Abstract
Psoriasis is a chronic immune-mediated skin disease in which the symptom-free, uninvolved skin carries alterations in gene expression, serving as a basis for lesion formation. Histones and histone acetylation-related processes are key regulators of gene expression, controlling cell proliferation and immune responses. Dysregulation of these processes is likely to play an important role in the pathogenesis of psoriasis. To gain a complete overview of these potential alterations, we performed a meta-analysis of a psoriatic uninvolved skin dataset containing differentially expressed transcripts from nearly 300 individuals and screened for histones and histone acetylation-related molecules. We identified altered expression of the replication-dependent histones HIST2H2AA3 and HIST2H4A and the replication-independent histones H2AFY, H2AFZ, and H3F3A/B. Eight histone chaperones were also identified. Among the histone acetyltransferases, ELP3 and KAT5 and members of the ATAC, NSL, and SAGA acetyltransferase complexes are affected in uninvolved skin. Histone deacetylation-related alterations were found to affect eight HDACs and members of the NCOR/SMRT, NURD, SIN3, and SHIP HDAC complexes. In this article, we discuss how histone and histone acetylation-related expression changes may affect proliferation and differentiation, as well as innate, macrophage-mediated, and T cell-mediated pro- and anti-inflammatory responses, which are known to play a central role in the development of psoriasis.
1. Introduction
Psoriasis is an inflammatory skin disease with an exaggerated response to external and internal stress reactions, resulting in keratinocyte hyperproliferation, abnormal differentiation, and massive immune cell infiltration [1,2]. The combined interaction of abnormal genetic, epigenetic, environmental, and microbiome-related factors is believed to be responsible for the development of psoriasis [3]. In this disease, the macroscopically healthy, uninvolved skin carries multiple molecular changes that lead to the appearance of symptoms [4,5]. Large-scale analyses comparing healthy, uninvolved, and psoriatic skin samples have found that the expression levels of many genes differ [6]. Epigenetic changes related to histones through their post-translational modification are partly behind these processes [7].
Chromatin is composed of DNA and histones, of which two main types can be distinguished: the gene-poor, transcriptionally less active heterochromatin and the gene-rich euchromatin, which is accessible for transcription [8]. The basic unit of chromatin is the nucleosome, composed of DNA and a core histone octamer [9], while the higher-order chromatin structures are promoted by the H1 linker histone [10]. On the basis of their role in replication, replication-dependent canonical [11] and replication-independent non-canonical histone variants [12] have been distinguished, encoded by 75 and 20 genes, respectively [13].
There are four classes of histone chaperones. Class I contains single chaperones, class II is a multichaperone complex, class III is enzymatic, and class IV is a multiclass chaperone complex [14]. These chaperones regulate the assembly, deposition, removal, exchange, and transport of histones, thereby modulating proliferation [15] and inflammatory responses [16,17].
Histone acetylation, carried out by histone acetyltransferases (HATs), leads to transcriptional activation [18,19]. There are two major types of HATs, A- and B-type [20]. A-type HATs acetylate chromatin-incorporated histones, whereas B-type HATs acetylate newly synthesized histones [21,22]. By contrast, histone deacetylation by histone deacetylases (HDACs) results in transcriptional repression [23].
Regarding psoriasis pathogenesis, histone acetylation in general and H3K27Ac in particular show a different pattern in lesional skin compared with healthy skin, as seen in heat images [7]. Histone H3 acetylation plays a role in Th17 cell differentiation and keratinocyte proliferation, both of which are known to play a central role in the pathogenesis of psoriasis [24]. Elevated expression of the epigenetic modifier CREMα has been detected in psoriatic T cells [25], which has been suggested to be partially responsible for the development of the abnormal expression of IL2 and IL17 [26].
On the one hand, histone variants can replace and substitute each other. On the other hand, they differ in the number and position of post-transcription modification sites at their globular core and N-terminal tail, allowing them to carry out distinct and specialized roles, including the regulation of tissue- and cell-type-specific functions. These functions include the regulation of proliferation [27,28], cell fate commitment [29], hematopoiesis [30], differentiation [31], macrophage [32] and T cell immune responses, and mutagenesis of immunoglobins [33,34].
To gain insight into how these processes are affected in uninvolved psoriatic skin, we screened for gene expression alterations in histones, histone chaperones, and histone acetylation-related molecules. We used a psoriatic transcriptome dataset containing nearly 300 published individual patient data (99 psoriatic lesional, 27 uninvolved psoriatic, and 172 healthy samples) to determine how these alterations may affect key processes in the pathogenesis of this common skin disease, as well as proliferation and immune responses.
2. Results and Discussion
2.1. Histones
To the best of our knowledge, there are no studies on the role of histones regarding the development of psoriasis. Among the canonical histones, we found that H2AC18 (also known as HIST2H2AA3) and H4C14 (also known asHIST2H4A) showed different expression in psoriatic uninvolved skin compared with the skin of healthy individuals (Figure 1 and Supplementary Table S2).
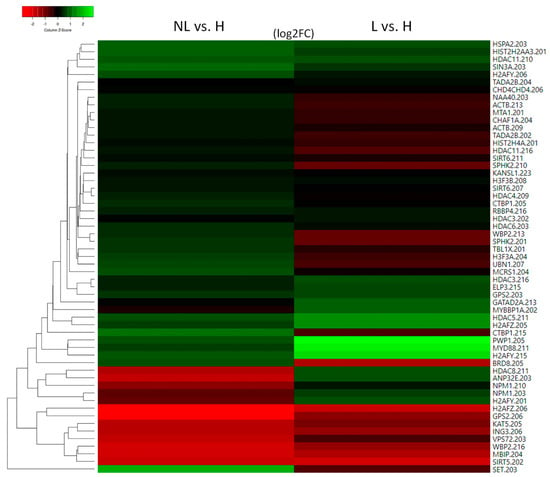
Figure 1.
Heatmap of histone and histone acetylation-related molecules with altered expression in uninvolved psoriatic skin (NL) and their expression in lesional skin (L) compared with healthy skin (H).
In non-dividing cells, HIST2H2AA3 participates in the terminal differentiation program [31] (Figure 2). HIST2H4A is commonly used as a marker for proliferation [27,28] (Figure 2). Therefore, differential expression of these histones may contribute to proliferation and differentiation-related alterations in psoriasis.
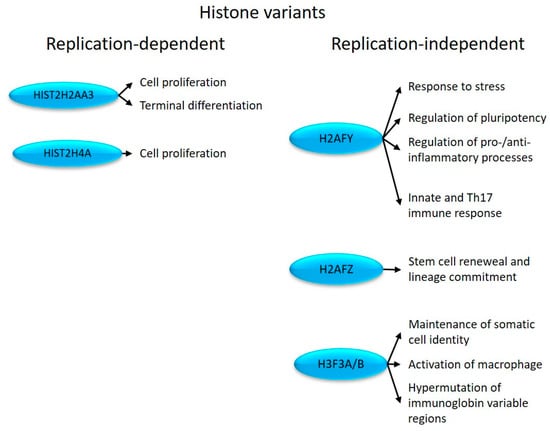
Figure 2.
Replication-dependent and -independent histones with altered expression in psoriatic uninvolved skin and their effects on cell proliferation and immune system-related processes.
Among the replication-independent histones that modulate nucleosome plasticity [12,35], MACROH2A1 (also known as H2AFY), H2AZ1 (also known as H2AFZ), and H3F3A/B show altered expression in uninvolved skin (Figure 1).
H2AFY plays a role in transcriptional repression [36] by regulating the transition between activating and inhibitory chromatin remodeling complexes [37]. It is also involved in the repression of pluripotent and bivalent developmental genes, thereby maintaining cell faith commitment [29] (Figure 2).
The H2AFY-PARP1 axis determines the cellular stress responses to DNA damage, heat shock, and aging [38]. H2AFY can suppress IFNB1 [39] and the proinflammatory cytokine IL-8 (CXCL8) [40], as well as CCL2 [41] transcription [42]. IFNB1 regulates the Th17 immune response [33], and IL-17A induces the production of IL-8 [43], while CCL2 promotes inflammatory processes in psoriasis [44]. Through the canonical JAK/STAT signaling pathway, the IFNB1-initiated response regulates cell proliferation [45], which is known to be dysregulated in psoriasis. Therefore, the differential expression of H2AFY in uninvolved psoriatic skin is likely to play a massive role in triggering psoriasis-related dysregulation in innate immune and proliferation-related responses (Figure 2).
H2AFZ ubiquitination regulates the transition between eu- and facultative heterochromatin, distinguishing constitutive from facultative heterochromatin [46] during the cell cycle (G1/S phase cMYC, Ki67) [47], and influencing lineage commitment [48,49] (Figure 2). Elevated expression of Ki67 and cMYC has been detected in psoriatic lesions, contributing to keratinocyte hyperproliferation [50]. Therefore, H2AFZ may contribute to the development of the disease by regulating stress response and proliferation, both of which are known to be involved in the pathogenesis of psoriasis.
H3F3A/B encodes histone H3.3, which is located at the euchromatin borders [51,52] and maintains heterochromatin structures [53]. As bifunctional histones, they can act as both transcriptional activators and repressors [54]. H3F3A/B is required in somatic cells to maintain their identity, for normal chromosome segregation [55], to maintain the balance of hematopoiesis [30], to activate macrophages [32], and to regulate the mutagenesis in the variable regions of immunoglobins [34] (Figure 2). In line with our results, an increased (hyper)mutation rate of IgE was detected in psoriasis patients [56].
In the development of abnormal differentiation, pluripotency, cell line commitment, and the differential expression of histones involved in terminal differentiation (H2AFY, H2AFZ, and HIST2H2AA3, respectively) may play a role during the development of the disease.
2.2. Histone Chaperones
Among the class I single chaperones, abnormal expression of the NPM1 and SET was identified in the non-lesioned skin (Figure 1).
NPM1 is an H1 and H3/H4 chaperone that participates in heterochromatin (re)arrangement [57]. NPM1 promotes cell proliferation [15] and is required for the maintenance of cell identity by maintaining a cell type-specific gene expression pattern [57]. The expression of NPM1 is elevated in proliferating keratinocytes of psoriatic lesions [58] and activates inflammatory responses when released into the extracellular space [16] (Figure 3).
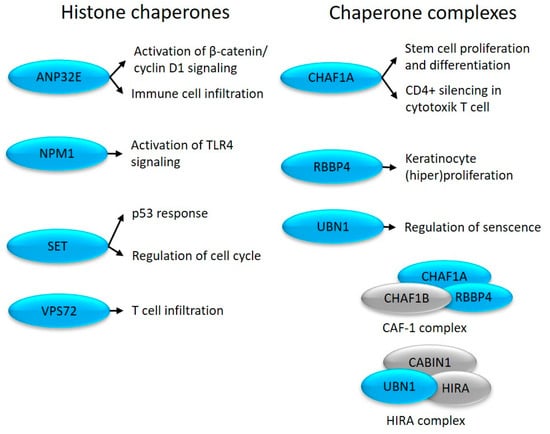
Figure 3.
Altered expression of histone chaperones in uninvolved skin (depicted in blue) and their role in cell proliferation and immune system-related processes.
The histone chaperone SET inhibits nucleosome acetylation and regulates p53-mediated cell cycle arrest [59]. SET regulates the G1/S and G2/M transition [60] via E-CDK2 and B-CDK1 [61], respectively (Figure 3), and inhibits cytotoxic T-cell-mediated apoptosis (www.genecards.org (accessed on 24 June 2023.)). These changes, characteristic of non-lesional skin, may be important in the development of the disease, as previous studies have shown increased activity of CDK1 and CDK2 in the psoriatic epidermis [62,63].
Among the class II. multi-chaperone complex members, CHAF1A, RBBP4, and UBN1 showed alterations in the non-lesioned skin (Figure 1).
