 36% | 12.82 | 21.31 | 0.602 | Not reported | [17] |
| DAEase from Bacillus sp. KCTC 13,219 (DaeB) | B. subtilis RIK 1285 | 8 | 55 | Mn2+ | 28% | 367 | 130.6 | 2.810 | 36,000 (50 °C + Mn2+) 1320 (55 °C + Mn2+) | [18] |
| DAEase from Clostridium cellulolyticum H10 | E. coli BL21(DE3) | 8 | 55 | Co2+ | 32% | 54.05 | 17.4 | 3.106 | 570 (55 °C + Co2+) | [19] |
| DAEase from Dorea sp. CAG317 | E. coli BL21(DE3) | 6 | 70 | Co2 | 30% | 507.4 | 153 | 3.316 | 36 (60 °C) | [20] |
| Enzyme Source | Expression Strain | Engineering Strategy | Functional Improvements | Reference |
|---|---|---|---|---|
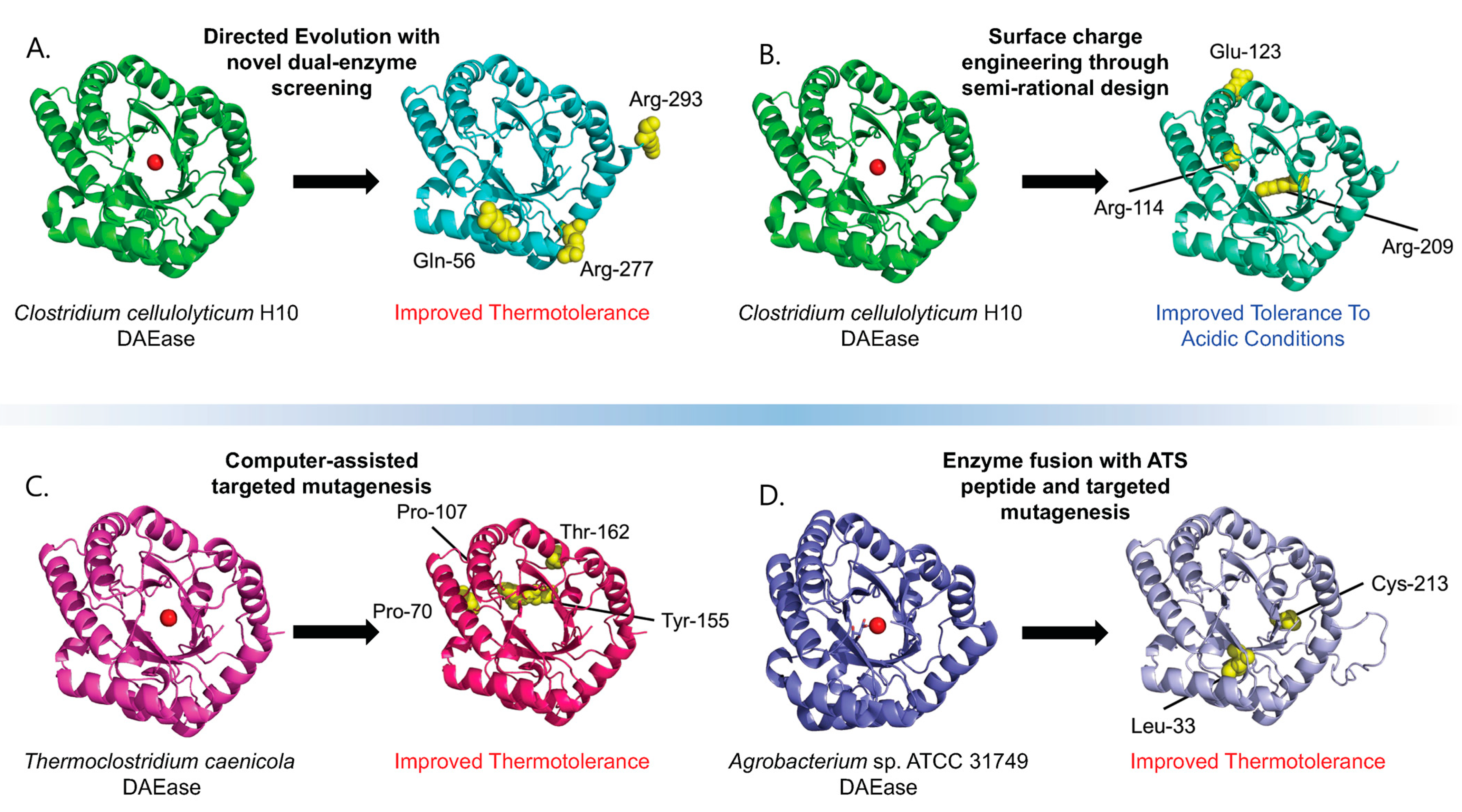
| DAEase from Clostridium cellulolyticum H10 | E. coli BL21(DE3) | Directed evolution supported by a novel dual-enzyme screening system | Improved thermostability | [22] |
| DAEase from Thermoclostridium caenicola | E. coli BL21(DE3) | Computer-assisted targeted mutagenesis | Improved thermostability | [23] |
| DAEase from Agrobacterium sp. | E. coli BL21(DE3) | Enzyme fusion with peptide derived from the acidic tail of alpha-synuclein (ATS) | Improved thermostability | [24] |
| DAEase from Clostridium bolteae | E. coli BL21(DE3) | Proline residue substitution to reduce unfolded enzyme entropy | Improved thermostability | [25] |
| DAEase from Clostridium cellulolyticum H10 | E. coli BL21(DE3) | Directed evolution, combinatorial mutagenesis | Improved enzyme specificity, improved thermotolerance | [21] |
| DAEase from Clostridium cellulolyticum H10 | E. coli BL21(DE3) | Protein surface charge engineering through semirational design and mutagenic library | Improved tolerance to acidic conditions | [26] |
