Analyzing Current Trends and Possible Strategies to Improve Sucrose Isomerases’ Thermostability
Abstract
1. Introduction
2. Sucrose Isomerases, Structure and Reaction Mechanism
3. Thermolability of Sucrose Isomerases
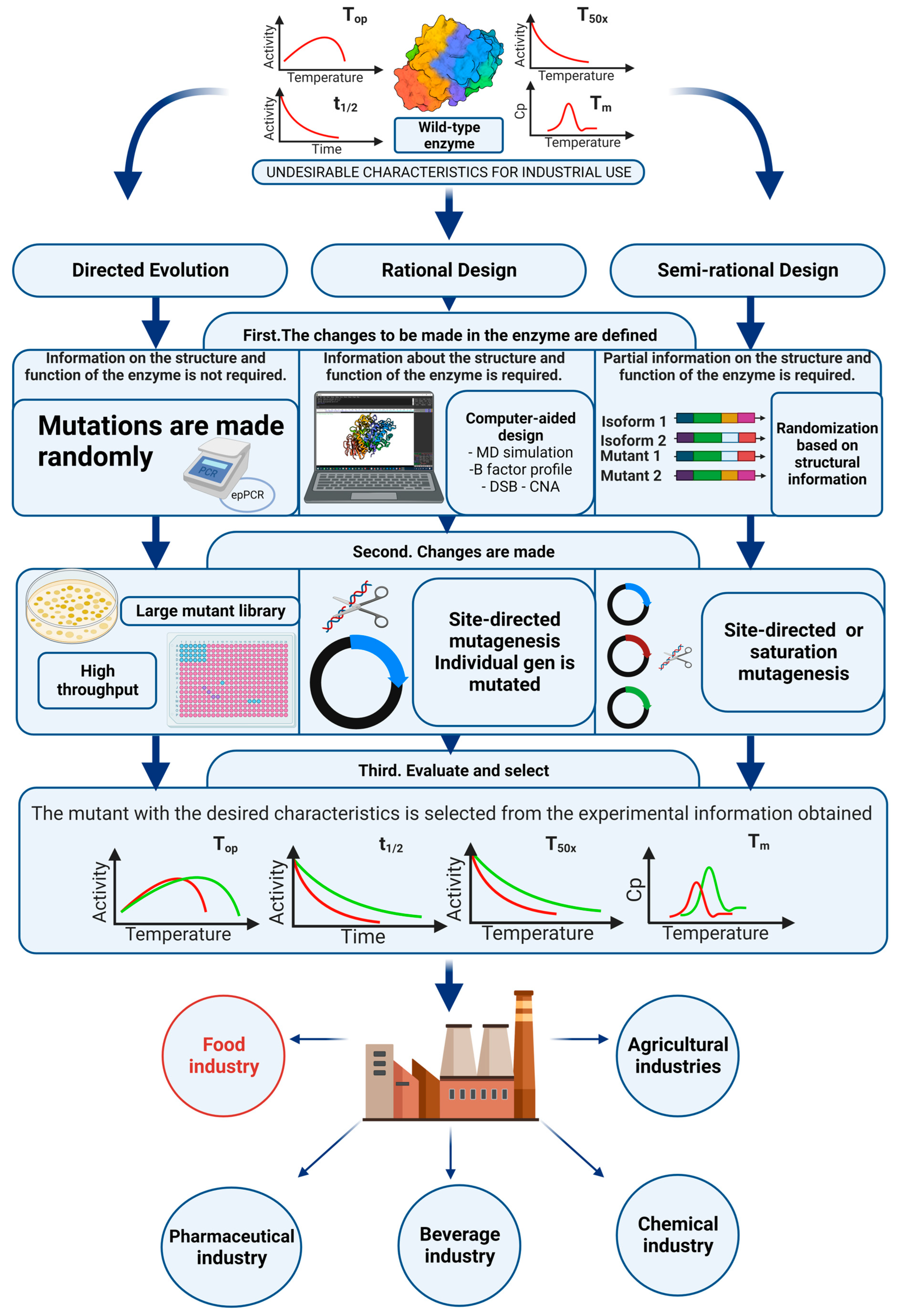
4. Protein Engineering, Thermostabilization of Sucrose Isomerases
4.1. Directed Evolution
4.2. Rational Design
4.3. Semi-Rational Design
4.4. Characterization of Thermostability
4.5. Thermostabilization of Sucrose Isomerases
5. Glycosylation of Sucrose Isomerases
6. Future Perspectives
6.1. Thermostable Sucrose Isomerases Based on Homology Models and Chimerization
6.2. Thermostable Sucrose Isomerases Based on Improving the Entropy of the Folded State
6.3. Thermostable Sucrose Isomerases, Other Alternative Strategies
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, A.; Gudiukaite, R.; Gricajeva, A.; Sadauskas, M.; Malunavicius, V.; Kamyab, H.; Sharma, S.; Sharma, T.; Pant, D. Microbial lipolytic enzymes–promising energy-efficient biocatalysts in bioremediation. Energy 2020, 192, 116674. [Google Scholar] [CrossRef]
- Lu, L.; Guo, L.; Wang, K.; Liu, Y.; Xiao, M. β-Galactosidases: A great tool for synthesizing galactose-containing carbohydrates. Biotechnol. Adv. 2020, 39, 107465. [Google Scholar] [CrossRef]
- Bornscheuer, U.T.; Huisman, G.W.; Kazlauskas, R.J.; Lutz, S.; Moore, J.C.; Robins, K. Engineering the third wave of biocatalysis. Nature 2012, 485, 185–194. [Google Scholar] [CrossRef]
- Clouthier, C.M.; Pelletier, J.N. Expanding the organic toolbox: A guide to integrating biocatalysis in synthesis. Chem. Soc. Rev. 2012, 41, 1585–1605. [Google Scholar] [CrossRef]
