miRNA Studies in Glaucoma: A Comprehensive Review of Current Knowledge and Future Perspectives
Abstract
1. Introduction
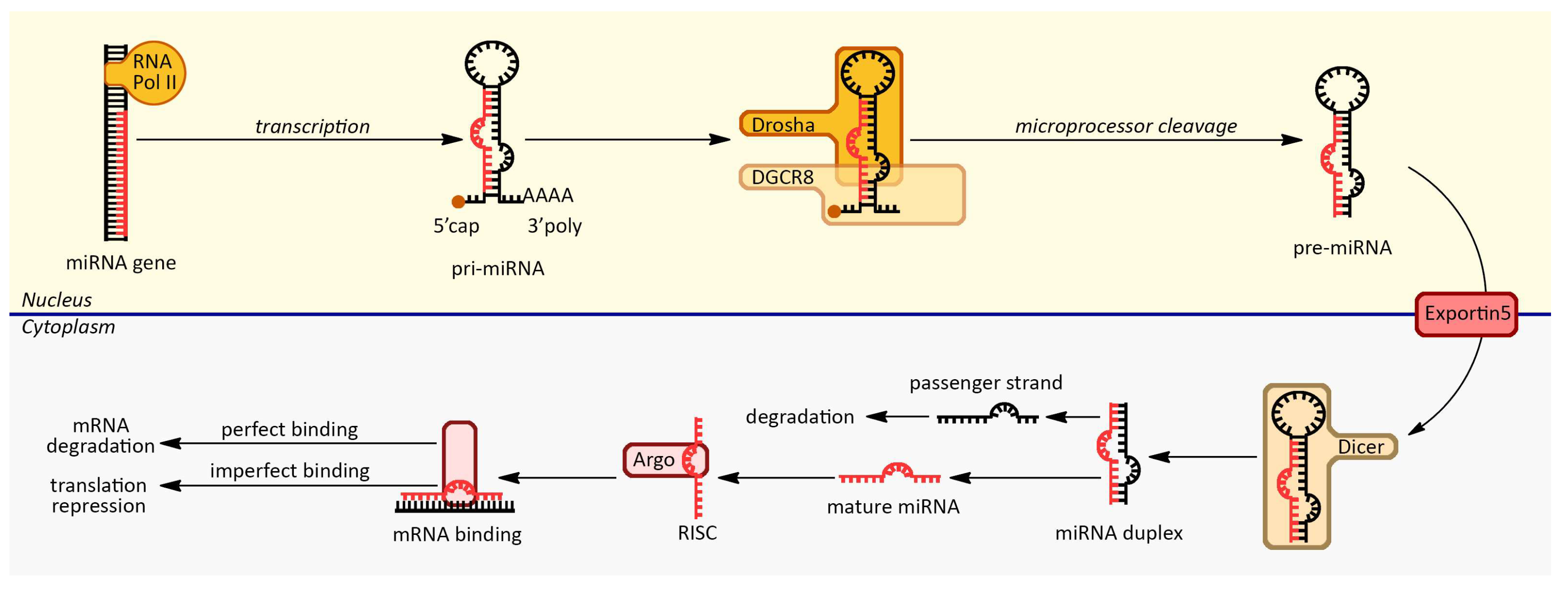
2. MiRNA Biogenesis and Silencing Mechanisms
3. miRNA Profiling Tools in Glaucoma
3.1. Real-Time Polymerase Chain Reaction (RT-PCR)
3.2. Digital Droplet PCR (ddPCR)
3.3. miRNA Arrays
3.4. NanoString nCounter-Based miRNA Assays
3.5. Small RNA Sequencing
3.6. miRNA Luciferase Reporter Assay
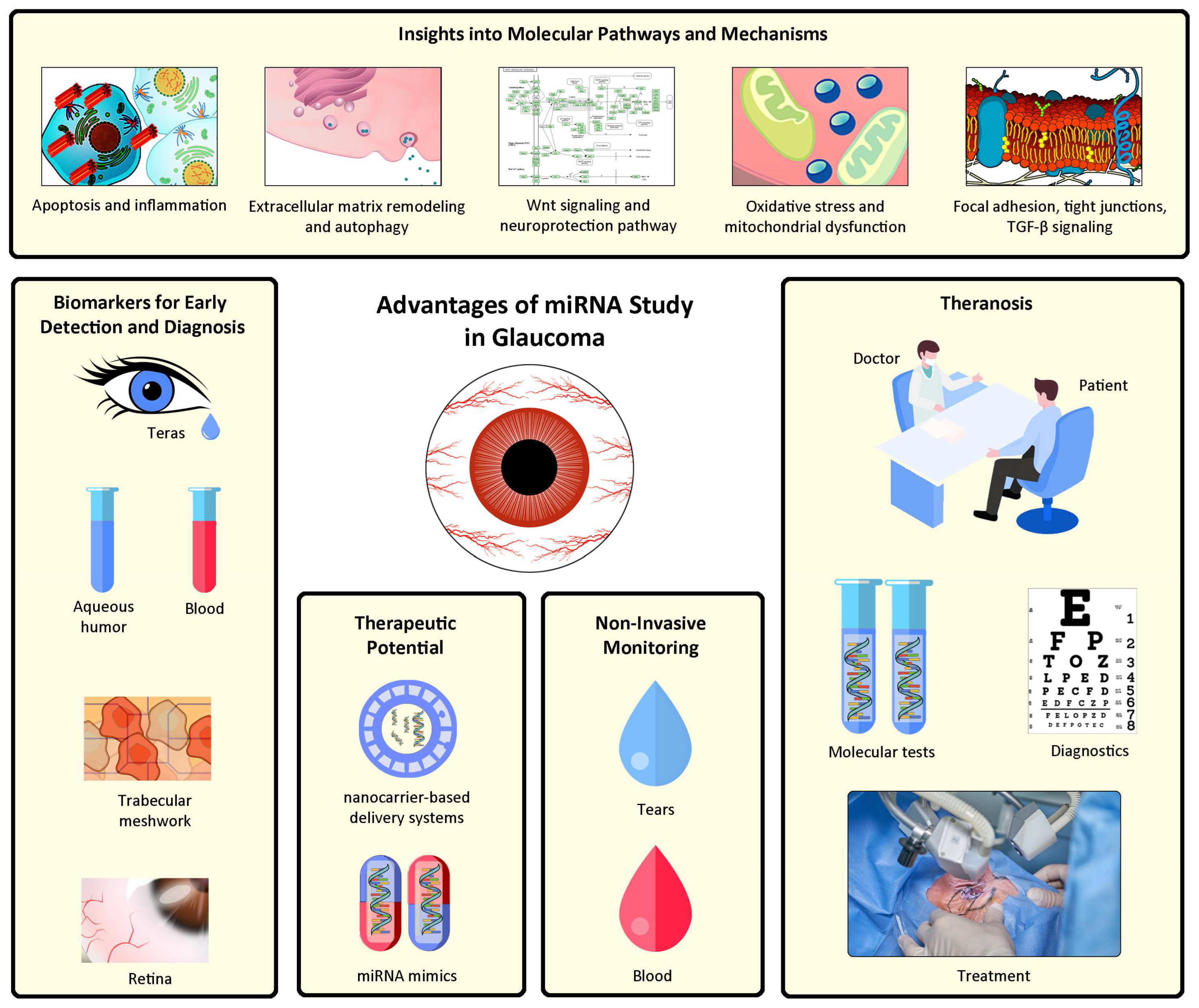
4. Advantages of miRNA in Glaucoma
4.1. Biomarkers for Early Detection and Diagnosis
4.1.1. miRNA Stability
4.1.2. Sources of miRNAs for Glaucoma Diagnosis
4.1.3. Storage, Extraction, and Quantification of miRNAs in Glaucoma Research
4.1.4. miRNA Expression Patterns in Glaucoma
4.2. Insights into Molecular Pathways and Mechanisms
4.3. Personalized Medicine
4.4. Therapeutic Potential
4.5. Non-Invasive Monitoring
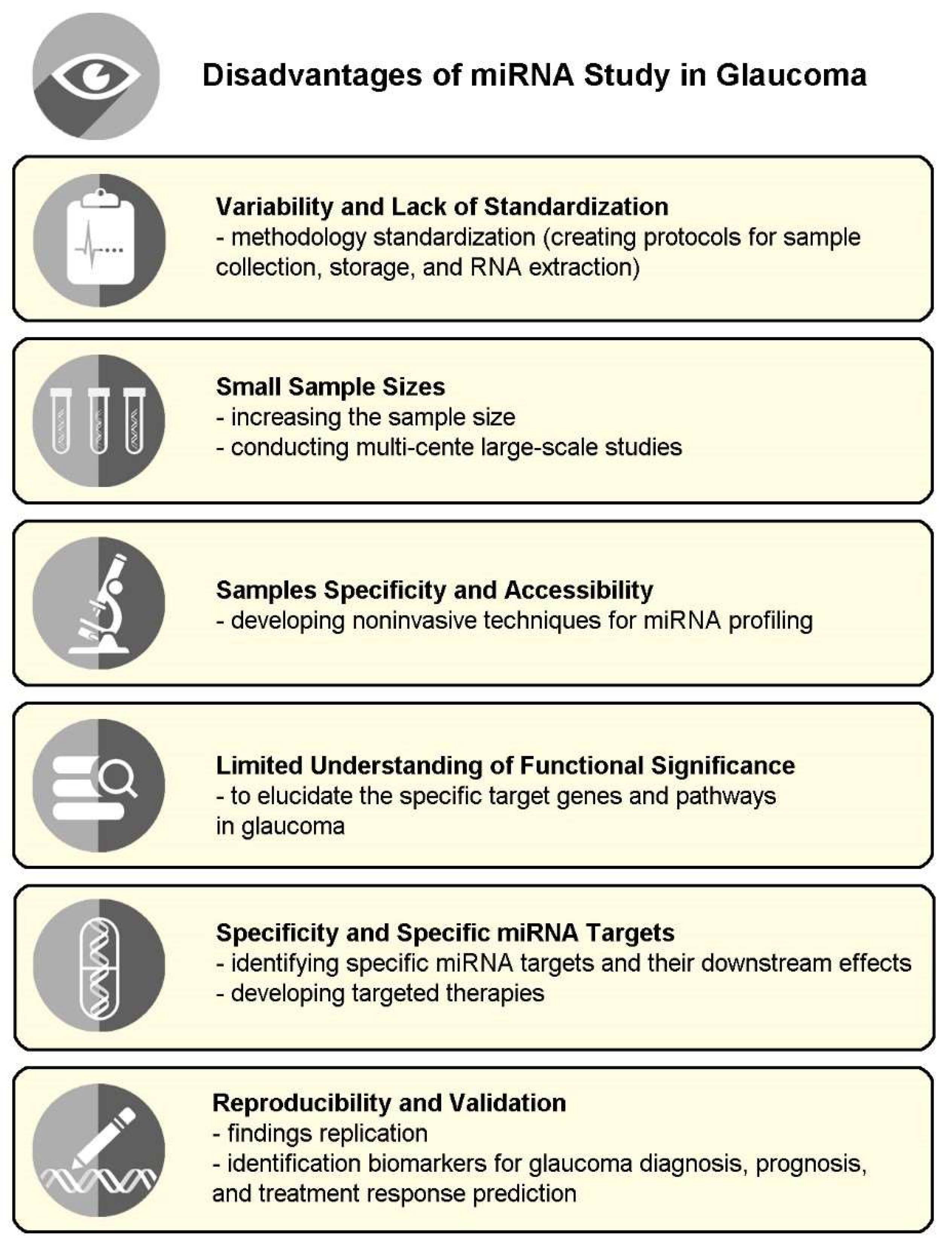
5. Disadvantages of miRNA in Glaucoma
5.1. Variability and Lack of Standardization
5.2. Small Sample Sizes
5.3. Samples Specificity and Accessibility
5.4. Limited Understanding of Functional Significance
5.5. Specificity and Specific miRNA Targets
5.6. Reproducibility and Validation
6. miRNA Studies in Glaucoma: Prospects and Limitations