CHAF1A is a component of the CAF1 complex that maintains Cd4 silencing in cytotoxic T cells [17]. The CAF1 complex is linked to DNA replication [64] and determines the proliferation–differentiation switch in stem cells [65], which is known to be abnormally regulated in psoriasis.
RBBP4 levels are upregulated in psoriasis by skin-derived mesenchymal stem cells, contributing to epidermal hyperplasia [66].
UBN1 is part of the bifunctional chaperone HIRA complex and participates in both transcriptional activation and inhibition [67]. By repressing proliferation-promoting genes, UBN1 regulates tissue aging-associated cellular senescence [68]. Consistent with our results, middle and upper epidermal keratinocytes of psoriatic plaques are characterized by a special state of aging, which is manifested by cell cycle arrest, as well as the release of inflammatory effectors and other molecules characteristic of aging [69].
Class III enzymatic complex members ANP32E and VPS72 also show altered expression. As part of the INO80 family, they regulate histone exchange [70]. ANP32E can remove H2AFZ from the nucleosome [71], while VPS72 deposits H2AFZ during mitosis [72], and immune cell infiltration [73,74] (Figure 3) that are known to be affected in the disease.
2.3. Histone Acetylation
Only type A HATs or their modulators showed abnormal expression in uninvolved psoriatic skin. Type A HATs can be classified into three subfamilies: the CBP/CREBBP, GNAT, and MYST families.
Members of the CBP/CREBBP family did not show transcriptional changes in uninvolved psoriatic skin. However, abnormal expression of EP300 modulators, such as the sequence-specific DNA binding protein MYBBP1A, the EP300 coactivator WBP2, and the EP300 corepressor CTBP1, was observed in the same samples (Figure 1). Elevated levels of CTBP1 have been demonstrated in psoriatic plaques, and mice overexpressing CTBP1 in epidermal keratinocytes show severe skin inflammation with increased expression of Th1 and Th17 cytokines [75], while WBP2 regulates epidermal [76] and T cell proliferation [77,78].
Abnormal expression of the GNAT family member ELP3 was also observed in uninvolved skin (Figure 1). ELP3 inhibits M1 and promotes M2 macrophage polarization [79].
We identified that the MYST family member KAT5 had altered expression in uninvolved skin (Figure 1). KAT5 modulates the differentiation and tissue infiltration of Th17 and Treg cells via FOXP3 [80]. As a cofactor of STAT3, KAT5 regulates IL-9 signaling [81] and hematopoietic stem cell maintenance [82] (Figure 4). KAT5 is also a catalytic subunit of the Tip60 histone acetyltransferase complex.
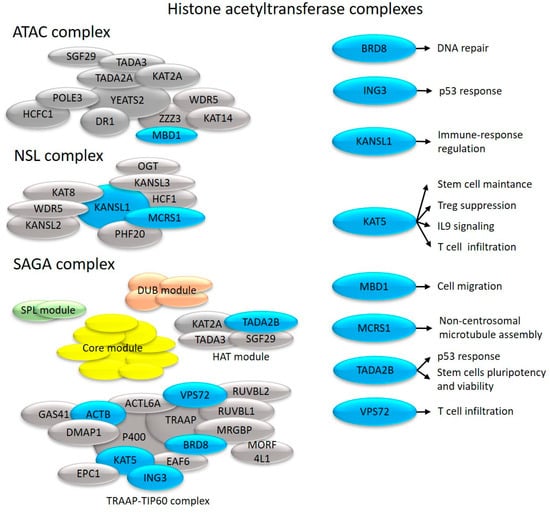
Figure 4.
Effects on cell proliferation and immune system-related processes of histone acetyltransferase complex components with altered transcription in uninvolved skin (depicted in blue).
The H4 and H2A histone-specific acetyltransferase [83] and the lipid synthesis regulator [84,85] NAA40 are also differentially expressed in uninvolved psoriatic skin (Figure 1).
Type A histone acetyltransferases are components of several complexes that exert specific or universal effects [86]. As a result of their analysis, we found differential transcriptional expression of individual subunits of the NSL acetyltransferase complex and the SAGA deubiquitinase and histone acetyltransferase multicomplex with various transcription factor-interacting proteins [87,88], including TRRAP [88] (Figure 1).
The NSL complex regulates many mitochondrial processes, as well as transcription, RNA splicing, and telomere elongation [89]. As components of this complex, KANSL1 and MCRS1 show transcriptional alterations in uninvolved psoriatic skin (Figure 1).
KANSL1 is a master regulator of immune gene expression [90] (Figure 4), whereas MCRS1 protects chromosome-associated microtubules from depolymerization during mitosis [91] (Figure 4).
We identified a change in the expression of TADA2B in uninvolved skin. This is a part of the HAT module of the SAGA complex (Figure 1), which regulates p53 responses [92], stem cell pluripotency, and viability [93] (Figure 4).
TRRAP, which is responsible for recruiting transcription factors and histone acetyltransferases to chromatin, is required for transcriptional activation [94]. TRRAP [95] regulates the entry from the G0 to G1 phase and transitions between the different phases throughout the cell cycle [96], and by regulating critical differentiation markers, it maintains stem cells self-renewal and prevents their differentiation, both of which are known to be affected in psoriasis [97]. TRRAP represses the master regulator of interferon genes, IRF9 [95], whose expression is elevated in psoriasis [98]. TRRAP is also a component of the Tip60 complex, which promotes histone acetyltransferase activity [95]. Among Tip60 complex members, we identified abnormal expression of ACTB, BRD8, ING3, and KAT5 (discussed above) in uninvolved psoriatic skin (Figure 1). The TIP60 complex coactivators BRD8 and ING3 regulate p53-dependent gene suppression and the cell cycle [99,100] (Figure 4).
Members of the inhibitor of histone acetyltransferases (INHAT) complex ANP32A and SET inhibit p300/CBP (CREBBP)- and KAT2B (PCAF)-mediated histone acetylation [101] resulting in the silencing of HAT-dependent transcription. The SET protein (described above among histone chaperones) can inhibit histone H4 and H1 acetylation-dependent transcription [102].
The HAT module of the SAGA complex shares several components with the large acetyltransferase ATAC complex [103], which is one of the main regulators of mitosis through the acetylation of histone H3 and H4 [104]. The ATAC complex component MBIP shows altered expression in non-lesioned skin. Splice variations of this gene have been described in psoriasis [105] pathogenesis, in which they contribute to immune cell infiltration [106] and/or keratinocyte hyperproliferation.
2.4. Histone Deacetylation
Among the members of the HDACI histone deacetylase family, HDAC3 and HDAC8 showed altered expression in uninvolved skin (Figure 1). HDAC3 inhibition results in the reduced expression of AQP3 [107], contributing to skin dryness in uninvolved and lesional psoriatic skin [108] and a decrease in LPS-induced inflammatory gene expression in macrophages [109] (Figure 5A). HDAC3 is part of the NCOR/SMRT complex, which is responsible for nuclear receptor-mediated transcriptional repression [110]. From this complex, we observed the abnormal expression of the GPS2 and TBL1X genes (Figure 1).
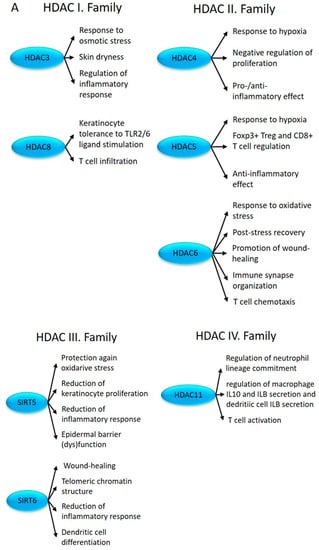
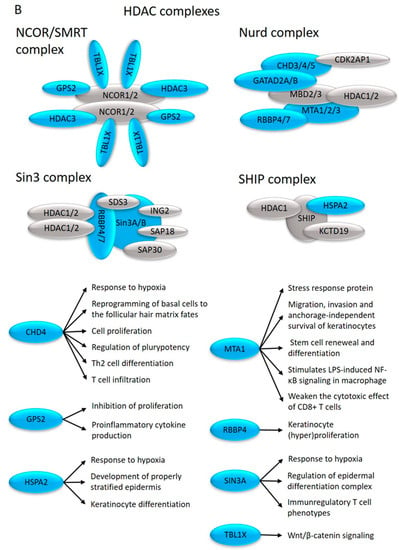
Figure 5.
The impact of differentially expressed HDACs (A) and HDAC complexes (B) (depicted in blue) on proliferation, differentiation, and immune regulation in uninvolved skin.
GPS2 regulates proinflammatory cytokine production in macrophages [111] and inhibits proliferation by suppressing mitogen-activated protein kinase-mediated signaling [112].
TBL1X modulates Wnt/β-catenin and TNFA-regulated transcription [110] (Figure 5B). Elevated levels of TBL1X have been described previously in psoriasis [113].
Altered expression of HDAC8 in uninvolved skin may modulate (keratinocyte) tolerance to TLR2/6 ligand stimulation [114,115] and may increase T cell infiltration [116] (Figure 5A).
HDAC I family members HDAC1 and HDAC2 are normally expressed in uninvolved skin, but the expression of their repressor SPHK2 is altered (Figure 1). SPHK2 inhibits HDAC1/2 activity [117], thus altering the differentiation of Th17 cells in psoriasis [118].
In addition, we observed altered expression levels of several members of the HDAC1/2 protein complexes, which affect the function of NURD, SHIP, and SIN3 complexes (Figure 1).
The NURD complex is a multi-functional complex, playing a role in remodeling chromatin; regulating histone deacetylase activities; and controlling the development of T cells [119], their cell cycle progression, and progenitor cell maintenance [120]. The NURD complex contains an ATP-dependent CHD3/4 chromodomain helicase; a transcriptional repressor adaptor macromolecule GATAD2A; the histone tail and promoter-reading transcriptional coregulator MTA1; a histone-binding, chromatin-remodeling factor RBBP4; and a DNA-binding MBD2/3, which connects the complex with DNA methylation processes [121] (Figure 5B). Among these molecules, the expression of CHD4, GATAD2A, and MTA1 is altered in uninvolved skin (Figure 1).
CHD4 plays an important role in the early development of the basal epidermal layer and regulates the induction and development of hair follicles by destabilizing the interactions between DNA and histones [122]. In keratinocytes, CHD4 can increase tolerance to stress by limiting the expression of stress response genes [123]. CHD4 also regulates Th2 cell differentiation [124], CD8+ T-cell infiltration [125], and self-antigen expression in epithelial cells [126] (Figure 5B).
GATAD2A regulates proliferation [127] and naive pluripotency [128] in association with CHD4. MTA1 regulates the balance between hematopoietic cell renewal and differentiation [129] via the MyD88 pathway [130]. The overexpression of MTA1 triggers the downregulation of the macrophage-attracting chemokine receptor (CCR2) and ligands, leading to M2 polarization and impairing the cytotoxic effect of T cells, resulting in CD8+ T cell enrichment [131] (Figure 5B).
The HDAC1/2-containing SHIP complex exhibits DNA binding and chromatin remodeling capabilities [132]. We found that HSPA2, a member of the SHIP complex, exhibited altered expression in uninvolved psoriatic skin compared with the skin of healthy individuals (Figure 1). HSPA2 acts as a molecular chaperone and provides protection against the cytotoxic effects of heat shock [132], and its expression in keratinocytes increases with hypoxia [133]. This molecule contributes to early keratinocyte differentiation [134] and acts as an important factor in the establishment and maintenance of the properly layered epidermis [135] (Figure 5B).