- Sandoval, B.A.; Hyster, T.K. Emerging strategies for expanding the toolbox of enzymes in biocatalysis. Curr. Opin. Chem. Biol. 2020, 55, 45–51. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Brady, D. Broadening the Scope of Biocatalysis in Sustainable Organic Synthesis. ChemSusChem 2019, 12, 2859–2881. [Google Scholar] [CrossRef]
- Breuer, M.; Ditrich, K.; Habicher, T.; Hauer, B.; Keßeler, M.; Stürmer, R.; Zelinski, T. Industrielle Verfahren zur Herstellung von optisch aktiven Zwischenprodukten. Angew. Chem. 2004, 116, 806–843. [Google Scholar] [CrossRef]
- Xue, Y.-P.; Cao, C.-H.; Zheng, Y.-G. Enzymatic asymmetric synthesis of chiral amino acids. Chem. Soc. Rev. 2018, 47, 1516–1561. [Google Scholar] [CrossRef]
- Thompson, M.P.; Peñafiel, I.; Cosgrove, S.C.; Turner, N.J. Biocatalysis Using Immobilized Enzymes in Continuous Flow for the Synthesis of Fine Chemicals. Org. Process. Res. Dev. 2018, 23, 9–18. [Google Scholar] [CrossRef]
- Bezbradica, D.; Crovic, M.; JTanaskovic, S.; Lukovic, N.; Carevic, M.; Milivojevic, A.; Knezevic-Jugovic, Z. Enzymatic syntheses of esters-Green chemistry for valuable food, fuel and fine chemicals. Curr. Org. Chem. 2017, 21, 104–138. [Google Scholar] [CrossRef]
- Schoemaker, H.E.; Mink, D.; Wubbolts, M.G. Dispelling the Myths--Biocatalysis in Industrial Synthesis. Science 2003, 299, 1694–1697. [Google Scholar] [CrossRef]
- Serra, S.; Fuganti, C.; Brenna, E. Biocatalytic preparation of natural flavours and fragrances. Trends Biotechnol. 2005, 23, 193–198. [Google Scholar] [CrossRef]
- Aravindan, R.; Anbumathi, P.; Viruthagiri, T. Lipase Applications in Food Industry. 2007. Available online: https://nopr.niscpr.res.in/handle/123456789/3016 (accessed on 21 November 2022).