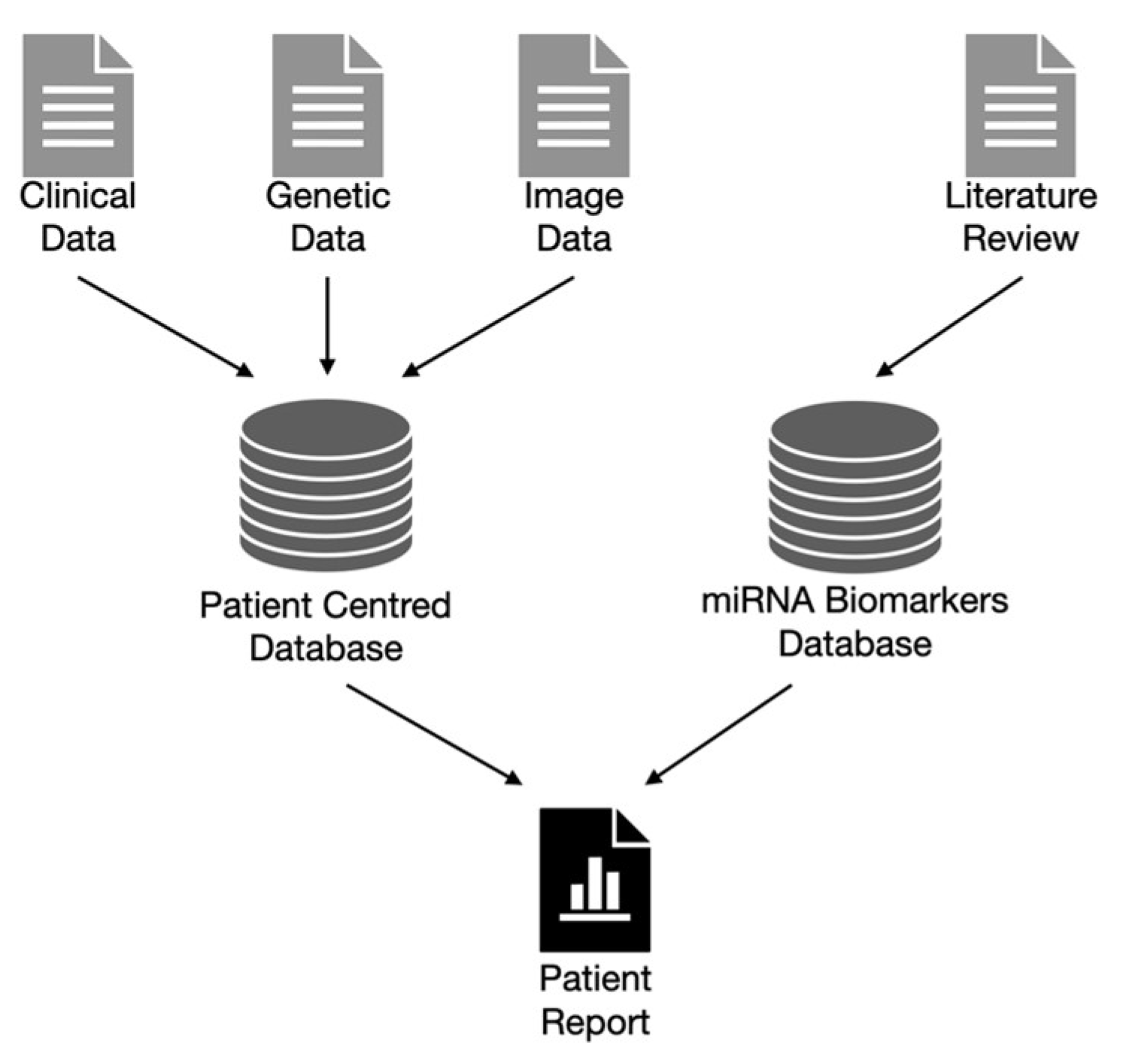
7. Bioinformatic Pipeline and Data Governance
7.1. Bioinformatic Pipeline
7.2. Data Governance
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weinreb, R.N.; Aung, T.; Medeiros, F.A. The pathophysiology and treatment of glaucoma: A review. JAMA 2014, 311, 1901–1911. [Google Scholar] [CrossRef] [PubMed]
- Zukerman, R.; Harris, A.; Vercellin, A.V.; Siesky, B.; Pasquale, L.R.; Ciulla, T.A. Molecular genetics of glaucoma: Subtype and ethnicity considerations. Genes 2020, 12, 55. [Google Scholar] [CrossRef] [PubMed]
- Greene, K.M.; Stamer, W.D.; Liu, Y. The role of microRNAs in glaucoma. Exp. Eye Res. 2022, 215, 108909. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Cannell, I.G.; Kong, Y.W.; Bushell, M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008, 36, 1224–1231. [Google Scholar] [CrossRef]
- Martinez, B.; Peplow, P.V. MicroRNAs as biomarkers in glaucoma and potential therapeutic targets. Neural Regen. Res. 2022, 17, 2368–2375. [Google Scholar] [CrossRef]
- Thermo Fisher Scientific Incorp. miRNA Quantitation. Available online: https://www.thermofisher.com/pl/en/home/life-science/pcr/real-time-pcr/real-time-pcr-applications/microrna-noncoding-rna-with-real-time-pcr/mirna-quantitation.html (accessed on 23 May 2023).