The SIN3 multiprotein complex influences protein stability, transcriptional activity, aging and heterochromatinization events, cell proliferation/cell cycle progression, cell survival [136], and pluripotency maintenance [137]. Among the complex components, SIN3A showed abnormal expression in uninvolved skin (Figure 1). Sin3A regulated T cell development [138], in particular Th17 cell differentiation, and the establishment of their inflammatory potential [139]. While in the skin, the same molecule is known to regulate terminal differentiation and the maintenance of epidermis homeostasis [140] (Figure 5B).
Among the members of the HDACII family, HDAC4, HDAC5, and HDAC6 show altered expression in uninvolved skin (Figure 1).
The histone deacetylase HDAC4 acts as a transcriptional repressor, but it may exhibit both pro- and anti-inflammatory effects depending on the target gene. While HDAC4-induced NF-κB gene expression inhibition results in the decreased production of proinflammatory cytokines [141], when inflammatory processes are initiated, it can also increase inflammation by indirectly activating Foxo3a [142]. HDAC4 also inhibits keratinocyte proliferation [143] (Figure 5A).
On the one hand, the overexpression of HDAC5 contributes to the initiation of apoptotic processes in keratinocyte stem cells [143]. On the other hand, it also regulates the transformation of CD4+ T cells into Tregs and the cytokine production of CD8+ T cells [144]. Fluid shear stress stimulates the phosphorylation and nuclear export of HDAC5, which plays an important role in the establishment and maintenance of flow-regulated anti-inflammatory processes [145] (Figure 5A).
Another HDACII family member, HDAC6, promotes cell motility [146] during wound healing [147] and chemotaxis of T lymphocytes [148], and it regulates the organization of immune synapses [149] (Figure 5A).
Among the HDACIII family members, SIRT5 and SIRT6 showed abnormal expression in uninvolved skin (Figure 1).
SIRT5 negatively regulates keratinocyte proliferation and inflammation (TNFA induction [150]) and improves epidermal barrier dysfunction [151] (Figure 5A).
SIRT6-mediated histone H3 deacetylation at the N-terminal tail (H3K9Ac) and during the cell cycle at the globular core (H3K56Ac) regulates telomeric chromatin structure, which is necessary to maintain genomic stability and lifespan [152]. By contrast, SIRT6-mediated deacetylation at H3K18 of the pericentric chromatin prevents proliferation-related (replicative) cellular senescence [153]. Changes in SIRT6 expression were also reported in association with the adaptive immune responses [150]. It regulates the balance between the M1 and M2 macrophages, influences wound healing [154], inhibits skin inflammation [155], and plays a role in cDC differentiation and function [156] (Figure 5A).
The HDACIV family member HDAC11 was also differentially expressed in uninvolved skin (Figure 1). The biological function of this family is incomplete. HDAC11 plays an important role in immune regulation, neutrophil lineage commitment, and inflammatory responses, including the regulation of macrophage IL10 and IL1B secretion, dendritic cell IL1B secretion, and T cell activation [157] (Figure 5A).
3. Materials and Methods
3.1. Establishment of the Psoriatic Transcriptome Sequencing Data Set
The dataset we used for these investigations was successfully used in another study that screened for psoriasis-related alterations affecting the peripheral nervous system in psoriatic uninvolved and lesional skin [158]. The combined transcriptome sequencing data were obtained from three studies [159,160,161] that randomly enrolled individuals with chronic plaque-type psoriasis and healthy donors (number of samples: lesional psoriatic: 99, uninvolved psoriatic: 27, healthy: 172). Skin punch biopsies were collected with no gender or age (>18) preferences for RNA sequencing. Psoriatic patients (PASI: min. 1% of total body surface area) on topical and systemic anti-psoriatic treatments had a washout period (the time between the last treatment and sample collection intended to exclude the interference of medication-related effects) of 1 and 2 weeks, respectively, prior to biopsy collection in all studies.
3.2. RNA Sequencing, Data Processing, and Differential Expression Analysis
RNA sequencing data processing and analysis were performed as described previously [158]. Briefly, the three RNA sequencing datasets [159,160] (ID Accession numbers: SRP035988, SRP050971, and SRP055813) were downloaded from the Sequence Read Archive using SRA tools (v2.9.2), and all available samples were uniformly reprocessed. Transcript levels were quantified using Kallisto (version 0.43.0) [162] and full transcriptome annotation GENCODE [163] v27 software. Transcript-level, length-scaled TPM (Transcripts Per Million) expression estimates from Kallisto were imported into the R statistical environment (v3.4.3) using the tximport [164] package (v1.6.0). The data were TMM-normalized [165] (edgeR [166] v3.20.9) and voom-transformed (limma [167,168] v3.34.9). voomWithQualityWeights() was used to combine the observation-level weight of the transcripts with the sample-specific weight, retaining lower-quality samples but down-weighing them in the analysis. Differential expression between uninvolved and healthy sample groups was tested using Limma. A linear model was fitted (limma lmFit), and the moderated t-statistics were calculated (eBayes). Differentially expressed transcripts (DETs) were defined if they had an FDR [168,169] corrected p-value of <0.05.
3.3. Screening for Histones and Histone Acetylation-Related DETs
Differentially expressed transcripts (DETs) from the NL vs. H (non-lesional/uninvolved and healthy, respectively) comparison were analyzed using libraries of datasets downloaded from https://amigo.geneontology.org/amigo/term/ (accessed on 24–29 June 2023) and supplemented with literature data. A complete list of libraries is shown in Supplementary Table S1, in which the GO database and literature datasets [13,14,73,87,101,103,132,170,171,172,173] are listed separately. The filtering used to determine matches between NL vs. H and the downloaded dataset was performed in Python by applying intersection analysis. Detailed information on all methodological steps and processes of the study is provided in Supplementary Methodology Figure S1.
4. Conclusions
On the basis of our findings, we identified complex expression abnormalities in histones and genes with functions in histone acetylation-related processes.
There are already some therapies available to alleviate the clinical manifestation of the symptoms, which are based on a significant number of genetic/epigenetic studies [174]. The regulatory effect of HDAC inhibitors on T cells has been reported. According to these studies, in the presence of histone deacetylase inhibitors, the release of Th1 cytokines and the polarization of Th17 cells decreases, while the formation of Treg cells increases [175]. In addition, HDAC inhibition also modulates pigmentation by reducing MITF expression [176]. A study on the pan-HDAC inhibitor vorinostat found that it induced the apoptosis and differentiation of keratinocytes—consistent with the inhibition of keratinocyte proliferation in psoriasis [177]. According to recent research, the HDAC1 inhibitor (entinostat) reduced the infiltration of IL-17A+ γδT cells into the skin [178].
The pan-BET bromodomain HAT inhibitor (JQ1) reduced the ratio of IL17A+/IFNY+ T cells and IL17A secretion in both psoriatic arthritis patients and healthy individuals [179]. The CREBBP and P300-specific (type A HATs) inhibitor (CBP30) reduced the induced Th17 response in patients with psoriatic arthritis [180].
The described alterations are likely to contribute to the dysregulation of proliferation and differentiation, pro- and anti-inflammatory processes mediated by innate and professional immune cells in uninvolved psoriatic skin, leading to disease flare-ups. Further experimental confirmation of their functional modification may represent new points of intervention.
5. Limitations of the Study
It is important to note that no fold change cut-off was used as a criterion for differential expression; therefore, minor differences between uninvolved and healthy skin (FDR < 0.05) are also included in the study. These minor differences (as well as all others) in the expression were observed at the level of RNS transcripts, some of which may not have manifested at the protein level due to post-transcriptional, translational, and post-translational events, including the processing and degradation of proteins. In addition, skin biopsies contain both the epidermis and the dermis. These two layers of the skin contain different cell types, including keratinocytes, melanocytes, Merkel cells, fibroblasts, and several resident immune cells, like T cells, dendritic cells, Langerhans cells, NK cells, and macrophages. Therefore, the cell type in which the mRNA expression differences manifest could not be determined with certainty, and further experimental confirmation is required to support them, for which these results provide a strong basis.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms241914551/s1.
Author Contributions