- Tan, T.; Lu, J.; Nie, K.; Deng, L.; Wang, F. Biodiesel production with immobilized lipase: A review. Biotechnol. Adv. 2010, 28, 628–634. [Google Scholar] [CrossRef]
- Gog, A.; Roman, M.; Toşa, M.; Paizs, C.; Irimie, F.D. Biodiesel production using enzymatic transesterification–Current state and perspectives. Renew. Energy 2012, 39, 10–16. [Google Scholar] [CrossRef]
- Volpato, G.; CRodrigues, R.; Fernandez-Lafuente, R. Use of enzymes in the production of semi-synthetic penicillins and cephalosporins: Drawbacks and perspectives. Curr. Med. Chem. 2010, 17, 3855–3873. [Google Scholar] [CrossRef]
- Fei, Q.; Guarnieri, M.T.; Tao, L.; Laurens, L.M.; Dowe, N.; Pienkos, P.T. Bioconversion of natural gas to liquid fuel: Opportunities and challenges. Biotechnol. Adv. 2014, 32, 596–614. [Google Scholar] [CrossRef]
- Basso, A.; Serban, S. Industrial applications of immobilized enzymes—A review. Mol. Catal. 2019, 479, 110607. [Google Scholar] [CrossRef]
- de Souza, W.F.C.; Almeida, F.L.C.; de Castro, R.J.S.; Sato, H.H. Isomaltulose: From origin to application and its beneficial properties–A bibliometric approach. Food Res. Int. 2022, 155, 111061. [Google Scholar] [CrossRef]
- Liu, L.; Bilal, M.; Luo, H.; Zhao, Y.; Duan, X. Studies on Biological Production of Isomaltulose Using Sucrose Isomerase: Current Status and Future Perspectives. Catal. Lett. 2020, 151, 1868–1881. [Google Scholar] [CrossRef]
- Shyam, S.; Ramadas, A.; Chang, S.K. Isomaltulose: Recent evidence for health benefits. J. Funct. Foods 2018, 48, 173–178. [Google Scholar] [CrossRef]
- Sawale, P.D.; Shendurse, A.M.; Mohan, M.S.; Patil, G.R. Isomaltulose (palatinose)—An emerging carbohydrate. Food Biosci. 2017, 18, 46–52. [Google Scholar] [CrossRef]
- Schiweck, H.; Munir, M.; Rapp, K.M.; Schneider, B.; Vogel, M. New developments in the use of sucrose as an industrial bulk chemical. Zuckerindustrie 1990, 115, 555–565. [Google Scholar]
- Holub, I.; Gostner, A.; Theis, S.; Nosek, L.; Kudlich, T.; Melcher, R.; Scheppach, W. Novel findings on the metabolic effects of the low glycaemic carbohydrate isomaltulose (Palatinose™). Br. J. Nutr. 2010, 103, 1730–1737. [Google Scholar] [CrossRef]
- Sentko, A.B.J. Isomaltulose. In Alternative Sweeteners; O’Brien-Nabors, L., Ed.; CRC Press: Boca Raton, FL, USA, 2011. [Google Scholar]
- Maresch, C.C.; Petry, S.F.; Theis, S.; Bosy-Westphal, A.; Linn, T. Low Glycemic Index Prototype Isomaltulose—Update of Clinical Trials. Nutrients 2017, 9, 381. [Google Scholar] [CrossRef]
- Kawai, K.; Okuda, Y.; Yamashita, K. Changes in blood glucose and insulin after an oral palatinose administration in normal subjects. Endocrinol. Jpn. 1985, 32, 933–936. [Google Scholar] [CrossRef]
- Lina, B.A.R.; Jonker, D.; Kozianowski, G. Isomaltulose (Palatinose®): A review of biological and toxicological studies. Food Chem. Toxicol. 2002, 40, 1375–1381. [Google Scholar] [CrossRef]
- Mu, W.; Li, W.; Wang, X.; Zhang, T.; Jiang, B. Current studies on sucrose isomerase and biological isomaltulose production using sucrose isomerase. Appl. Microbiol. Biotechnol. 2014, 98, 6569–6582. [Google Scholar] [CrossRef]
- Häberer, D.; Thibault, L.; Langhans, W.; Geary, N. Beneficial Effects on Glucose Metabolism of Chronic Feeding of Isomaltulose versus Sucrose in Rats. Ann. Nutr. Metab. 2009, 54, 75–82. [Google Scholar] [CrossRef]
- Kawai, K.; Yoshikawa, H.; Murayama, Y.; Okuda, Y.; Yamashita, K. Usefulness of Palatinose as a Caloric Sweetener for Diabetic Patients. Horm. Metab. Res. 1989, 21, 338–340. [Google Scholar] [CrossRef]