| Enzyme Source | Support Material | Immobilization Method | pH | Temperature (°C) | Metal Ion | Reference |
|---|---|---|---|---|---|---|
| DPEase from Agrobacterium tumefaciens | Glutaraldehyde functionalised TiO2 | D | 6 | 60 | Mn2+ | [34] |
| DAEase from Agrobacterium tumefaciens | Cobalt-based magnetic MOF ZIF-67@Fe3O4 | B | 8 | 50 | Co2+ (in ZIF-67) | [35] |
| DAEase from Ruminiclostridium papyrosolvens | ZiF-67 | B | 7.5 | 60 | Co2+ (in ZIF-67) | [36] |
| DAEase from Clostridium cellulolyticum | Epoxy resin ES-103B with or without SpyCatcher-SpyTag | D | 8 | 55 | - | [37] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, J.H.; Chen, A.; Bi, J.; Lim, Y.H.; Wong, F.T.; Ow, D.S.-W. The Engineering, Expression, and Immobilization of Epimerases for D-allulose Production. Int. J. Mol. Sci. 2023, 24, 12703. https://doi.org/10.3390/ijms241612703
Tan JH, Chen A, Bi J, Lim YH, Wong FT, Ow DS-W. The Engineering, Expression, and Immobilization of Epimerases for D-allulose Production. International Journal of Molecular Sciences. 2023; 24(16):12703. https://doi.org/10.3390/ijms241612703
Chicago/Turabian StyleTan, Jin Hao, Anqi Chen, Jiawu Bi, Yee Hwee Lim, Fong Tian Wong, and Dave Siak-Wei Ow. 2023. "The Engineering, Expression, and Immobilization of Epimerases for D-allulose Production" International Journal of Molecular Sciences 24, no. 16: 12703. https://doi.org/10.3390/ijms241612703
APA StyleTan, J. H., Chen, A., Bi, J., Lim, Y. H., Wong, F. T., & Ow, D. S.-W. (2023). The Engineering, Expression, and Immobilization of Epimerases for D-allulose Production. International Journal of Molecular Sciences, 24(16), 12703. https://doi.org/10.3390/ijms241612703