Conceptualization, G.G.; methodology, G.G. and D.R.; software, G.G. and D.R.; formal analysis, G.G. and D.R.; resources, G.G. and D.R.; data curation, G.G. and D.R.; writing—original draft preparation, D.R. and G.G.; writing—review and editing, D.R., K.S., L.K. and G.G.; visualization, D.R.; supervision, G.G.; project administration, G.G. and D.R.; funding acquisition, G.G. and D.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Research, Development, and Innovation Office of Hungary, GINOP-2.2.1-15-2016-00007, and by the Géza Hetényi (5S 269 A202) Research Grants. The project also has received funding from the EU’s Horizon 2020 research and innovation program under a grant agreement (number 739593). D.R. was supported by the Gedeon Richter Talentum Foundation (number 5A090) (H-1103 Budapest, Gyömrői str. 19-21). K.S. was supported by the Albert Szent-Györgyi Research Grant of the University of Szeged, Albert Szent-Györgyi Medical School, and the OTKA K143576 Research Grant from the National Research, Development and Innovation Office.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Only publicly available data was used in the study (Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accessed on 15 November 2021); study ID: SRP035988, SRP050971, and SRP055813).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ACTB | Actin Beta |
| ANP32A | Acidic Nuclear Phosphoprotein 32 Family Member A |
| ANP32E | Acidic Nuclear Phosphoprotein 32 Family Member E |
| AQP3 | Aquaporin 3 |
| ATAC | Ada-two-A-containing |
| B-CDK1 | Cyclin Dependent Kinase 1 |
| BRD8 | Bromodomain Containing 8 |
| CAF1 | Chromatin Assembly Factor-1 |
| CBP/CREBBP | CREB-binding Protein |
| CCL2 | C-C Motif Chemokine Ligand 2 |
| CCR2 | C-C Motif Chemokine Receptor 2 |
| cDC | classical Dendritic Cells |
| CHAF1A | Chromatin Assembly Factor 1 Subunit A |
| CHD3 | Chromodomain Helicase DNA-Binding Protein 3 |
| CHD4 | Chromodomain Helicase DNA-Binding Protein 4 |
| cMYC | MYC Proto-Oncogene, BHLH Transcription Factor |
| CREMα | cAMP-responsive Element Modulator α |
| CTBP1 | C-Terminal Binding Protein 1 |
| DET | Differentially Expressed Transcript |
| DNA | Deoxyribonucleic Acid |
| DUB | Deubiquitinating |
| E-CDK2 | Cyclin-Dependent Kinase 2 |
| ELP3 | Elongator Acetyltransferase Complex Subunit 3 |
| EP300 | E1A Binding Protein P300 |
| FDR | False Discovery Rate |
| FOXP3 | Forkhead Box P3 |
| GATAD2A | GATA Zinc Finger Domain-Containing 2A |
| GO | Gene Ontology |
| GNAT | GCN5-related N-acetyltransferases |
| GPS2 | G Protein Pathway Suppressor 2 |
| H | Healthy |
| H2AFY | MacroH2A.1 Histone |
| H2AFZ | H2A.Z Variant Histone 1 |
| H3F3A | H3.3 Histone A |
| H3F3B | H3.3 Histone B |
| HAT | Histone Acetyltransferase |
| HDAC | Histone Deacetylase |
| HDAC11 | Histone Deacetylase 11 |
| HDAC3 | Histone Deacetylase 3 |
| HDAC4 | Histone Deacetylase 4 |
| HDAC5 | Histone Deacetylase 5 |
| HDAC6 | Histone Deacetylase 6 |
| HDAC8 | Histone Deacetylase 8 |
| HIRA | Histone Cell Cycle Regulator |
| HIST2H2AA3 | H2A Clustered Histone 18 |
| HIST2H4A | H4 Clustered Histone 14 |
| HSPA2 | Heat Shock Protein Family A (Hsp70) Member 2 |
| ID | Identification |
| IFNB1 | Interferon Beta 1 |
| IFNG | Interferon Gamma |
| IL10 | Interleukin 10 |
| IL-17A | Interleukin 17A |
| IL-8 | Interleukin 8 |
| IL-9 | Interleukin 9 |
| ILB | Interleukin 1 Beta |
| ING3 | Inhibitor of Growth Family Member 3 |
| INHAT | Inhibitor of Acetyltransferases |
| JAK/STAT | Janus Kinase/Signal Transducers and Activators of Transcription |
| KANSL1 | KAT8 Regulatory NSL Complex Subunit 1 |
| KAT2B | Lysine Acetyltransferase 2B |
| KAT5 | Lysine Acetyltransferase 5 |
| Ki67 | Marker Of Proliferation Ki-67 |
| L | Lesional |
| LPS | Lipopolysaccharide |
| MBD2 | Methyl-CpG Binding Domain Protein 2 |
| MBD3 | Methyl-CpG Binding Domain Protein 3 |
| MBIP | MAP3K12 Binding Inhibitory Protein 1 |
| MITF | Melanocyte Inducing Transcription Factor |
| MCRS1 | Microspherule Protein 1 |
| MTA1 | Metastasis Associated 1 |
| MYBBP1A | MYB Binding Protein 1a |
| MYD88 | MYD88 Innate Immune Signal Transduction Adaptor |
| MYST | Moz, Ybf2/Sas3, Sas2, Tip60 |
| NAA40 | N-Alpha-Acetyltransferase 40, NatD Catalytic Subunit |
| NCOR | Nuclear Receptor—Co-repressor |
| NF-κB | Nuclear Factor Kappa-light-chain-enhancer of Activated B Cells |
| NL | Non-lesional |
| NPM1 | Nucleophosmin 1 |
| NSL | Non-specific Lethal |
| NURD | NUcleosome Remodeling and Deacetylase |
| P53 | Tumor Protein P53 |
| PASI | Psoriasis Area and Severity Index |
| PARP1 | Poly(ADP-Ribose) Polymerase 1 |
| PWP1 | PWP1 Homolog, Endonuclein |
| RBBP4 | RB Binding Protein 4, Chromatin Remodeling Factor |
| RNA | Ribonucleic Acid |
| SAGA | Spt-Ada-Gcn5 Acetyltransferase |
| SET | SET Nuclear Proto-Oncogene |
| SIN3A | SIN3 Transcription Regulator Family Member A |
| SIRT5 | Sirtuin 5 |
| SIRT6 | Sirtuin 6 |
| SMRT | Silencing Mediator for Retinoid and Thyroid Receptor |
| SPHK2 | Sphingosine Kinase 2 |
| SPL | Splicing |
| STAT3 | Signal Transducer and Activator of Transcription 3 |
| TADA2B | Transcriptional Adaptor 2B |
| TBL1X | Transducin Beta-Like 1 X-Linked |
| Th17 | T helper 17 |
| TLR2 | Toll-Like Receptor 2 |
| TLR4 | Toll-Like Receptor 4 |
| TLR6 | Toll-Like Receptor 6 |
| TNFA | Tumor Necrosis Factor |
| TMM | Trimmed mean of M-values |
| TPM | Transcripts per million |
| Treg | Regulatory T cell |
| TRRAP | Transformation/Transcription Domain-Associated Protein |
| UBN1 | Ubinuclein 1 |
| VPS72 | Vacuolar Protein-Sorting 72 Homolog |
| WBP2 | WW Domain-Binding Protein 2 |
References
- Chiricozzi, A.; Romanelli, P.; Volpe, E.; Borsellino, G.; Romanelli, M. Scanning the Immunopathogenesis of Psoriasis. Int. J. Mol. Sci. 2018, 19, 179. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, Y.; Cui, L.; Shi, Y.; Guo, C. Advances in the pathogenesis of psoriasis: From keratinocyte perspective. Cell Death Dis. 2022, 13, 81. [Google Scholar] [CrossRef]
- Caputo, V.; Strafella, C.; Termine, A.; Dattola, A.; Mazzilli, S.; Lanna, C.; Cosio, T.; Campione, E.; Novelli, G.; Giardina, E.; et al. Overview of the molecular determinants contributing to the expression of Psoriasis and Psoriatic Arthritis phenotypes. J. Cell. Mol. Med. 2020, 24, 13554–13563. [Google Scholar] [CrossRef]
- Szabó, K.; Bata-Csörgő, Z.; Dallos, A.; Bebes, A.; Francziszti, L.; Dobozy, A.; Kemény, L.; Széll, M. Regulatory Networks Contributing to Psoriasis Susceptibility. Acta Dermato-Venereologica 2014, 94, 380–385. [Google Scholar] [CrossRef]
- Gudjonsson, J.E.; Ding, J.; Li, X.; Nair, R.P.; Tejasvi, T.; Qin, Z.S.; Ghosh, D.; Aphale, A.; Gumucio, D.L.; Voorhees, J.J.; et al. Global Gene Expression Analysis Reveals Evidence for Decreased Lipid Biosynthesis and Increased Innate Immunity in Uninvolved Psoriatic Skin. J. Investig. Dermatol. 2009, 129, 2795–2804. [Google Scholar] [CrossRef]
- Szél, E.; Bozó, R.; Hunyadi-Gulyás, É.; Manczinger, M.; Szabó, K.; Kemény, L.; Bata-Csörgő, Z.; Groma, G. Comprehensive Proteomic Analysis Reveals Intermediate Stage of Non-Lesional Psoriatic Skin and Points out the Importance of Proteins Outside this Trend. Sci. Rep. 2019, 9, 11382. [Google Scholar] [CrossRef]
- Masalha, M.; Ben-Dov, I.Z.; Ram, O.; Meningher, T.; Jacob-Hirsch, J.; Kassem, R.; Sidi, Y.; Avni, D. H3K27Ac modification and gene expression in psoriasis. J. Dermatol. Sci. 2021, 103, 93–100. [Google Scholar] [CrossRef]
- Tamaru, H. Confining euchromatin/heterochromatin territory: Jumonji crosses the line. Genes Dev. 2010, 24, 1465–1478. [Google Scholar] [CrossRef]
- Peterson, C.L.; Laniel, M.-A. Histones and histone modifications. Curr. Biol. 2004, 14, R546–R551. [Google Scholar] [CrossRef]
- Fyodorov, D.V.; Zhou, B.-R.; Skoultchi, A.I.; Bai, Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 2018, 19, 192–206. [Google Scholar] [CrossRef]
- Marzluff, W.F.; Wagner, E.J.; Duronio, R.J. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat. Rev. Genet. 2008, 9, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Martire, S.; Banaszynski, L.A. The roles of histone variants in fine-tuning chromatin organization and function. Nat. Rev. Mol. Cell Biol. 2020, 21, 522–541. [Google Scholar] [CrossRef] [PubMed]
- Amatori, S.; Tavolaro, S.; Gambardella, S.; Fanelli, M. The dark side of histones: Genomic organization and role of oncohistones in cancer. Clin. Epigenet. 2021, 13, 71. [Google Scholar] [CrossRef]
- De Koning, L.; Corpet, A.E.; Haber, J.; Almouzni, G. Histone chaperones: An escort network regulating histone traffic. Nat. Struct. Mol. Biol. 2007, 14, 997–1007. [Google Scholar] [CrossRef]
- Wang, D.; Li, Y.; Liu, Y.; Cheng, S.; Liu, F.; Zuo, R.; Ding, C.; Shi, S.; Liu, G. NPM1 promotes cell proliferation by targeting PRDX6 in colorectal cancer. Int. J. Biochem. Cell Biol. 2022, 147, 106233. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, M.; Beji, S.; Sileno, S.; Lulli, D.; Mercurio, L.; Madonna, S.; Cirielli, C.; Pallotta, S.; Albanesi, C.; Capogrossi, M.C.; et al. Extracellular Nucleophosmin Is Increased in Psoriasis and Correlates with the Determinants of Cardiovascular Diseases. Front. Cardiovasc. Med. 2022, 9, 867813. [Google Scholar] [CrossRef]
- Ng, C.; Aichinger, M.; Nguyen, T.; Au, C.; Najar, T.; Wu, L.; Mesa, K.R.; Liao, W.; Quivy, J.-P.; Hubert, B.; et al. The histone chaperone CAF-1 cooperates with the DNA methyltransferases to maintain Cd4 silencing in cytotoxic T cells. Genes Dev. 2019, 33, 669–683. [Google Scholar] [CrossRef]
- Mehta, S.; Jeffrey, K. Chapter 12—Immune System Disorders and Epigenetics. In Medical Epigenetics; Tollefsbol, T.O., Ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 199–219. [Google Scholar] [CrossRef]
- Ellenbroek, B.; Youn, J. Chapter 5—Environment Challenges and the Brain. In Gene-Environment Interactions in Psychiatry; Ellenbroek, B., Youn, J., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 107–139. [Google Scholar] [CrossRef]
- Milazzo, G.; Mercatelli, D.; Di Muzio, G.; Triboli, L.; De Rosa, P.; Perini, G.; Giorgi, F.M. Histone Deacetylases (HDACs): Evolution, Specificity, Role in Transcriptional Complexes, and Pharmacological Actionability. Genes 2020, 11, 556. [Google Scholar] [CrossRef]