- Thermo Fisher Scientific Incorp. microRNA & Noncoding RNA Analysis Using Real-Time PCR. Available online: https://www.thermofisher.com/pl/en/home/life-science/pcr/real-time-pcr/real-time-pcr-applications/microrna-noncoding-rna-with-real-time-pcr.html (accessed on 20 May 2023).
- Ferracin, M.; Negrini, M. Quantification of circulating microRNAs by droplet digital PCR. Methods Mol. Biol. 2018, 1768, 445–457. [Google Scholar] [CrossRef]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: From variable nonsense to publication quality data. Sci. Rep. 2017, 7, 2409. [Google Scholar] [CrossRef]
- Stein, E.V.; Duewer, D.L.; Farkas, N.; Romsos, E.L.; Wang, L.; Cole, K.D. Steps to achieve quantitative measurements of microRNA using two step droplet digital PCR. PLoS ONE 2017, 12, e0188085. [Google Scholar] [CrossRef]
- Thermo Fisher Scientific Incorp. miRNA Profiling with Microarrays. Available online: https://www.thermofisher.com/pl/en/home/life-science/microarray-analysis/mirna-profiling-microarrays.html (accessed on 25 May 2023).
- Siddika, T.; Heinemann, I.U. Bringing microRNAs to light: Methods for microRNA quantification and visualization in live cells. Front. Bioeng. Biotechnol. 2020, 8, 619583. [Google Scholar] [CrossRef] [PubMed]
- NanoString Technologies Inc. nCounter® miRNA Expression Panels. Available online: https://nanostring.com/products/ncounter-assays-panels/immunology/mirna/ (accessed on 23 May 2023).
- Foye, C.; Yan, I.K.; David, W.; Shukla, N.; Habboush, Y.; Chase, L.; Ryland, K.; Kesari, V.; Patel, T. Comparison of miRNA quantitation by Nanostring in serum and plasma samples. PLoS ONE 2017, 12, e0189165. [Google Scholar] [CrossRef] [PubMed]
- Illumina, I. A Targeted Method for Both Small RNA Profiling and Discovery Applications. Available online: https://www.illumina.com/techniques/sequencing/rna-sequencing/small-rna-seq.html (accessed on 27 May 2022).
- Benesova, S.; Kubista, M.; Valihrach, L. Small RNA-Sequencing: Approaches and considerations for miRNA analysis. Diagnostics 2021, 11, 964. [Google Scholar] [CrossRef] [PubMed]
- Creative Biogene miRNA Luciferase Reporter Assay. Available online: https://integraterna.creative-biogene.com/service/mirna-luciferase-array-service.html (accessed on 19 May 2023).
- Jin, Y.; Chen, Z.; Liu, X.; Zhou, X. Evaluating the microRNA targeting sites by luciferase reporter gene assay. Methods Mol. Biol. 2013, 936, 117–127. [Google Scholar] [CrossRef]
- Hindle, A.G.; Thoonen, R.; Jasien, J.V.; Grange, R.M.H.; Amin, K.; Wise, J.; Ozaki, M.; Ritch, R.; Malhotra, R.; Buys, E.S. Identification of candidate miRNA biomarkers for glaucoma. Investig. Ophthalmol. Vis. Sci. 2019, 60, 134–146. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as biomarkers in disease: Latest findings regarding their role in diagnosis and prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Li, Z.; Chen, D.; Wang, Q.; Tian, H.; Tan, M.; Peng, D.; Tan, Y.; Zhu, J.; Liang, W.; Zhang, L. mRNA and microRNA stability validation of blood samples under different environmental conditions. Forensic Sci. Int. Genet. 2021, 55, 102567. [Google Scholar] [CrossRef]
- Kotar, A.; Ma, S.; Keane, S.C. pH dependence of C•A, G•A and A•A mismatches in the stem of precursor microRNA-31. Biophys. Chem. 2022, 283, 106763. [Google Scholar] [CrossRef]
- Matias-Garcia, P.R.; Wilson, R.; Mussack, V.; Reischl, E.; Waldenberger, M.; Gieger, C.; Anton, G.; Peters, A.; Kuehn-Steven, A. Impact of long-term storage and freeze-thawing on eight circulating microRNAs in plasma samples. PLoS ONE 2020, 15, e0227648. [Google Scholar] [CrossRef]
- Sulewska, A.; Pilz, L.; Manegold, C.; Ramlau, R.; Charkiewicz, R.; Niklinski, J. A systematic review of progress toward unlocking the power of epigenetics in NSCLC: Latest updates and perspectives. Cells 2023, 12, 905. [Google Scholar] [CrossRef]
- Gareev, I.; Beylerli, O.; Yang, G.; Sun, J.; Pavlov, V.; Izmailov, A.; Shi, H.; Zhao, S. The current state of MiRNAs as biomarkers and therapeutic tools. Clin. Exp. Med. 2020, 20, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, Y.; Gao, Q.; Ping, D.; Wang, Y.; Wu, W.; Lin, X.; Fang, Y.; Zhang, J.; Shao, A. The role of exosomal microRNAs and oxidative stress in neurodegenerative diseases. Oxid. Med. Cell. Longev. 2020, 2020, 3232869. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Z.; Zhou, S.; Wen, J.; Geng, B.; Yang, J.; Cui, Q. Genome-wide analysis of human microRNA stability. BioMed. Res. Int. 2013, 2013, 368975. [Google Scholar] [CrossRef] [PubMed]
- Pasquinelli, A.E. MicroRNAs and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012, 13, 271–282. [Google Scholar] [CrossRef]
- Stavast, C.J.; Erkeland, S.J. The non-canonical aspects of microRNAs: Many roads to gene regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef]
- Nik Mohamed Kamal, N.N.S.B.; Shahidan, W.N.S. Non-exosomal and exosomal circulatory microRNAs: Which are more valid as biomarkers? Front. Pharmacol. 2020, 10, 1500. [Google Scholar] [CrossRef]
- Konno, M.; Koseki, J.; Asai, A.; Yamagata, A.; Shimamura, T.; Motooka, D.; Okuzaki, D.; Kawamoto, K.; Mizushima, T.; Eguchi, H.; et al. Distinct methylation levels of mature microRNAs in gastrointestinal cancers. Nat. Commun. 2019, 10, 3888. [Google Scholar] [CrossRef]
- Salzman, D.W.; Nakamura, K.; Nallur, S.; Dookwah, M.T.; Metheetrairut, C.; Slack, F.J.; Weidhaas, J.B. miR-34 activity is modulated through 5’-end phosphorylation in response to DNA damage. Nat. Commun. 2016, 7, 10954. [Google Scholar] [CrossRef]
- Kosior-Jarecka, E.; Czop, M.; Gasińska, K.; Wróbel-Dudzińska, D.; Zalewski, D.P.; Bogucka-Kocka, A.; Kocki, J.; Żarnowski, T. MicroRNAs in the aqueous humor of patients with different types of glaucoma. Graefes Arch. Clin. Exp. Ophthalmol. 2021, 259, 2337–2349. [Google Scholar] [CrossRef]
- Seong, H.; Cho, H.K.; Kee, C.; Song, D.H.; Cho, M.C.; Kang, S.S. Profiles of microRNA in aqueous humor of normal tension glaucoma patients using RNA sequencing. Sci. Rep. 2021, 11, 19024. [Google Scholar] [CrossRef]