- Camilo, V.; Jerónimo, C. Chapter 17—Present and future perspectives for targeting histone modifications in therapy. In Histone Modifications in Therapy; Castelo-Branco, P., Jeronimo, C., Eds.; Academic Press: Cambridge, MA, USA, 2020; Volume 20, pp. 415–457. [Google Scholar] [CrossRef]
- Herceg, Z.; Murr, R. Chapter 3—Mechanisms of Histone Modifications. In Handbook of Epigenetics; Tollefsbol, T., Ed.; Academic Press: Cambridge, MA, USA, 2011; pp. 25–45. [Google Scholar] [CrossRef]
- Lai, F.; Jin, L.; Gallagher, S.; Mijatov, B.; Zhang, X.D.; Hersey, P. Chapter Two—Histone Deacetylases (HDACs) as Mediators of Resistance to Apoptosis in Melanoma and as Targets for Combination Therapy with Selective BRAF Inhibitors. In Advances in Pharmacology; Smalley, K.S.M., Ed.; Academic Press: Cambridge, MA, USA, 2012; Volume 65, pp. 27–43. [Google Scholar] [CrossRef]
- Xia, X.; Cao, G.; Sun, G.; Zhu, L.; Tian, Y.; Song, Y.; Guo, C.; Wang, X.; Zhong, J.; Zhou, W.; et al. GLS1-mediated glutaminolysis unbridled by MALT1 protease promotes psoriasis pathogenesis. J. Clin. Investig. 2020, 130, 5180–5196. [Google Scholar] [CrossRef]
- Hofmann, S.R.; Carlsson, E.; Kapplusch, F.; Carvalho, A.L.; Liloglou, T.; Schulze, F.; Abraham, S.; Northey, S.; Russ, S.; Surace, A.E.A.; et al. Cyclic AMP Response Element Modulator-α Suppresses PD-1 Expression and Promotes Effector CD4+ T Cells in Psoriasis. J. Immunol. 2021, 207, 55–64. [Google Scholar] [CrossRef]
- Rauen, T.; Hedrich, C.M.; Tenbrock, K.; Tsokos, G.C. cAMP responsive element modulator: A critical regulator of cytokine production. Trends Mol. Med. 2013, 19, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Kubrova, E.; Su, M.; Galeano-Garces, C.; Galvan, M.L.; Jerez, S.; Dietz, A.B.; Smith, J.; Qu, W.; van Wijnen, A.J. Differences in Cytotoxicity of Lidocaine, Ropivacaine, and Bupivacaine on the Viability and Metabolic Activity of Human Adipose-Derived Mesenchymal Stem Cells. Am. J. Phys. Med. Rehabil. 2021, 100, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Kubrova, E.; Wu, T.; Denbeigh, J.M.; Hunt, C.; Dietz, A.B.; Smith, J.; Qu, W.; Wijnen, A.J. Effect of Lidocaine on Viability and Gene Expression of Human Adipose–derived Mesenchymal Stem Cells: An in vitro Study. PM R J. Inj. Funct. Rehabil. 2019, 11, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Barrero, M.J.; Sese, B.; Kuebler, B.; Bilic, J.; Boue, S.; Martí, M.; Belmonte, J.C.I. Macrohistone Variants Preserve Cell Identity by Preventing the Gain of H3K4me2 during Reprogramming to Pluripotency. Cell Rep. 2013, 3, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Cipta, N.O.; Chen, Y.; Loh, Y.-H. H3.3 safeguards haematopoietic ERV-quilibrium. Nature 2022, 24, 7–9. [Google Scholar] [CrossRef]
- Lyons, S.M.; Cunningham, C.H.; Welch, J.D.; Groh, B.; Guo, A.Y.; Wei, B.; Whitfield, M.L.; Xiong, Y.; Marzluff, W.F. A subset of replication-dependent histone mRNAs are expressed as polyadenylated RNAs in terminally differentiated tissues. Nucleic Acids Res. 2016, 44, 9190–9205. [Google Scholar] [CrossRef]
- Armache, A.; Yang, S.; de Paz, A.M.; Robbins, L.E.; Durmaz, C.; Cheong, J.Q.; Ravishankar, A.; Daman, A.W.; Ahimovic, D.J.; Klevorn, T.; et al. Histone H3.3 phosphorylation amplifies stimulation-induced transcription. Nature 2020, 583, 852–857. [Google Scholar] [CrossRef]
- Axtell, R.C.; Raman, C.; Steinman, L. Interferon-β exacerbates Th17-mediated inflammatory disease. Trends Immunol. 2011, 32, 272–277. [Google Scholar] [CrossRef]
- Yu, G.; Zhang, Y.; Gupta, V.; Zhang, J.; MacCarthy, T.; Duan, Z.; Scharff, M.D. The role of HIRA-dependent H3.3 deposition and its modifications in the somatic hypermutation of immunoglobulin variable regions. Proc. Natl. Acad. Sci. USA 2021, 118, e2114743118. [Google Scholar] [CrossRef]
- Timinszky, G.; Till, S.O.; Hassa, P.; Hothorn, M.; Kustatscher, G.; Nijmeijer, B.; Colombelli, J.; Altmeyer, M.; Stelzer, E.H.K.; Scheffzek, K.; et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009, 16, 923–929. [Google Scholar] [CrossRef]
- Weber, C.M.; Henikoff, S. Histone variants: Dynamic punctuation in transcription. Genes Dev. 2014, 28, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Chang, E.Y.; Ferreira, H.; Somers, J.; Nusinow, D.A.; Owen-Hughes, T.; Narlikar, G.J. MacroH2A Allows ATP-Dependent Chromatin Remodeling by SWI/SNF and ACF Complexes but Specifically Reduces Recruitment of SWI/SNF. Biochemistry 2008, 47, 13726–13732. [Google Scholar] [CrossRef] [PubMed]
- Hurtado-Bagès, S.; Guberovic, I.; Buschbeck, M. The MacroH2A1.1–PARP1 Axis at the Intersection Between Stress Response and Metabolism. Front. Genet. 2018, 9, 417. [Google Scholar] [CrossRef]
- Haque, N.; Ouda, R.; Chen, C.; Ozato, K.; Hogg, J.R. ZFR coordinates crosstalk between RNA decay and transcription in innate immunity. Nat. Commun. 2018, 9, 1145. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, E.A.; Andersen, S.; Marqvorsen, M.H.S.; Skipper, K.A.; Paludan, S.R.; Mikkelsen, J.G. Single-Cell Monitoring of Activated Innate Immune Signaling by a d2eGFP-Based Reporter Mimicking Time-Restricted Activation of IFNB1 Expression. Front. Cell. Infect. Microbiol. 2022, 11, 784762. [Google Scholar] [CrossRef] [PubMed]
- Agelopoulos, M.; Thanos, D. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J. 2006, 25, 4843–4853. [Google Scholar] [CrossRef] [PubMed]
- Hussey, K.M.; Chen, H.; Yang, C.; Park, E.; Hah, N.; Erdjument-Bromage, H.; Tempst, P.; Gamble, M.J.; Kraus, W.L. The Histone Variant MacroH2A1 Regulates Target Gene Expression in Part by Recruiting the Transcriptional Coregulator PELP1. Mol. Cell. Biol. 2014, 34, 2437–2449. [Google Scholar] [CrossRef]
- Chen, H.-L.; Lo, C.-H.; Huang, C.-C.; Lu, M.-P.; Hu, P.-Y.; Chen, C.-S.; Chueh, D.-Y.; Chen, P.; Lin, T.-N.; Lo, Y.-H.; et al. Galectin-7 downregulation in lesional keratinocytes contributes to enhanced IL-17A signaling and skin pathology in psoriasis. J. Clin. Investig. 2021, 131, e130740. [Google Scholar] [CrossRef]
- Novoszel, P.; Holcmann, M.; Stulnig, G.; Fernandes, C.D.S.; Zyulina, V.; Borek, I.; Linder, M.; Bogusch, A.; Drobits, B.; Bauer, T.; et al. Psoriatic skin inflammation is promoted by c-Jun/AP-1-dependent CCL2 and IL-23 expression in dendritic cells. EMBO Mol. Med. 2021, 13, e12409. [Google Scholar] [CrossRef]
- Liu, S.; Chen, X.; Huang, K.; Xiong, X.; Shi, Y.; Wang, X.; Pan, X.; Cong, Y.; Sun, Y.; Ge, L.; et al. Long noncoding RNA RFPL1S-202 inhibits ovarian cancer progression by downregulating the IFN-β/STAT1 signaling. Exp. Cell Res. 2023, 422, 113438. [Google Scholar] [CrossRef]
- Sarcinella, E.; Zuzarte, P.C.; Lau, P.N.I.; Draker, R.; Cheung, P. Monoubiquitylation of H2A.Z Distinguishes Its Association with Euchromatin or Facultative Heterochromatin. Mol. Cell. Biol. 2007, 27, 6457–6468. [Google Scholar] [CrossRef] [PubMed]
- Sales-Gil, R.; Kommer, D.C.; de Castro, I.J.A.; Amin, H.; Vinciotti, V.; Sisu, C.; Vagnarelli, P. Non-redundant functions of H2A.Z.1 and H2A.Z.2 in chromosome segregation and cell cycle progression. EMBO Rep. 2021, 22, e52061. [Google Scholar] [CrossRef] [PubMed]
- Rangasamy, D.J.; Berven, L.; Ridgway, P.; Tremethick, D. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 2003, 22, 1599–1607. [Google Scholar] [CrossRef] [PubMed]
- Colino-Sanguino, Y.; Clark, S.J.; Valdes-Mora, F. The H2A.Z-nucleosome code in mammals: Emerging functions. Trends Genet. 2022, 38, 273–289. [Google Scholar] [CrossRef]
- Li, J.; Li, X.; Hou, R.; Liu, R.; Zhao, X.; Dong, F.; Wang, C.; Yin, G.; Zhang, K. Psoriatic T cells reduce epidermal turnover time and affect cell proliferation contributed from differential gene expression. J. Dermatol. 2015, 42, 874–880. [Google Scholar] [CrossRef]
- Hake, S.B.; Allis, C.D. Histone H3 variants and their potential role in indexing mammalian genomes: The “H3 barcode hypothesis”. Proc. Natl. Acad. Sci. USA 2006, 103, 6428–6435. [Google Scholar] [CrossRef]
- Jin, C.; Felsenfeld, G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007, 21, 1519–1529. [Google Scholar] [CrossRef]
- Jang, C.-W.; Shibata, Y.; Starmer, J.; Yee, D.; Magnuson, T. Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 2015, 29, 1377–1392. [Google Scholar] [CrossRef]
- Shi, L.; Wen, H.; Shi, X. The histone variant H3.3 in transcriptional regulation and human disease. J. Mol. Biol. 2017, 429, 1934–1945. [Google Scholar] [CrossRef]
- Gupta, S.; Reddy, D. Histone variant H3.3 and its future prospects in cancer clinic. J. Radiat. Cancer Res. 2017, 8, 77. [Google Scholar] [CrossRef]
- Luo, L.; Luo, Y.; Xu, J.; Zhu, R.; Wu, J.; Liu, X.; Li, W.; Yao, X. Heterogeneous origin of IgE in atopic dermatitis and psoriasis revealed by B cell receptor repertoire analysis. Allergy 2022, 77, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Escobar, T.M.; Yu, J.-R.; Liu, S.; Lucero, K.; Vasilyev, N.; Nudler, E.; Reinberg, D. Inheritance of repressed chromatin domains during S phase requires the histone chaperone NPM1. Sci. Adv. 2022, 8, eabm3945. [Google Scholar] [CrossRef] [PubMed]
- Szegedi, K.; Göblös, A.; Bacsa, S.; Antal, M.; Németh, I.B.; Bata-Csörgő, Z.; Kemény, L.; Dobozy, A.; Széll, M. Expression and Functional Studies on the Noncoding RNA, PRINS. Int. J. Mol. Sci. 2013, 14, 205–225. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Lee, K.-S.; Seol, J.-E.; Yu, K.; Chakravarti, D.; Seo, S.-B. Inhibition of p53 acetylation by INHAT subunit SET/TAF-Iβ represses p53 activity. Nucleic Acids Res. 2012, 40, 75–87. [Google Scholar] [CrossRef]
- Canela, N.; Rodriguez-Vilarrupla, A.; Estanyol, J.M.; Díaz, C.; Pujol, M.J.; Agell, N.; Bachs, O. The SET Protein Regulates G2/M Transition by Modulating Cyclin B-Cyclin-dependent Kinase 1 Activity. J. Biol. Chem. 2003, 278, 1158–1164. [Google Scholar] [CrossRef] [PubMed]
- Estanyol, J.M.; Jaumot, M.; Casanovas, O.; Rodriguez-Vilarrupla, A.; Agell, N.; Bachs, O. The Protein SET Regulates the Inhibitory Effect of p21Cip1 on Cyclin E-Cyclin-dependent Kinase 2 Activity. J. Biol. Chem. 1999, 274, 33161–33165. [Google Scholar] [CrossRef] [PubMed]
- Henri, P.; Prevel, C.; Pellerano, M.; Lacotte, J.; Stoebner, P.; Morris, M.; Meunier, L. Psoriatic epidermis is associated with upregulation of CDK2 and inhibition of CDK4 activity. Br. J. Dermatol. 2020, 182, 678–689. [Google Scholar] [CrossRef]
- Melero, J.L.; Andrades, S.; Arola, L.; Romeu, A. Deciphering psoriasis. A bioinformatic approach. J. Dermatol. Sci. 2018, 89, 120–126. [Google Scholar] [CrossRef]
- Yu, Z.; Liu, J.; Deng, W.-M.; Jiao, R. Histone chaperone CAF-1: Essential roles in multi-cellular organism development. Cell. Mol. Life Sci. 2015, 72, 327–337. [Google Scholar] [CrossRef]
- You, W.; Wang, Z. The Loss of Chaf1a Affects the Proliferation and Differentiation of Hematopoietic Stem Progenitor Cells. Int. Core J. Eng. 2022, 8, 606–610. [Google Scholar] [CrossRef]
- Li, J.; Xing, J.; Lu, F.; Chang, W.; Liang, N.; Wang, Y.; Li, X.; Zhao, X.; Hou, R.; Man, M.; et al. Psoriatic Dermal-derived Mesenchymal Stem Cells Reduce Keratinocyte Junctions, and Increase Glycolysis. Acta Dermato-Venereologica 2020, 100, adv00122. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.C.; Dilworth, F.J. Chapter Eight—Epigenetic Regulation of Adult Myogenesis. In Current Topics in Developmental Biology; Sassoon, D., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 126, pp. 235–284. [Google Scholar] [CrossRef]
- Banumathy, G.; Somaiah, N.; Zhang, R.; Tang, Y.; Hoffmann, J.; Andrake, M.; Ceulemans, H.; Schultz, D.; Marmorstein, R.; Adams, P.D. Human UBN1 Is an Ortholog of Yeast Hpc2p and Has an Essential Role in the HIRA/ASF1a Chromatin-Remodeling Pathway in Senescent Cells. Mol. Cell. Biol. 2009, 29, 758–770. [Google Scholar] [CrossRef] [PubMed]
- Mercurio, L.; Lulli, D.; Mascia, F.; Dellambra, E.; Scarponi, C.; Morelli, M.; Valente, C.; Carbone, M.L.; Pallotta, S.; Girolomoni, G.; et al. Intracellular Insulin-like growth factor binding protein 2 (IGFBP2) contributes to the senescence of keratinocytes in psoriasis by stabilizing cytoplasmic p21. Aging 2020, 12, 6823–6851. [Google Scholar] [CrossRef] [PubMed]
- Papamichos-Chronakis, M.; Watanabe, S.; Rando, O.J.; Peterson, C.L. Global Regulation of H2A.Z Localization by the INO80 Chromatin-Remodeling Enzyme Is Essential for Genome Integrity. Cell 2011, 144, 200–213. [Google Scholar] [CrossRef]
- Obri, A.; Ouararhni, K.; Papin, C.; Diebold, M.-L.; Padmanabhan, K.; Marek, M.; Stoll, I.; Roy, L.; Reilly, P.T.; Mak, T.W.; et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 2014, 505, 7485. [Google Scholar] [CrossRef]
- Zhang, J.; Lan, Z.; Qiu, G.; Ren, H.; Zhao, Y.; Gu, Z.; Li, Z.; Feng, L.; He, J.; Wang, C. Over-expression of ANP32E is associated with poor prognosis of pancreatic cancer and promotes cell proliferation and migration through regulating β-catenin. BMC Cancer 2020, 20, 1065. [Google Scholar] [CrossRef]
- Moreno-Andrés, D.; Yokoyama, H.; Scheufen, A.; Holzer, G.; Lue, H.; Schellhaus, A.K.; Weberruss, M.; Takagi, M.; Antonin, W. VPS72/YL1-Mediated H2A.Z Deposition Is Required for Nuclear Reassembly after Mitosis. Cells 2020, 9, 1702. [Google Scholar] [CrossRef]
- Gui, Y.; Liu, X.; Wang, C.; Yang, P. Bioinformatics Analysis of Prognostic Significance of VPS72 and Correlations with Immune Infiltrates in Hepatocellular Carcinoma. Eur. PMC, 2021; ahead of print. [Google Scholar] [CrossRef]
- Li, H.; Zhang, C.; Bian, L.; Deng, H.; Blevins, M.; Han, G.; Fan, B.; Yang, C.; Zhao, R.; High, W.; et al. Inhibition of CtBP-Regulated Proinflammatory Gene Transcription Attenuates Psoriatic Skin Inflammation. J. Investig. Dermatol. 2022, 142, 390–401. [Google Scholar] [CrossRef]
- Walko, G.; Woodhouse, S.; Pisco, A.O.; Rognoni, E.; Liakath-Ali, K.; Lichtenberger, B.M.; Mishra, A.; Telerman, S.B.; Viswanathan, P.; Logtenberg, M.; et al. A genome-wide screen identifies YAP/WBP2 interplay conferring growth advantage on human epidermal stem cells. Nat. Commun. 2017, 8, 14744. [Google Scholar] [CrossRef]
- Field, N.S.; Elbulok, O.A.; Dybas, J.M.; Moser, E.K.; Dar, A.A.; Spruce, L.A.; Fazelinia, H.; Seeholzer, S.H.; Oliver, P.M. Itch attenuates CD4 T-cell proliferation in mice by limiting WBP2 protein stability. Eur. J. Immunol. 2020, 50, 1468–1483. [Google Scholar] [CrossRef]
- Mori, S.; Bernardi, R.; Laurent, A.; Resnati, M.; Crippa, A.; Gabrieli, A.; Keough, R.; Gonda, T.J.; Blasi, F. Myb-Binding Protein 1A (MYBBP1A) Is Essential for Early Embryonic Development, Controls Cell Cycle and Mitosis, and Acts as a Tumor Suppressor. PLoS ONE 2012, 7, e39723. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Nemazanyy, I.; Peulen, O.; Shostak, K.; Xu, X.; Tang, S.C.; Wathieu, C.; Turchetto, S.; Tielens, S.; Nguyen, L.; et al. Elp3-mediated codon-dependent translation promotes mTORC2 activation and regulates macrophage polarization. EMBO J. 2022, 41, e109353. [Google Scholar] [CrossRef]
- Su, Q.; Jing, J.; Li, W.; Ma, J.; Zhang, X.; Wang, Z.; Zhou, Z.; Dai, L.; Shao, L. Impaired Tip60-mediated Foxp3 acetylation attenuates regulatory T cell development in rheumatoid arthritis. J. Autoimmun. 2019, 100, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Slivaab, D.; Zhu, Y.X.; Tsaic, S.; Kamined, J.; Yang, Y.-C. Tip60 Interacts with Human Interleukin-9 Receptor α-Chain. Biochem. Biophys. Res. Commun. 1999, 263, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Numata, A.; Kwok, H.S.; Zhou, Q.-L.; Li, J.; Tirado-Magallanes, R.; Angarica, V.E.; Hannah, R.L.; Park, J.; Wang, C.Q.; Krishnan, V.; et al. Lysine acetyltransferase Tip60 is required for hematopoietic stem cell maintenance. Blood 2020, 136, 1735–1747. [Google Scholar] [CrossRef]
- Demetriadou, C.; Raoukka, A.; Charidemou, E.; Mylonas, C.; Michael, C.; Parekh, S.; Koufaris, C.; Skourides, P.; Papageorgis, P.; Tessarz, P.; et al. Histone N-terminal acetyltransferase NAA40 links one-carbon metabolism to chemoresistance. Oncogene 2022, 41, 571–585. [Google Scholar] [CrossRef]
- Charidemou, E.; Tsiarli, M.A.; Theophanous, A.; Yilmaz, V.; Pitsouli, C.; Strati, K.; Griffin, J.L.; Kirmizis, A. Histone acetyltransferase NAA40 modulates acetyl-CoA levels and lipid synthesis. BMC Biol. 2022, 20, 22. [Google Scholar] [CrossRef]
- Pavlou, D.; Kirmizis, A. Depletion of histone N-terminal-acetyltransferase Naa40 induces p53-independent apoptosis in colorectal cancer cells via the mitochondrial pathway. Apoptosis Int. J. Program. Cell Death 2016, 21, 298–311. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Herbst, D.A.; Esbin, M.N.; Louder, R.K.; Dugast-Darzacq, C.; Dailey, G.M.; Fang, Q.; Darzacq, X.; Tjian, R.; Nogales, E. Structure of the human SAGA coactivator complex. Nat. Struct. Mol. Biol. 2021, 28, 989–996. [Google Scholar] [CrossRef]
- Cheon, Y.; Kim, H.; Park, K.; Kim, M.; Lee, D. Dynamic modules of the coactivator SAGA in eukaryotic transcription. Exp. Mol. Med. 2020, 52, 991–1003. [Google Scholar] [CrossRef]
- Radzisheuskaya, A.; Shliaha, P.V.; Grinev, V.V.; Shlyueva, D.; Damhofer, H.; Koche, R.; Gorshkov, V.; Kovalchuk, S.; Zhan, Y.; Rodriguez, K.L.; et al. Complex-dependent histone acetyltransferase activity of KAT8 determines its role in transcription and cellular homeostasis. Mol. Cell 2021, 81, 1749–1765.e8. [Google Scholar] [CrossRef]
- Fejzo, M.S.; Chen, H.-W.; Anderson, L.; McDermott, M.S.; Karlan, B.; Konecny, G.E.; Slamon, D.J. Analysis in epithelial ovarian cancer identifies KANSL1 as a biomarker and target gene for immune response and HDAC inhibition. Gynecol. Oncol. 2021, 160, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Meunier, S.; Vernos, I. K-fibre minus ends are stabilized by a RanGTP-dependent mechanism essential for functional spindle assembly. Nat. Cell Biol. 2011, 13, 1406–1414. [Google Scholar] [CrossRef]
- Gamper, A.M.; Kim, J.; Roeder, R.G. The STAGA Subunit ADA2b Is an Important Regulator of Human GCN5 Catalysis. Mol. Cell. Biol. 2009, 29, 266–280. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef]
- Yin, B.-K.; Wang, Z.-Q. Beyond HAT Adaptor: TRRAP Liaisons with Sp1-Mediated Transcription. Int. J. Mol. Sci. 2021, 22, 12445. [Google Scholar] [CrossRef] [PubMed]
- Detilleux, D.; Raynaud, P.; Pradet-Balade, B.; Helmlinger, D. The TRRAP transcription cofactor represses interferon-stimulated genes in colorectal cancer cells. eLife 2022, 11, e69705. [Google Scholar] [CrossRef]
- Murr, R.; Vaissière, T.; Sawan, C.; Shukla, V.; Herceg, Z. Orchestration of chromatin-based processes: Mind the TRRAP. Oncogene 2007, 26, 37. [Google Scholar] [CrossRef]
- Sawan, C.; Hernandez-Vargas, H.; Murr, R.; Lopez, F.; Vaissière, T.; Ghantous, A.Y.; Cuenin, C.; Imbert, J.; Wang, Z.-Q.; Ren, B.; et al. Histone Acetyltransferase Cofactor Trrap Maintains Self-Renewal and Restricts Differentiation of Embryonic Stem Cells. Stem Cells 2013, 31, 979–991. [Google Scholar] [CrossRef]
- Swindell, W.R.; Johnston, A.; Xing, X.; Voorhees, J.J.; Elder, J.T.; Gudjonsson, J.E. Modulation of Epidermal Transcription Circuits in Psoriasis: New Links between Inflammation and Hyperproliferation. PLoS ONE 2013, 8, e79253. [Google Scholar] [CrossRef] [PubMed]