- Errera, M.H.; Pratas, A.; Fisson, S.; Manicom, T.; Boubaya, M.; Sedira, N.; Héron, E.; Merabet, L.; Kobal, A.; Levy, V.; et al. Cytokines, chemokines and growth factors profile in human aqueous humor in idiopathic uveitis. PLoS ONE 2022, 17, e0254972. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Bradley, J.C.; Reid, T.W.; McCartney, D.L. Growth factors in aqueous humor. Ophthalmology 2011, 118, 1003–1003.e1. [Google Scholar] [CrossRef] [PubMed]
- Van Zyl, T.; Yan, W.; McAdams, A.; Peng, Y.R.; Shekhar, K.; Regev, A.; Juric, D.; Sanes, J.R. Cell atlas of aqueous humor outflow pathways in eyes of humans and four model species provides insight into glaucoma pathogenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 10339–10349. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.K.; Seong, H.; Kee, C.; Song, D.H.; Kim, S.J.; Seo, S.W.; Kang, S.S. MicroRNA profiles in aqueous humor between pseudoexfoliation glaucoma and normal tension glaucoma patients in a Korean population. Sci. Rep. 2022, 12, 6217. [Google Scholar] [CrossRef] [PubMed]
- Goel, M.; Picciani, R.G.; Lee, R.K.; Bhattacharya, S.K. Aqueous humor dynamics: A review. Open Ophthalmol. J. 2010, 4, 52–59. [Google Scholar] [CrossRef]
- Tanaka, Y.; Tsuda, S.; Kunikata, H.; Sato, J.; Kokubun, T.; Yasuda, M.; Nishiguchi, K.M.; Inada, T.; Nakazawa, T. Profiles of extracellular miRNAs in the aqueous humor of glaucoma patients assessed with a microarray system. Sci. Rep. 2014, 4, 5089. [Google Scholar] [CrossRef]
- Jayaram, H.; Phillips, J.I.; Lozano, D.C.; Choe, T.E.; Cepurna, W.O.; Johnson, E.C.; Morrison, J.C.; Gattey, D.M.; Saugstad, J.A.; Keller, K.E. Comparison of microRNA expression in aqueous humor of normal and primary open-angle glaucoma patients using PCR arrays: A pilot study. Investig. Ophthalmol. Vis. Sci. 2017, 58, 2884–2890. [Google Scholar] [CrossRef] [PubMed]
- Hubens, W.H.G.; Krauskopf, J.; Beckers, H.J.M.; Kleinjans, J.C.S.; Webers, C.A.B.; Gorgels, T.G.M.F. Small RNA Sequencing of aqueous humor and plasma in patients with primary open-angle glaucoma. Investig. Ophthalmol. Vis. Sci. 2021, 62, 24. [Google Scholar] [CrossRef]
- Chen, S.; Yuan, M.; Liu, Y.; Zhao, X.; Lian, P.; Chen, Y.; Liu, B.; Lu, L. Landscape of microRNA in the aqueous humour of proliferative diabetic retinopathy as assessed by next-generation sequencing. Clin. Exp. Ophthalmol. 2019, 47, 925–936. [Google Scholar] [CrossRef]
- Altman, J.; Jones, G.; Ahmed, S.; Sharma, S.; Sharma, A. Tear film microRNAs as potential biomarkers: A review. Int. J. Mol. Sci. 2019, 47, 3694. [Google Scholar] [CrossRef]
- Raga-Cervera, J.; Bolarin, J.M.; Millan, J.M.; Garcia-Medina, J.J.; Pedrola, L.; Abellán-Abenza, J.; Valero-Vello, M.; Sanz-González, S.M.; O’Connor, J.E.; Galarreta-Mira, D.; et al. miRNAs and genes involved in the interplay between ocular hypertension and primary open-angle glaucoma. Oxidative stress, inflammation, and apoptosis networks. J. Clin. Med. 2021, 10, 2227. [Google Scholar] [CrossRef] [PubMed]
- Youngblood, H.; Cai, J.; Drewry, M.D.; Helwa, I.; Hu, E.; Liu, S.; Yu, H.; Mu, H.; Hu, Y.; Perkumas, K.; et al. Expression of mRNAs, miRNAs, and lncRNAs in human trabecular meshwork cells upon mechanical stretch. Investig. Ophthalmol. Vis. Sci. 2020, 61, 2. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, P.; Li, G.; Qiu, J.; Wu, J.; Luna, C. Role of microRNAs in the trabecular meshwork. J. Ocul. Pharmacol. Ther. 2014, 30, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Lewczuk, K.; Jabłońska, J.; Konopińska, J.; Mariak, Z.; Rękas, M. Schlemm’s canal: The outflow ‘vessel’. Acta Ophthalmol. 2022, 100, e881–e890. [Google Scholar] [CrossRef]
- Guo, R.; Shen, W.; Su, C.; Jiang, S.; Wang, J. Relationship between the pathogenesis of glaucoma and miRNA. Ophthal. Res. 2017, 57, 194–199. [Google Scholar] [CrossRef]
- Sutherland, C.; Wang, Y.; Brown, R.V.; Foley, J.; Mahler, B.; Janardhan, K.S.; Kovi, R.C.; Jetten, A.M. Laser capture microdissection of highly pure trabecular meshwork from mouse eyes for gene expression analysis. J. Vis. Exp. 2018, 136, 57576. [Google Scholar] [CrossRef]
- Keller, K.E.; Bhattacharya, S.K.; Borrás, T.; Brunner, T.M.; Chansangpetch, S.; Clark, A.F.; Dismuke, W.M.; Du, Y.; Elliott, M.H.; Ethier, C.R.; et al. Consensus recommendations for trabecular meshwork cell isolation, characterization and culture. Exp. Eye Res. 2018, 171, 164–173. [Google Scholar] [CrossRef]
- Mochizuki, H.; Murphy, C.J.; Brandt, J.D.; Kiuchi, Y.; Russell, P. Altered stability of mRNAs associated with glaucoma progression in human trabecular meshwork cells following oxidative stress. Investig. Ophthalmol. Vis. Sci. 2012, 53, 1734–1741. [Google Scholar] [CrossRef]
- Zuzic, M.; Rojo Arias, J.E.; Wohl, S.G.; Busskamp, V. Retinal miRNA Functions in health and disease. Genes 2019, 10, 377. [Google Scholar] [CrossRef]
- Mure, L.S. Intrinsically photosensitive retinal ganglion cells of the human retina. Front. Neurol. 2021, 12, 636330. [Google Scholar] [CrossRef]
- Do, M.T.H.; Yau, K.W. Intrinsically photosensitive retinal ganglion cells. Physiol. Rev. 2010, 90, 1547–1581. [Google Scholar] [CrossRef] [PubMed]
- Hackler, L.; Masuda, T.; Oliver, V.F.; Merbs, S.L.; Zack, D.J. Use of laser capture microdissection for analysis of retinal mRNA/miRNA expression and DNA methylation. Methods Mol. Biol. 2012, 884, 289–304. [Google Scholar] [PubMed]
- Schnichels, S.; Paquet-Durand, F.; Löscher, M.; Tsai, T.; Hurst, J.; Joachim, S.C.; Klettner, A. Retina in a dish: Cell cultures, retinal explants and animal models for common diseases of the retina. Prog. Retin. Eye Res. 2021, 81, 100880. [Google Scholar] [CrossRef] [PubMed]
- Hurst, J.; Fietz, A.; Tsai, T.; Joachim, S.C.; Schnichels, S. Organ cultures for retinal diseases. Front. Neurosci. 2020, 14, 583392. [Google Scholar] [CrossRef]
- Gooding, S.W.; Chrenek, M.A.; Ferdous, S.; Nickerson, J.M.; Boatright, J.H. Set screw homogenization of murine ocular tissue, including the whole eye. Mol. Vis. 2018, 24, 690–699. [Google Scholar]