- Lashgari, A.; Fauteux, M.; Maréchal, A.; Gaudreau, L. Cellular Depletion of BRD8 Causes p53-Dependent Apoptosis and Induces a DNA Damage Response in Non-Stressed Cells. Sci. Rep. 2018, 8, 14089. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, M.; Shiseki, M.; Pedeux, R.M.; Okamura, S.; Kitahama-Shiseki, M.; Miura, K.; Yokota, J.; Harris, C.C. A novel PHD-finger motif protein, p47ING3, modulates p53-mediated transcription, cell cycle control, and apoptosis. Oncogene 2003, 22, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.-B.; Macfarlan, T.; McNamara, P.; Hong, R.; Mukai, Y.; Heo, S.; Chakravarti, D. Regulation of Histone Acetylation and Transcription by Nuclear Protein pp32, a Subunit of the INHAT Complex. J. Biol. Chem. 2002, 277, 14005–14010. [Google Scholar] [CrossRef]
- Zhang, Q.; Giebler, H.A.; Isaacson, M.K.; Nyborg, J.K. Eviction of linker histone H1 by NAP-family histone chaperones enhances activated transcription. Epigenetics Chromatin 2015, 8, 30. [Google Scholar] [CrossRef]
- Arede, L.; Pina, C. Buffering noise: KAT2A modular contributions to stabilization of transcription and cell identity in cancer and development. Exp. Hematol. 2020, 93, 25–37. [Google Scholar] [CrossRef]
- Orpinell, M.; Fournier, M.; Riss, A.; Nagy, Z.; Krebs, A.R.; Frontini, M.; Tora, L. The ATAC acetyl transferase complex controls mitotic progression by targeting non-histone substrates. EMBO J. 2010, 29, 2381–2394. [Google Scholar] [CrossRef]
- Aggarwal, S.; Nayek, A.; Pradhan, D.; Verma, R.; Yadav, M.; Ponnusamy, K.; Jain, A.K. dbGAPs: A comprehensive database of genes and genetic markers associated with psoriasis and its subtypes. Genomics 2018, 110, 240–247. [Google Scholar] [CrossRef]
- Ochieng, J.K.; Kundu, S.T.; Bajaj, R.; Rodriguez, B.L.; Fradette, J.J.; Gibbons, D.L. MBIP (MAP3K12 binding inhibitory protein) drives NSCLC metastasis by JNK-dependent activation of MMPs. Oncogene 2020, 39, 43. [Google Scholar] [CrossRef]
- Choudhary, V.; Olala, L.O.; Kagha, K.; Pan, Z.-Q.; Chen, X.; Yang, R.; Cline, A.; Helwa, I.; Marshall, L.; Kaddour-Djebbar, I.; et al. Regulation of the Glycerol Transporter, Aquaporin-3, by Histone Deacetylase-3 and p53 in Keratinocytes. J. Investig. Dermatol. 2017, 137, 1935–1944. [Google Scholar] [CrossRef]
- Lee, Y.; Je, Y.-J.; Lee, S.-S.; Li, Z.J.; Choi, D.-K.; Kwon, Y.-B.; Sohn, K.-C.; Im, M.; Seo, Y.J.; Lee, J.H. Changes in Transepidermal Water Loss and Skin Hydration according to Expression of Aquaporin-3 in Psoriasis. Ann. Dermatol. 2012, 24, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Thatikonda, S.; Pooladanda, V.; Sigalapalli, D.K.; Godugu, C. Piperlongumine regulates epigenetic modulation and alleviates psoriasis-like skin inflammation via inhibition of hyperproliferation and inflammation. Cell Death Dis. 2020, 11, 21. [Google Scholar] [CrossRef] [PubMed]
- Pray, B.A.; Youssef, Y.; Alinari, L. TBL1X: At the crossroads of transcriptional and posttranscriptional regulation. Exp. Hematol. 2022, 116, 18–25. [Google Scholar] [CrossRef]
- Fan, R.; Toubal, A.; Goñi, S.; Drareni, K.; Huang, Z.; Alzaid, F.; Ballaire, R.; Ancel, P.; Liang, N.; Damdimopoulos, A.; et al. Loss of the co-repressor GPS2 sensitizes macrophage activation upon metabolic stress induced by obesity and type 2 diabetes. Nat. Med. 2016, 22, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Corbalan-Garcia, S.; Yang, S.-S.; Degenhardt, K.R.; Bar-Sagi, D. Identification of the mitogen-activated protein kinase phosphorylation sites on human Sosl that regulate interaction with Grb2. Mol. Cell. Biol. 1996, 16, 5674–5682. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, Y.; Li, Y.; Yang, H.; Wang, H. Quantitative Analysis of the Global Proteome in Peripheral Blood Mononuclear Cells from Patients with New-Onset Psoriasis. Proteomics 2018, 18, 1800003. [Google Scholar] [CrossRef]
- Sawada, Y.; Nakatsuji, T.; Dokoshi, T.; Kulkarni, N.N.; Liggins, M.C.; Sen, G.; Gallo, R.L. Cutaneous innate immune tolerance is mediated by epigenetic control of MAP2K3 by HDAC8/9. Sci. Immunol. 2021, 6, eabe1935. [Google Scholar] [CrossRef]
- Sanford, J.A.; Zhang, L.-J.; Williams, M.R.; Gangoiti, J.A.; Huang, C.-M.; Gallo, R.L. Inhibition of HDAC8 and HDAC9 by microbial short-chain fatty acids breaks immune tolerance of the epidermis to TLR ligands. Sci. Immunol. 2016, 1, eaah4609. [Google Scholar] [CrossRef]
- Yang, W.; Feng, Y.; Zhou, J.; Cheung, O.K.-W.; Cao, J.; Wang, J.; Tang, W.; Tu, Y.; Xu, L.; Wu, F.; et al. A selective HDAC8 inhibitor potentiates antitumor immunity and efficacy of immune checkpoint blockade in hepatocellular carcinoma. Sci. Transl. Med. 2021, 13, eaaz6804. [Google Scholar] [CrossRef]
- Hait, N.C.; Allegood, J.; Maceyka, M.; Strub, G.M.; Harikumar, K.B.; Singh, S.K.; Luo, C.; Marmorstein, R.; Kordula, T.; Milstien, S.; et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 2009, 325, 1254–1257. [Google Scholar] [CrossRef]
- Shin, S.; Cho, K.; Hahn, S.; Lee, Y.; Kim, Y.; Woo, S.; Ryu, K.; Park, W.; Park, J. Inhibiting Sphingosine Kinase 2 Derived-sphingosine-1-phosphate Ameliorates Psoriasis-like Skin Disease via Blocking Th17 Differentiation of Naïve CD4 T Lymphocytes in Mice. Acta Derm.-Venereol. 2019, 99, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Dege, C.; Hagman, J. Mi-2/NuRD chromatin remodeling complexes regulate B and T-lymphocyte development and function. Immunol. Rev. 2014, 261, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Basta, J.; Rauchman, M. The nucleosome remodeling and deacetylase complex in development and disease. Transl. Res. 2015, 165, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Torchy, M.P.; Hamiche, A.; Klaholz, B.P. Structure and function insights into the NuRD chromatin remodeling complex. Cell. Mol. Life Sci. 2015, 72, 2491–2507. [Google Scholar] [CrossRef]
- Kashiwagi, M.; Morgan, B.A.; Georgopoulos, K. The chromatin remodeler Mi-2β is required for establishment of the basal epidermis and normal differentiation of its progeny. Development 2007, 134, 1571–1582. [Google Scholar] [CrossRef]
- Shibata, S.; Kashiwagi, M.; Morgan, B.A.; Georgopoulos, K. Functional interactions between Mi-2β and AP1 complexes control response and recovery from skin barrier disruption. J. Exp. Med. 2019, 217, e20182402. [Google Scholar] [CrossRef]
- Hosokawa, H.; Tanaka, T.; Suzuki, Y.; Iwamura, C.; Ohkubo, S.; Endoh, K.; Kato, M.; Endo, Y.; Onodera, A.; Tumes, D.J.; et al. Functionally distinct Gata3/Chd4 complexes coordinately establish T helper 2 (Th2) cell identity. Proc. Natl. Acad. Sci. USA 2013, 110, 4691–4696. [Google Scholar] [CrossRef]
- Shao, S.; Cao, H.; Wang, Z.; Zhou, D.; Wu, C.; Wang, S.; Xia, D.; Zhang, D. CHD4/NuRD complex regulates complement gene expression and correlates with CD8 T cell infiltration in human hepatocellular carcinoma. Clin. Epigenetics 2020, 12, 31. [Google Scholar] [CrossRef]
- Tomofuji, Y.; Takaba, H.; Suzuki, H.I.; Benlaribi, R.; Martinez, C.D.P.; Abe, Y.; Morishita, Y.; Okamura, T.; Taguchi, A.; Kodama, T.; et al. Chd4 choreographs self-antigen expression for central immune tolerance. Nat. Immunol. 2020, 21, 892–901. [Google Scholar] [CrossRef]
- Wang, Z.; Kang, J.; Deng, X.; Guo, B.; Wu, B.; Fan, Y. Knockdown of GATAD2A suppresses cell proliferation in thyroid cancer in vitro. Oncol. Rep. 2017, 37, 2147–2152. [Google Scholar] [CrossRef]
- Kempermann, G.; Gage, F.H.; Aigner, L.; Song, H.; Curtis, M.A.; Thuret, S.; Kuhn, H.G.; Jessberger, S.; Frankland, P.W.; Cameron, H.A.; et al. Human Adult Neurogenesis: Evidence and Remaining Questions. Cell Stem Cell 2018, 23, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Zemaitis, K.; Prosz, A.; Subramaniam, A.; Galeev, R.; Jassinskaja, M.; Hansson, J.; Larsson, J. 3020—RNA-INTERFERENCE SCREEN IDENTIFIES MTA1 as a Regulator of Human Hematopoietic Stem and Progenitor cells. Exp. Hematol. 2022, 111, S54–S55. [Google Scholar] [CrossRef]
- Pakala, S.B.; Reddy, S.D.N.; Bui-Nguyen, T.M.; Rangparia, S.S.; Bommana, A.; Kumar, R. MTA1 Coregulator Regulates LPS Response via MyD88-dependent Signaling. J. Biol. Chem. 2010, 285, 32787–32792. [Google Scholar] [CrossRef]
- Zhou, Y.; Nan, P.; Li, C.; Mo, H.; Zhang, Y.; Wang, H.; Xu, D.; Ma, F.; Qian, H. Upregulation of MTA1 in Colon Cancer Drives A CD8+ T Cell-Rich but Classical Macrophage-Lacking Immunosuppressive Tumor Microenvironment. Front. Oncol. 2022, 12, 825783. [Google Scholar] [CrossRef]
- Fang, Z.; Wang, X.; Sun, X.; Hu, W.; Miao, Q.R. The Role of Histone Protein Acetylation in Regulating Endothelial Function. Front. Cell Dev. Biol. 2021, 9, 672447. [Google Scholar] [CrossRef]
- Habryka, A.; Gogler-Pigłowska, A.; Sojka, D.; Kryj, M.; Krawczyk, Z.; Scieglinska, D. Cell type-dependent modulation of the gene encoding heat shock protein HSPA2 by hypoxia-inducible factor HIF-1: Down-regulation in keratinocytes and up-regulation in HeLa cells. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2015, 1849, 1155–1169. [Google Scholar] [CrossRef] [PubMed]
- Scieglinska, D.; Krawczyk, Z.; Sojka, D.R.; Gogler-Pigłowska, A. Heat shock proteins in the physiology and pathophysiology of epidermal keratinocytes. Cell Stress Chaperon 2019, 24, 1027–1044. [Google Scholar] [CrossRef]
- Gogler-Pigłowska, A.; Klarzyńska, K.; Sojka, D.R.; Habryka, A.; Głowala-Kosińska, M.; Herok, M.; Kryj, M.; Halczok, M.; Krawczyk, Z.; Scieglinska, D. Novel role for the testis-enriched HSPA2 protein in regulating epidermal keratinocyte differentiation. J. Cell. Physiol. 2017, 233, 2629–2644. [Google Scholar] [CrossRef]
- Kadamb, R.; Mittal, S.; Bansal, N.; Batra, H.; Saluja, D. Sin3: Insight into its transcription regulatory functions. Eur. J. Cell Biol. 2013, 92, 237–246. [Google Scholar] [CrossRef]
- Saunders, A.; Huang, X.; Fidalgo, M.; Reimer, M.H.; Faiola, F.; Ding, J.; Sánchez-Priego, C.; Guallar, D.; Sáenz, C.; Li, D.; et al. The SIN3A/HDAC Corepressor Complex Functionally Cooperates with NANOG to Promote Pluripotency. Cell Rep. 2017, 18, 1713–1726. [Google Scholar] [CrossRef]