- Malik, K.J.; Chen, C.D.; Olsen, T.W. Stability of RNA from the retina and retinal pigment epithelium in a porcine model simulating human eye bank conditions. Investig. Ophthalmol. Vis. Sci. 2003, 44, 2730–2735. [Google Scholar] [CrossRef][Green Version]
- Kallestad, L.; Blackshaw, S.; Khalil, A.M.; Palczewski, K. Tissue- and Species-Specific Patterns of RNA metabolism in Post-Mortem Mammalian retina and Retinal Pigment epithelium. Sci. Rep. 2019, 9, 14821. [Google Scholar] [CrossRef]
- Hackler, L.; Wan, J.; Swaroop, A.; Qian, J.; Zack, D.J. MicroRNA profile of the developing mouse retina. Investig. Ophthalmol. Vis. Sci. 2010, 51, 1823–1831. [Google Scholar] [CrossRef]
- Intartaglia, D.; Giamundo, G.; Conte, I. The impact of miRNAs in health and disease of retinal pigment epithelium. Front. Cell Dev. Biol. 2020, 8, 589985. [Google Scholar] [CrossRef]
- Karali, M.; Persico, M.; Mutarelli, M.; Carissimo, A.; Pizzo, M.; Singh Marwah, V.S.; Ambrosio, C.; Pinelli, M.; Carrella, D.; Ferrari, S.; et al. High-resolution analysis of the human retina miRNome reveals isomiR variations and novel microRNAs. Nucleic Acids Res. 2016, 44, 1525–1540. [Google Scholar] [CrossRef]
- Dean, L. Blood and the Cells It Contains; National Center for Biotechnology Information: Bethesda, MD, USA, 2005; pp. 1–8. [Google Scholar]
- Mathew, J.; Sankar, P.; Varacallo, M. Physiology, Blood Plasma; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Flores, C.F.Y.; Pineda, Á.; De las, M.H.; Bonilla, V.M.C.; Sáenz-Flor, K. Sample Management: Stability of Plasma and Serum on Different Storage Conditions. Electron. J. Int. Fed. Clin. Chem. Lab. Med. 2020, 31, 46–55. [Google Scholar]
- Wu, D.W.; Li, Y.M.; Wang, F. How long can we store blood samples: A systematic review and meta-analysis. eBiomedicine 2017, 24, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Heyer, N.J.; Derzon, J.H.; Winges, L.; Shaw, C.; Mass, D.; Snyder, S.R.; Epner, P.; Nichols, J.H.; Gayken, J.A.; Ernst, D.; et al. Effectiveness of practices to reduce blood sample hemolysis in EDs: A laboratory medicine best practices systematic review and meta-analysis. Clin. Biochem. 2012, 45, 1012–1032. [Google Scholar] [CrossRef] [PubMed]
- Linskens, E.A.; Devreese, K.M.J. Pre-analytical stability of coagulation parameters in plasma stored at room temperature. Int. J. Lab. Hematol. 2018, 40, 292–303. [Google Scholar] [CrossRef]
- Zürcher, M.; Sulzer, I.; Barizzi, G.; Lämmle, B.; Alberio, L. Stability of coagulation assays performed in plasma from citrated whole blood transported at ambient temperature. Thromb. Haemost. 2008, 99, 416–426. [Google Scholar] [CrossRef]
- Bryzgunova, O.; Konoshenko, M.; Zaporozhchenko, I.; Yakovlev, A.; Laktionov, P. Isolation of cell-free miRNA from biological fluids: Influencing factors and methods. Diagnostics 2021, 11, 865. [Google Scholar] [CrossRef]
- Roest, H.P.; Ijzermans, J.N.M.; van der Laan, L.J.W. Evaluation of RNA isolation methods for microRNA quantification in a range of clinical biofluids. BMC Biotechnol. 2021, 21, 48. [Google Scholar] [CrossRef]
- Khamina, K.; Diendorfer, A.B.; Skalicky, S.; Weigl, M.; Pultar, M.; Krammer, T.L.; Aquino, C.; Fournier, C.A.; Schofield, A.L.; Otto, C.; et al. A microRNA next-generation-sequencing discovery assay (miND) for genome-scale analysis and absolute quantitation of circulating microRNA biomarkers. Int. J. Mol. Sci. 2022, 23, 1226. [Google Scholar] [CrossRef]
- Potla, P.; Ali, S.A.; Kapoor, M. A bioinformatics approach to microRNA-sequencing analysis. Osteoarthr. Cartil. 2020, 3, 100131. [Google Scholar] [CrossRef]
- Forero, D.A.; González-Giraldo, Y.; Castro-Vega, L.J.; Barreto, G.E. qPCR-based methods for expression analysis of miRNAs. BioTechniques 2019, 67, 192–199. [Google Scholar] [CrossRef]
- Chen, C.; Tan, R.; Wong, L.; Fekete, R.; Halsey, J. Quantitation of microRNAs by real-time RT-qPCR. Methods Mol. Biol. 2011, 687, 113–134. [Google Scholar] [CrossRef]
- Drewry, M.D.; Challa, P.; Kuchtey, J.G.; Navarro, I.; Helwa, I.; Hu, Y.; Mu, H.; Stamer, W.D.; Kuchtey, R.W.; Liu, Y. Differentially expressed microRNAs in the aqueous humor of patients with exfoliation glaucoma or primary open-angle glaucoma. Hum. Mol. Genet. 2018, 27, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.; Chakraborty, M.; Roy, A.; Sahay, P.; Pradhan, A.; Raj, N. Differential miRNA expression: Signature for glaucoma in pseudoexfoliation. Clin. Ophthalmol. 2020, 14, 3025–3038. [Google Scholar] [CrossRef] [PubMed]
- Czop, M.; Gasińska, K.; Kosior-Jarecka, E.; Wróbel-Dudzińska, D.; Kocki, J.; Żarnowski, T. Twenty novel microRNAs in the aqueous humor of pseudoexfoliation glaucoma patients. Cells 2023, 12, 737. [Google Scholar] [CrossRef] [PubMed]
- Zhai, R.; Xu, H.; Kong, X.; Wang, J. Differentially Expressed microRNAs Associated with Primary Open-angle Glaucoma Based on Bioinformatics analysis of microRNA microarray Data. Investig. Ophthalmol. Vis. Sci. 2018, 59, 3520. [Google Scholar]
- Romano, G.L.; Platania, C.B.M.; Forte, S.; Salomone, S.; Drago, F.; Bucolo, C. MicroRNA target prediction in glaucoma. Prog. Brain Res. 2015, 220, 217–240. [Google Scholar] [CrossRef]
- Su, W.; Li, Z.; Jia, Y.; Zhu, Y.; Cai, W.; Wan, P.; Zhang, Y.; Zheng, S.G.; Zhuo, Y. microRNA-21a-5p/PDCD4 axis regulates mesenchymal stem cell-induced neuroprotection in acute glaucoma. J. Mol. Cell Biol. 2017, 9, 289–301. [Google Scholar] [CrossRef]
- Tabak, S.; Schreiber-Avissar, S.; Beit-Yannai, E. Crosstalk between microRNA and oxidative stress in primary open-angle glaucoma. Int. J. Mol. Sci. 2021, 22, 2421. [Google Scholar] [CrossRef]
- Li, G.; Luna, C.; Qiu, J.; Epstein, D.L.; Gonzalez, P. Modulation of inflammatory markers by miR-146a during replicative senescence in trabecular meshwork cells. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2976–2985. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, H.; Liu, X.; Han, Y.; Pan, S.; Wang, Y. MiR-181a inhibits human trabecular meshwork cell apoptosis induced by H₂O₂ through the suppression of NF-κB and JNK pathways. Adv. Clin. Exp. Med. 2018, 27, 577–582. [Google Scholar] [CrossRef]
- Ou-Yang, Y.; Liu, Z.L.; Xu, C.L.; Wu, J.L.; Peng, J.; Peng, Q.H. miR-223 induces retinal ganglion cells apoptosis and inflammation via decreasing HSP-70 in vitro and in vivo. J. Chem. Neuroanat. 2020, 104, 101747. [Google Scholar] [CrossRef] [PubMed]
- Reinehr, S.; Mueller-Buehl, A.M.; Tsai, T.; Joachim, S.C. Specific biomarkers in the aqueous humour of glaucoma patients. Klin. Monbl. Augenheilkd. 2022, 239, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Lerner, N.; Schreiber-Avissar, S.; Beit-Yannai, E. Extracellular vesicle-mediated crosstalk between NPCE cells and TM cells result in modulation of Wnt signalling pathway and ECM remodelling. J. Cell. Mol. Med. 2020, 24, 4646–4658. [Google Scholar] [CrossRef]
- Tan, C.; Jia, F.; Zhang, P.; Sun, X.; Qiao, Y.; Chen, X.; Wang, Y.; Chen, J.; Lei, Y. A miRNA stabilizing polydopamine nano-platform for intraocular delivery of miR-21-5p in glaucoma therapy. J. Mater. Chem. B 2021, 9, 3335–3345. [Google Scholar] [CrossRef]
- Zaharia, A.C.; Dumitrescu, O.M.; Radu, M.; Rogoz, R.E. Adherence to therapy in glaucoma treatment—A review. J. Pers. Med. 2022, 12, 514. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, Y.; Wang, Y.; Zhang, X.; Gao, K.; Chen, S.; Zhang, X. microRNA profiling in glaucoma eyes with varying degrees of optic neuropathy by using next-generation sequencing. Investig. Ophthalmol. Vis. Sci. 2018, 59, 2955–2966. [Google Scholar] [CrossRef]
- Wagner, I.V.; Stewart, M.W.; Dorairaj, S.K. Updates on the diagnosis and management of glaucoma. Mayo Clin. Proc. Innov. Qual. Outcomes 2022, 6, 618–635. [Google Scholar] [CrossRef]
- Schuster, A.K.; Erb, C.; Hoffmann, E.M.; Dietlein, T.; Pfeiffer, N. The diagnosis and treatment of glaucoma. Dtsch. Arztebl. Int. 2020, 117, 225–234. [Google Scholar] [CrossRef]
- Jayaram, H.; Lozano, D.C.; Johnson, E.C.; Morrison, J.C. Investigation of microRNA expression in experimental glaucoma. Methods Mol. Biol. 2018, 1695, 287–297. [Google Scholar] [CrossRef]
- Wang, Y.; Niu, L.; Zhao, J.; Wang, M.; Li, K.; Zheng, Y. An update: Mechanisms of microRNA in primary open-angle glaucoma. Brief. Funct. Genom. 2021, 20, 19–27. [Google Scholar] [CrossRef]
- Lo Faro, V.; Ten Brink, J.B.; Snieder, H.; Jansonius, N.M.; Bergen, A.A. Genome-wide CNV investigation suggests a role for cadherin, Wnt, and p53 pathways in primary open-angle glaucoma. BMC Genom. 2021, 22, 590. [Google Scholar] [CrossRef] [PubMed]
- Mak, H.K.; Leung, C.K.S. MicroRNA-based therapeutics for optic neuropathy: Opportunities and challenges. Neural Regen. Res. 2021, 16, 1996–1997. [Google Scholar] [CrossRef] [PubMed]
- Mead, B.; Kerr, A.; Nakaya, N.; Tomarev, S.I. miRNA changes in retinal ganglion cells after optic nerve crush and glaucomatous damage. Cells 2021, 10, 1564. [Google Scholar] [CrossRef] [PubMed]
- Rong, R.; Wang, M.; You, M.; Li, H.; Xia, X.; Ji, D. Pathogenesis and prospects for therapeutic clinical application of noncoding RNAs in glaucoma: Systematic perspectives. J. Cell. Physiol. 2021, 236, 7097–7116. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yang, X.; Liang, Q.; Xue, X.; Huang, J.; Wang, J.; Xu, Y.; Tong, R.; Liu, M.; Zhou, Q.; et al. Drugs for the treatment of glaucoma: Targets, structure-activity relationships and clinical research. Eur. J. Med. Chem. 2021, 226, 113842. [Google Scholar] [CrossRef]
- Lampis, A.; Hahne, J.C.; Hedayat, S.; Valeri, N. MicroRNAs as mediators of drug resistance mechanisms. Curr. Opin. Pharmacol. 2020, 54, 44–50. [Google Scholar] [CrossRef]
- Rezaei, M.; Faramarzpour, M.; Shobeiri, P.; Seyedmirzaei, H.; Sarasyabi, M.S.; Dabiri, S.S. A systematic review, meta-analysis, and network analysis of diagnostic microRNAs in glaucoma. Eur. J. Med. Res. 2023, 28, 137. [Google Scholar] [CrossRef]
- Molasy, M.; Walczak, A.; Szaflik, J.; Szaflik, J.P.; Majsterek, I. MicroRNAs in glaucoma and neurodegenerative diseases. J. Human Genet. 2017, 62, 105–112. [Google Scholar] [CrossRef]
- Tan, C.; Song, M.; Stamer, W.D.; Qiao, Y.; Chen, X.; Sun, X.; Lei, Y.; Chen, J. miR-21-5p: A viable therapeutic strategy for regulating intraocular pressure. Exp. Eye Res. 2020, 200, 108197. [Google Scholar] [CrossRef]
- Aggio-Bruce, R.; Chu-Tan, J.A.; Wooff, Y.; Cioanca, A.V.; Schumann, U.S.R.; Natoli, R. Inhibition of microRNA-155 protects retinal function through attenuation of inflammation in retinal degeneration. Mol. Neurobiol. 2021, 58, 835–854. [Google Scholar] [CrossRef]
- Krol, J.; Busskamp, V.; Markiewicz, I.; Stadler, M.B.; Ribi, S.; Richter, J.; Duebel, J.; Bicker, S.; Fehling, H.J.; Schübeler, D.; et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 2010, 141, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Gorantla, S.; Rapalli, V.K.; Waghule, T.; Singh, P.P.; Dubey, S.K.; Saha, R.N.; Singhvi, G. Nanocarriers for ocular drug delivery: Current status and translational opportunity. RSC Adv. 2020, 10, 27835–27855. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, Y.; Chen, J.; Gao, X.; Pan, S.; Su, Y.; Zhou, X. Co-delivery of brinzolamide and miRNA-124 by biodegradable nanoparticles as a strategy for glaucoma therapy. Drug Deliv. 2020, 27, 410–421. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Li, S.; Xu, M.; Zhong, Y.; Fan, W.; Xu, J.; Zhou, T.; Ji, J.; Ye, J.; Yao, K. Polymer- and lipid-based nanocarriers for ocular drug delivery: Current status and future perspectives. Adv. Drug Deliv. Rev. 2023, 196, 114770. [Google Scholar] [CrossRef]
- Wu, K.Y.; Ashkar, S.; Jain, S.; Marchand, M.; Tran, S.D. Breaking barriers in eye treatment: Polymeric nano-based drug-delivery system for anterior segment diseases and glaucoma. Polymers 2023, 15, 1373. [Google Scholar] [CrossRef]
- Mead, B.; Cullather, E.; Nakaya, N.; Niu, Y.; Kole, C.; Ahmed, Z.; Tomarev, S. Viral delivery of multiple miRNAs promotes retinal ganglion cell survival and functional preservation after optic nerve crush injury. Exp. Eye Res. 2020, 197, 108071. [Google Scholar] [CrossRef]