- Cowley, S.M.; Iritani, B.M.; Mendrysa, S.M.; Xu, T.; Cheng, P.F.; Yada, J.; Liggitt, H.D.; Eisenman, R.N. The mSin3A Chromatin-Modifying Complex Is Essential for Embryogenesis and T-Cell Development. Mol. Cell. Biol. 2005, 25, 6990–7004. [Google Scholar] [CrossRef] [PubMed]
- Perucho, L.; Icardi, L.; Simone, E.D.; Basso, V.; Zornoza, A.V.; Lozano, T.; Prosper, F.; Lasarte, J.J.; Mondino, A. The transcriptional regulator Sin3A balances IL-17A and Foxp3 expression in primary CD4 T cells. EMBO Rep. 2022, 24, e55326. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, E.M.; Cox, C.L.; MacArthur, S.; Hussain, S.; Trotter, M.; Blanco, S.; Suraj, M.; Nichols, J.; Kübler, B.; Benitah, S.A.; et al. The opposing transcriptional functions of Sin3a and c-Myc are required to maintain tissue homeostasis. Nat. Cell Biol. 2011, 13, 1395–1405. [Google Scholar] [CrossRef] [PubMed]
- Luan, B.; Goodarzi, M.O.; Phillips, N.G.; Guo, X.; Chen, Y.-D.I.; Yao, J.; Allison, M.; Rotter, J.I.; Shaw, R.; Montminy, M. Leptin-Mediated Increases in Catecholamine Signaling Reduce Adipose Tissue Inflammation via Activation of Macrophage HDAC4. Cell Metab. 2014, 19, 1058–1065. [Google Scholar] [CrossRef]
- Yang, D.; Xiao, C.; Long, F.; Su, Z.; Jia, W.; Qin, M.; Huang, M.; Wu, W.; Suguro, R.; Liu, X.; et al. HDAC4 regulates vascular inflammation via activation of autophagy. Cardiovasc. Res. 2018, 114, 1016–1028. [Google Scholar] [CrossRef]
- Calenic, B.; Yaegaki, K.; Ishkitiev, N.; Kumazawa, Y.; Imai, T.; Tanaka, T. P53-Pathway activity and apoptosis in hydrogen sulfide-exposed stem cells separated from human gingival epithelium. J. Periodontal Res. 2012, 48, 322–330. [Google Scholar] [CrossRef]
- Xiao, H.; Jiao, J.; Wang, L.; O’Brien, S.; Newick, K.; Wang, L.-C.S.; Falkensammer, E.; Liu, Y.; Han, R.; Kapoor, V.; et al. HDAC5 controls the functions of Foxp3+T-regulatory and CD8+T cells. Int. J. Cancer 2016, 138, 2477–2486. [Google Scholar] [CrossRef]
- Wang, W.; Ha, C.H.; Jhun, B.S.; Wong, C.; Jain, M.K.; Jin, Z.-G. Fluid shear stress stimulates phosphorylation-dependent nuclear export of HDAC5 and mediates expression of KLF2 and eNOS. Blood 2010, 115, 2971–2979. [Google Scholar] [CrossRef]
- Gao, Y.-S.; Hubbert, C.C.; Lu, J.; Lee, Y.-S.; Lee, J.-Y.; Yao, T.-P. Histone deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Mol. Cell. Biol. 2007, 27, 8637–8647. [Google Scholar] [CrossRef]
- Qin, Y.-M.; Li, P.; Mu, X.-P.; Li, Z.-M.; Sun, C.; Xue, W.-L.; Sun, J.; Bai, J.-J.; Zhu, Y.-C.; Wang, M.-J. Histone deacetylase 6 promotes skin wound healing by regulating fibroblast migration and differentiation in aged mice. Sheng Li Xue Bao 2022, 74, 979–992. [Google Scholar]
- Cabrero, J.R.; Serrador, J.M.; Barreiro, O.; Mittelbrunn, M.; Naranjo-Suárez, S.; Martín-Cófreces, N.; Vicente-Manzanares, M.; Mazitschek, R.; Bradner, J.E.; Ávila, J.; et al. Lymphocyte chemotaxis is regulated by histone deacetylase 6, independently of its deacetylase activity. Mol. Biol. Cell 2006, 17, 3435–3445. [Google Scholar] [CrossRef] [PubMed]
- Serrador, J.M.; Cabrero, J.R.; Sancho, D.; Mittelbrunn, M.; Urzainqui, A.; Sánchez-Madrid, F. hdac6 deacetylase activity links the tubulin cytoskeleton with immune synapse organization. Immunity 2004, 20, 417–428. [Google Scholar] [CrossRef]
- Fan, X.; Yan, K.; Meng, Q.; Sun, R.; Yang, X.; Yuan, D.; Li, F.; Deng, H. Abnormal expression of SIRTs in psoriasis: Decreased expression of SIRT 1-5 and increased expression of SIRT 6 and 7. Int. J. Mol. Med. 2019, 44, 157–171. [Google Scholar] [CrossRef]
- Wang, C.; He, D.; Shi, C. SIRT5 reduces the inflammatory response and barrier dysfunction in IL-17A-induced epidermal keratinocytes. Allergol. Immunopathol. 2023, 51, 30–36. [Google Scholar] [CrossRef]
- Michishita, E.; McCord, R.A.; Boxer, L.D.; Barber, M.F.; Hong, T.; Gozani, O.; Chua, K.F. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 2009, 8, 2664–2666. [Google Scholar] [CrossRef]
- Toiber, D.; Erdel, F.; Bouazoune, K.; Silberman, D.M.; Zhong, L.; Mulligan, P.; Sebastian, C.; Cosentino, C.; Martinez-Pastor, B.; Giacosa, S.; et al. SIRT6 recruits SNF2H to dna break sites, preventing genomic instability through chromatin remodeling. Mol. Cell 2013, 51, 454–468. [Google Scholar] [CrossRef]
- Koo, J.-H.; Jang, H.-Y.; Lee, Y.; Moon, Y.J.; Bae, E.J.; Yun, S.-K.; Park, B.-H. Myeloid cell-specific sirtuin 6 deficiency delays wound healing in mice by modulating inflammation and macrophage phenotypes. Exp. Mol. Med. 2019, 51, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, H.; Guo, Q.; Zhang, L.; Zhu, J.; Ji, J. Sirtuin6 inhibits c-triggered inflammation through TLR4 abrogation regulated by ROS and TRPV1/CGRP. J. Cell. Biochem. 2018, 119, 9141–9153. [Google Scholar] [CrossRef] [PubMed]
- Lasigliè, D.; Boero, S.; Bauer, I.; Morando, S.; Damonte, P.; Cea, M.; Monacelli, F.; Odetti, P.; Ballestrero, A.; Uccelli, A.; et al. Sirt6 regulates dendritic cell differentiation, maturation, and function. Aging 2016, 8, 34–47. [Google Scholar] [CrossRef]
- Yanginlar, C.; Logie, C. HDAC11 is a regulator of diverse immune functions. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2018, 1861, 54–59. [Google Scholar] [CrossRef]
- Romhányi, D.; Szabó, K.; Kemény, L.; Sebestyén, E.; Groma, G. Transcriptional Analysis-Based Alterations Affecting Neuritogenesis of the Peripheral Nervous System in Psoriasis. Life 2022, 12, 111. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Tsoi, L.C.; Swindell, W.R.; Gudjonsson, J.E.; Tejasvi, T.; Johnston, A.; Ding, J.; Stuart, P.E.; Xing, X.; Kochkodan, J.J.; et al. Transcriptome Analysis of Psoriasis in a Large Case–Control Sample: RNA-Seq Provides Insights into Disease Mechanisms. J. Investig. Dermatol. 2014, 134, 1828–1838. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Tsoi, L.C.; Xing, X.A.; Beamer, M.; Swindell, W.R.; Sarkar, M.K.; Berthier, C.C.E.; Stuart, P.; Harms, P.W.; Nair, R.P.; et al. A gene network regulated by the transcription factor VGLL3 as a promoter of sex-biased autoimmune diseases. Nat. Immunol. 2017, 18, 152–160. [Google Scholar] [CrossRef]
- Tsoi, L.C.; Iyer, M.K.E.; Stuart, P.; Swindell, W.R.E.; Gudjonsson, J.; Tejasvi, T.; Sarkar, M.K.; Li, B.; Ding, J.; Voorhees, J.J.; et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015, 16, 24. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2016, 4, 1521. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Schwämmle, V.; León, I.R.; Jensen, O.N. Assessment and Improvement of Statistical Tools for Comparative Proteomics Analysis of Sparse Data Sets with Few Experimental Replicates. J. Proteome Res. 2013, 12, 3874–3883. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Liu, R.; Holik, A.Z.; Su, S.; Jansz, N.; Chen, K.; Leong, H.S.; Blewitt, M.E.; Asselin-Labat, M.-L.; Smyth, G.K.; Ritchie, M.E. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015, 43, e97. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yang, Y.; Zhou, N.; Tang, K.; Lau, W.B.; Lau, B.; Wang, W.; Xu, L.; Yang, Z.; Huang, S.; et al. Epigenetics in ovarian cancer: Premise, properties, and perspectives. Mol. Cancer 2018, 17, 109. [Google Scholar] [CrossRef] [PubMed]
- Di Cerbo, V.; Schneider, R. Cancers with wrong HATs: The impact of acetylation. Briefings Funct. Genom. 2013, 12, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Lamaa, A.; Humbert, J.; Aguirrebengoa, M.; Cheng, X.; Nicolas, E.; Côté, J.; Trouche, D. Integrated analysis of H2A.Z isoforms function reveals a complex interplay in gene regulation. eLife 2020, 9, e53375. [Google Scholar] [CrossRef]
- Filipescu, D.; Szenker, E.; Almouzni, G. Developmental roles of histone H3 variants and their chaperones. Trends Genet. 2013, 29, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Caputo, V.; Strafella, C.; Cosio, T.; Lanna, C.; Campione, E.; Novelli, G.; Giardina, E.; Cascella, R. Pharmacogenomics: An Update on Biologics and Small-Molecule Drugs in the Treatment of Psoriasis. Genes 2021, 12, 1398. [Google Scholar] [CrossRef]
- Glauben, R.; Sonnenberg, E.; Wetzel, M.; Mascagni, P.; Siegmund, B. Histone Deacetylase Inhibitors Modulate Interleukin 6-dependent CD4+ T Cell Polarization in Vitro and in Vivo. J. Biol. Chem. 2014, 289, 6142–6151. [Google Scholar] [CrossRef]
- Yokoyama, S.; Feige, E.; Poling, L.L.; Levy, C.; Widlund, H.R.; Khaled, M.; Kung, A.L.; Fisher, D.E. Pharmacologic suppression of MITF expression via HDAC inhibitors in the melanocyte lineage. Pigment. Cell Melanoma Res. 2008, 21, 457–463. [Google Scholar] [CrossRef]
- Samuelov, L.; Bochner, R.; Magal, L.; Malovitski, K.; Sagiv, N.; Nousbeck, J.; Keren, A.; Fuchs-Telem, D.; Sarig, O.; Gilhar, A.; et al. Vorinostat, a histone deacetylase inhibitor, as a potential novel treatment for psoriasis. Exp. Dermatol. 2022, 31, 567–576. [Google Scholar] [CrossRef]
- Jiang, Y.; Lu, S.; Lai, Y.; Wang, L. Topical histone deacetylase 1 inhibitor Entinostat ameliorates psoriasiform dermatitis through suppression of IL-17A response. J. Dermatol. Sci. 2023, 110, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Hammitzsch, A.; de Wit, J.; Ridley, A.; Al-Mossawi, M.; Bowness, P. AB0022 Comparison of in Vitro Effects of Kinase and Epigenetic Inhibitors on TH17 Responses in Inflammatory Arthritis. Ann. Rheum. Dis. 2014, 73, 811. [Google Scholar] [CrossRef]
- Hammitzsch, A.; Tallant, C.; Fedorov, O.; O’mahony, A.; Brennan, P.E.; Hay, D.A.; Martinez, F.O.; Al-Mossawi, M.H.; de Wit, J.; Vecellio, M.; et al. CBP30, a selective CBP/p300 bromodomain inhibitor, suppresses human Th17 responses. Proc. Natl. Acad. Sci. USA 2015, 112, 10768–10773. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).