- Wang, M.; Li, J.; Zheng, Y. The potential role of nuclear factor erythroid 2-related Factor 2 (Nrf2) in glaucoma: A review. Med. Sci. Monit. 2020, 26, e921514. [Google Scholar] [CrossRef]
- Vallée, A.; Lecarpentier, Y.; Vallée, J.N. Cannabidiol and the canonical WNT/β-catenin pathway in glaucoma. Int. J. Mol. Sci. 2021, 22, 3798. [Google Scholar] [CrossRef]
- Wang, N.; Yang, W.; Xiao, T.; Miao, Z.; Luo, W.; You, Z.; Li, G. Possible role of miR-204 in optic nerve injury through the regulation of GAP-43. Mol. Med. Rep. 2018, 17, 3891–3897. [Google Scholar] [CrossRef]
- Giangiacomo, A.; Garway-Heath, D.; Caprioli, J. Diagnosing glaucoma progression: Current practice and promising technologies. Curr. Opin. Ophthalmol. 2006, 17, 153–162. [Google Scholar] [CrossRef]
- Tamkovich, S.; Grigor’eva, A.; Eremina, A.; Tupikin, A.; Kabilov, M.; Chernykh, V.; Vlassov, V.; Ryabchikova, E. What information can be obtained from the tears of a patient with primary open angle glaucoma? Clin. Chim. Acta 2019, 495, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Smyth, A.; Callaghan, B.; Willoughby, C.E.; O’Brien, C. The role of miR-29 family in TGF-β driven fibrosis in glaucomatous optic neuropathy. Int. J. Mol. Sci. 2022, 23, 10216. [Google Scholar] [CrossRef]
- Yang, J.; Wang, N.; Luo, X. Intraocular miR-211 exacerbates pressure-induced cell death in retinal ganglion cells via direct repression of FRS2 signaling. Biochem. Biophys. Res. Commun. 2018, 503, 2984–2992. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Y.; Chen, Y.; Fang, X.; Wen, T.; Xiao, M.; Chen, S.; Zhang, X. Discovery and validation of circulating Hsa-miR-210-3p as a potential biomarker for primary open-angle glaucoma. Investig. Ophthalmol. Vis. Sci. 2019, 60, 2925–2934. [Google Scholar] [CrossRef] [PubMed]
- Harris, T.A.; Yamakuchi, M.; Ferlito, M.; Mendell, J.T.; Lowenstein, C.J. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl. Acad. Sci. USA 2008, 105, 1516–1521. [Google Scholar] [CrossRef] [PubMed]
- Marzi, M.J.; Montani, F.; Carletti, R.M.; Dezi, F.; Dama, E.; Bonizzi, G.; Sandri, M.T.; Rampinelli, C.; Bellomi, M.; Maisonneuve, P.; et al. Optimization and standardization of circulating microRNA detection for clinical application: The miR-test case. Clin. Chem. 2016, 62, 743–754. [Google Scholar] [CrossRef]
- Kapnisis, K.; Doormaal, M.V.; Ross Ethier, C. Modeling aqueous humor collection from the human eye. J. Biomech. 2009, 42, 2454–2457. [Google Scholar] [CrossRef]
- Gonzales, J.A.; Chan, C.C. Biopsy techniques and yields in diagnosing primary intraocular lymphoma. Int. Ophthalmol. 2007, 27, 241–250. [Google Scholar] [CrossRef]
- Asgarpour, K.; Shojaei, Z.; Amiri, F.; Ai, J.; Mahjoubin-Tehran, M.; Ghasemi, F.; ArefNezhad, R.; Hamblin, M.R.; Mirzaei, H. Exosomal microRNAs derived from mesenchymal stem cells: Cell-to-cell messages. Cell Commun. Signal. 2020, 18, 149. [Google Scholar] [CrossRef]
- Liu, H.; Xiu, Y.; Zhang, Q.; Xu, Y.; Wan, Q.; Tao, L. Silencing microRNA-29b-3p expression protects human trabecular meshwork cells against oxidative injury via upregulation of RNF138 to activate the ERK pathway. Int. J. Mol. Med. 2021, 47, 101. [Google Scholar] [CrossRef]
- Meng, J.; Yang, X.; Huang, X.; Li, Q. Long Non-coding RNA GAS5 Knockdown Attenuates H2O2-Induced Human Trabecular Meshwork Cell Apoptosis and Promotes Extracellular Matrix Deposition by Suppressing miR-29b-3p and Upregulating STAT3. J. Mol. Neurosci. 2022, 72, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. 2019, 76, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Carrella, S.; Massa, F.; Indrieri, A. The role of microRNAs in mitochondria-mediated eye diseases. Front. Cell Dev. Biol. 2021, 9, 653522. [Google Scholar] [CrossRef]
- Rice, J.; Roberts, H.; Burton, J.; Pan, J.; States, V.; Rai, S.N.; Galandiuk, S. Assay reproducibility in clinical studies of plasma miRNA. PLoS ONE 2015, 10, e0121948. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W. Circulating microRNA biomarker studies: Pitfalls and potential solutions. Clin. Chem. 2015, 61, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yue, P.; Tong, X.; Bai, J.; Yang, J.; Guo, J. mRNA-seq and miRNA-seq profiling analyses reveal molecular mechanisms regulating induction of fruiting body in Ophiocordyceps sinensis. Sci. Rep. 2021, 11, 12944. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Shao, C.; Ye, X.; Meng, Y.; Zhou, Y.; Chen, M. miRNA Digger: A comprehensive pipeline for genome-wide novel miRNA mining. Sci. Rep. 2016, 6, 18901. [Google Scholar] [CrossRef]
- Bendifallah, S.; Dabi, Y.; Suisse, S.; Jornea, L.; Bouteiller, D.; Touboul, C.; Puchar, A.; Daraï, E. A Bioinformatics Approach to MicroRNA-Sequencing Analysis Based on Human Saliva Samples of Patients with Endometriosis. Int. J. Mol. Sci. 2022, 23, 8045. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, D.; Agís-Balboa, R.C.; López-Fernández, H. MyBrain-Seq: A pipeline for MiRNA-seq data analysis in neuropsychiatric disorders. Biomedicines 2023, 11, 1230. [Google Scholar] [CrossRef]
- Bisgin, H.; Gong, B.; Wang, Y.; Tong, W. Evaluation of bioinformatics approaches for next-generation sequencing analysis of microRNAs with a toxicogenomics study design. Front. Genet. 2018, 9, 22. [Google Scholar] [CrossRef]
- Li, B.; Zhang, K.; Ye, Y.; Xing, J.; Wu, Y.; Ma, Y.; Li, Y. Effects of castration on miRNA, lncRNA, and mRNA profiles in mice Thymus. Genes 2020, 11, 147. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef] [PubMed]
- Ziemann, M.; Kaspi, A.; El-Osta, A. Evaluation of microRNA alignment techniques. RNA 2016, 22, 1120–1138. [Google Scholar] [CrossRef]
- Stegmayer, G.; Di Persia, L.E.; Rubiolo, M.; Gerard, M.; Pividori, M.; Yones, C.; Bugnon, L.A.; Rodriguez, T.; Raad, J.; Milone, D.H. Predicting novel microRNA: A comprehensive comparison of machine learning approaches. Brief. Bioinform. 2019, 20, 1607–1620. [Google Scholar] [CrossRef] [PubMed]
- Fair Principles. Available online: https://www.go-fair.org/fair-principles/ (accessed on 23 May 2023).
- Grauslund, J.; Stokholm, L.; Ohm Kyvik, K.; Dornonville de la Cour, M.; Kessel, L.; Hass Rubin, K. Interactions between ocular and systemic disease using national register-based data in the Danish Excellence Centre in ophthalmic Epidemiology (DECODE-EYE): Study perspective. Acta Ophthalmol. 2020, 98, 573–578. [Google Scholar] [CrossRef]
- Chiang, M.F.; Sommer, A.; Rich, W.L.; Lum, F.; Parke, D.W. The 2016 American Academy of Ophthalmology IRIS® Registry (Intelligent Research in Sight) Database: Characteristics and Methods. Ophthalmology 2018, 125, 1143–1148. [Google Scholar] [CrossRef]
- Daien, V.; Korobelnik, J.F.; Delcourt, C.; Cougnard-Gregoire, A.; Delyfer, M.N.; Bron, A.M.; Carrière, I.; Villain, M.; Daures, J.P.; Lacombe, S.; et al. French medical-administrative database for epidemiology and safety in ophthalmology (EPISAFE): The EPISAFE collaboration program in cataract surgery. Ophthal. Res. 2017, 58, 67–73. [Google Scholar] [CrossRef]
- Wong, D.C.S.; Olivera, M.; Yu, J.; Szabo, A.; Moghul, I.; Balaskas, K.; Luben, R.; Khawaja, A.P.; Pontikos, N.; Keane, P.A. Cloud-based genomics pipelines for ophthalmology: Reviewed from research to clinical practice. Model. Artif. Intell. Ophthalmol. 2021, 3, 101–140. [Google Scholar] [CrossRef]
- Yuan, J.; Chen, F.; Fan, D.; Jiang, Q.; Xue, Z.; Zhang, J.; Yu, X.; Li, K.; Qu, J.; Su, J. EyeDiseases: An integrated resource for dedicating to genetic variants, gene expression and epigenetic factors of human eye diseases. NAR Genom. Bioinform. 2021, 3, lqab050. [Google Scholar] [CrossRef]
- Zhang, X.; Kong, L.; Liu, S.; Zhang, X.; Shang, X.; Zhu, Z.; Huang, Y.; Ma, S.; Jason, H.; Kiburg, K.V.; et al. EBD: An eye biomarker database. Bioinformatics 2023, 39, btad194. [Google Scholar] [CrossRef]
- Karali, M.; Peluso, I.; Gennarino, V.A.; Bilio, M.; Verde, R.; Lago, G.; Dollé, P.; Banfi, S. miRNeye: A microRNA expression atlas of the mouse eye. BMC Genom. 2010, 11, 715. [Google Scholar] [CrossRef] [PubMed]
- Kakrana, A.; Yang, A.; Anand, D.; Djordjevic, D.; Ramachandruni, D.; Singh, A.; Huang, H.; Ho, J.W.K.; Lachke, S.A. iSyTE 2.0: A database for expression-based gene discovery in the eye. Nucleic Acids Res. 2018, 46, D875–D885. [Google Scholar] [CrossRef] [PubMed]
- Duraisamy, A.; Suganthi, M.; Shanmugam, G.; Suryadevara, N. GluDB: A glaucoma associated gene database. Mater. Today Proc. 2019, 16, 1590–1595. [Google Scholar] [CrossRef]
- Jin, K.; Huang, X.; Zhou, J.; Li, Y.; Yan, Y.; Sun, Y.; Zhang, Q.; Wang, Y.; Ye, J. FIVES: A fundus Image Dataset for Artificial Intelligence based Vessel Segmentation. Sci. Data 2022, 9, 475. [Google Scholar] [CrossRef]
- Panchal, S.; Naik, A.; Kokare, M.; Pachade, S.; Naigaonkar, R.; Phadnis, P.; Bhange, A. Retinal fundus multi-disease image dataset (RFMiD) 2.0: A dataset of frequently and rarely identified diseases. Data 2023, 8, 29. [Google Scholar] [CrossRef]
- Khan, S.M.; Liu, X.; Nath, S.; Korot, E.; Faes, L.; Wagner, S.K.; Keane, P.A.; Sebire, N.J.; Burton, M.J.; Denniston, A.K. A global review of publicly available datasets for ophthalmological imaging: Barriers to access, usability, and generalisability. Lancet Digit. Health 2021, 3, e51–e66. [Google Scholar] [CrossRef]

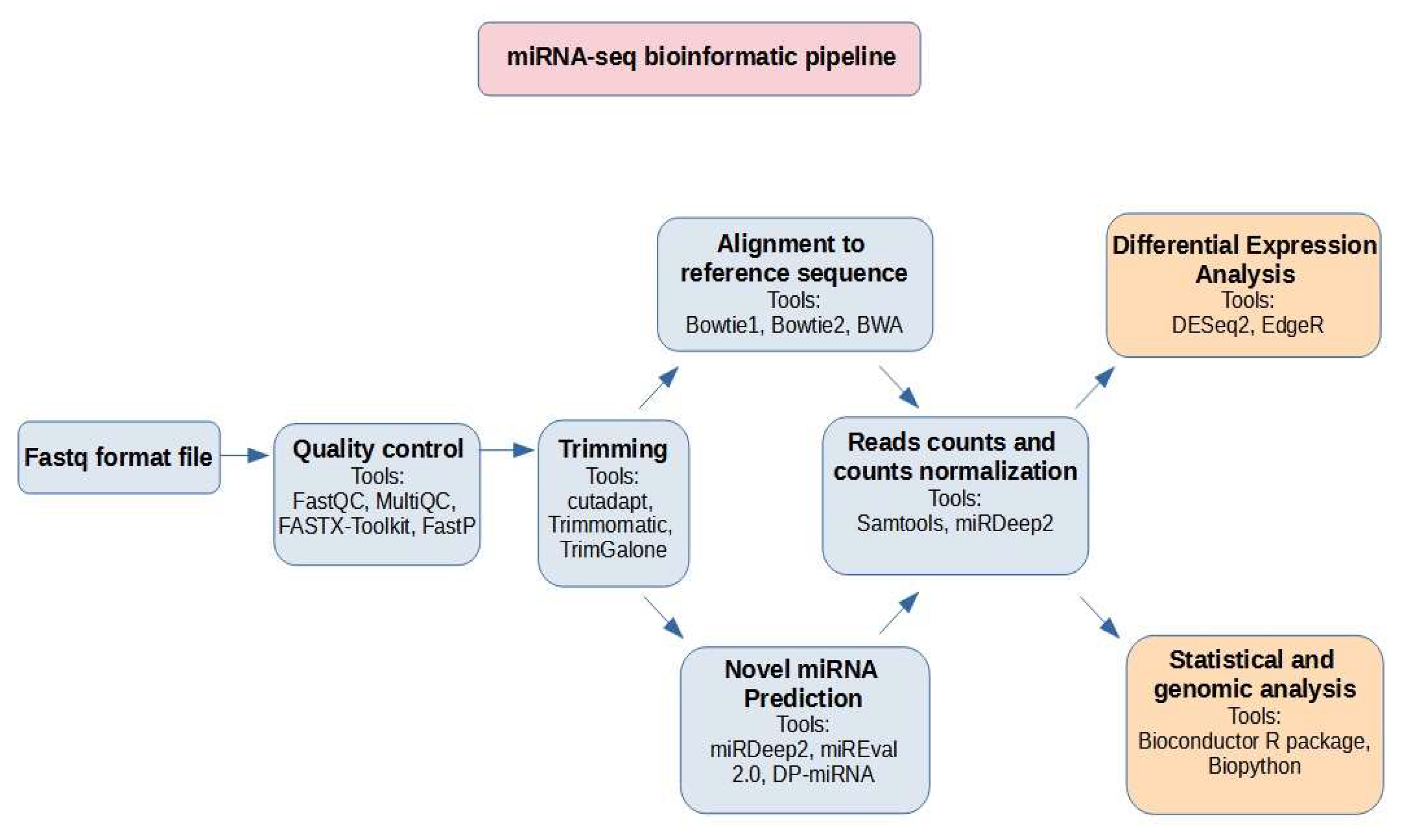
| Pipeline Step | Proposed Tools |
|---|---|
| Quality control | FastQC, MultiQC, FASTX-Toolkit, FastP |
| Trimming | Cutadapt, Trimmomatic, TrimGalone |
| Alignment | Bowtie1, Bowtie2, BWA |
| Novel miRNA prediction | miRDeep2, miREval 2.0, DP-miRNA |
| Differential Expression Analysis | DESeq2, EdgeR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dobrzycka, M.; Sulewska, A.; Biecek, P.; Charkiewicz, R.; Karabowicz, P.; Charkiewicz, A.; Golaszewska, K.; Milewska, P.; Michalska-Falkowska, A.; Nowak, K.; et al. miRNA Studies in Glaucoma: A Comprehensive Review of Current Knowledge and Future Perspectives. Int. J. Mol. Sci. 2023, 24, 14699. https://doi.org/10.3390/ijms241914699
Dobrzycka M, Sulewska A, Biecek P, Charkiewicz R, Karabowicz P, Charkiewicz A, Golaszewska K, Milewska P, Michalska-Falkowska A, Nowak K, et al. miRNA Studies in Glaucoma: A Comprehensive Review of Current Knowledge and Future Perspectives. International Journal of Molecular Sciences. 2023; 24(19):14699. https://doi.org/10.3390/ijms241914699
Chicago/Turabian StyleDobrzycka, Margarita, Anetta Sulewska, Przemyslaw Biecek, Radoslaw Charkiewicz, Piotr Karabowicz, Angelika Charkiewicz, Kinga Golaszewska, Patrycja Milewska, Anna Michalska-Falkowska, Karolina Nowak, and et al. 2023. "miRNA Studies in Glaucoma: A Comprehensive Review of Current Knowledge and Future Perspectives" International Journal of Molecular Sciences 24, no. 19: 14699. https://doi.org/10.3390/ijms241914699
APA StyleDobrzycka, M., Sulewska, A., Biecek, P., Charkiewicz, R., Karabowicz, P., Charkiewicz, A., Golaszewska, K., Milewska, P., Michalska-Falkowska, A., Nowak, K., Niklinski, J., & Konopińska, J. (2023). miRNA Studies in Glaucoma: A Comprehensive Review of Current Knowledge and Future Perspectives. International Journal of Molecular Sciences, 24(19), 14699. https://doi.org/10.3390/ijms241914699