- König, D.; Theis, S.; Kozianowski, G.; Berg, A. Postprandial substrate use in overweight subjects with the metabolic syndrome after isomaltulose (Palatinose™) ingestion. Nutrition 2012, 28, 651–656. [Google Scholar] [CrossRef]
- Achten, J.; Jentjens, R.L.; Brouns, F.; Jeukendrup, A.E. Exogenous Oxidation of Isomaltulose Is Lower than That of Sucrose during Exercise in Men. J. Nutr. 2007, 137, 1143–1148. [Google Scholar] [CrossRef]
- van Can, J.G.; van Loon, L.J.; Brouns, F.; Blaak, E.E. Reduced glycaemic and insulinaemic responses following trehalose and isomaltulose ingestion: Implications for postprandial substrate use in impaired glucose-tolerant subjects. Br. J. Nutr. 2012, 108, 1210–1217. [Google Scholar] [CrossRef]
- Siddiqui, I.R.; Furgala, B. Isolation and Characterization of Oligosaccharides from Honey. Part I. Disaccharides. J. Apic. Res. 1967, 6, 139–145. [Google Scholar] [CrossRef]
- Eggleston, G.; Grisham, M. Oligosaccharides in Cane and Their Formation on Cane Deterioration; ACS Symposium Series; ACS Publications: Washington, DC, USA, 2003; pp. 211–234. [Google Scholar]
- Kawaguti, H.Y.; Sato, H.H. Palatinose production by free and Ca-alginate gel immobilized cells of Erwinia sp. Biochem. Eng. J. 2007, 36, 202–208. [Google Scholar] [CrossRef]
- Lee, G.-Y.; Jung, J.-H.; Seo, D.-H.; Hansin, J.; Ha, S.-J.; Cha, J.; Kim, Y.-S.; Park, C.-S. Isomaltulose production via yeast surface display of sucrose isomerase from Enterobacter sp. FMB-1 on Saccharomyces cerevisiae. Bioresour. Technol. 2011, 102, 9179–9184. [Google Scholar] [CrossRef]
- Li, S.; Cai, H.; Qing, Y.; Ren, B.; Xu, H.; Zhu, H.; Yao, J. Cloning and Characterization of a Sucrose Isomerase from Erwinia rhapontici NX-5 for Isomaltulose Hyperproduction. Appl. Biochem. Biotechnol. 2011, 163, 52–63. [Google Scholar] [CrossRef]
- Kawaguti, H.Y.; Celestino, M.; Moraes, A.L.; Yim, D.K.; Yamamoto, L.K.; Sato, H.H. Characterization of a glucosyltransferase from Erwinia sp. D12 and the conversion of sucrose into isomaltulose by immobilized cells. Biochem. Eng. J. 2010, 48, 211–217. [Google Scholar] [CrossRef]
- Véronèse, T.; Perlot, P. Mechanism of sucrose conversion by the sucrose isomerase of Serratia plymuthica ATCC 15928. Enzym. Microb. Technol. 1999, 24, 263–269. [Google Scholar] [CrossRef]
- Ravaud, S.; Watzlawick, H.; Haser, R.; Mattes, R.; Aghajari, N. Overexpression, purification, crystallization and preliminary diffraction studies of the Protaminobacter rubrum sucrose isomerase SmuA. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006, 62, 74–76. [Google Scholar] [CrossRef]
- Wu, L.; Birch, R. Characterization of Pantoea dispersa UQ68J: Producer of a highly efficient sucrose isomerase for isomaltulose biosynthesis. J. Appl. Microbiol. 2004, 97, 93–103. [Google Scholar] [CrossRef]
- Huang, J.-H.; Hsu, L.-H.; Su, Y.-C. Conversion of sucrose to isomaltulose by Klebsiella planticola CCRC 19112. J. Ind. Microbiol. Biotechnol. 1998, 21, 22–27. [Google Scholar] [CrossRef]
- Cho, M.-H.; Park, S.-E.; Lim, J.K.; Kim, J.-S.; Kim, J.H.; Kwon, D.Y.; Park, C.-S. Conversion of sucrose into isomaltulose by Enterobacter sp. FMB1, an isomaltulose-producing microorganism isolated from traditional Korean food. Biotechnol. Lett. 2007, 29, 453–458. [Google Scholar] [CrossRef]
- Nagai-Miyata, Y.; Tsuyuki, K.-I.; Sugitani, T.; Ebashi, T.; Nakajima, Y. Isolation and Characterization of a Trehalulose-producing Strain of Agrobacterium. Biosci. Biotechnol. Biochem. 1993, 57, 2049–2053. [Google Scholar] [CrossRef]
- Miyata, Y.; Sugitani, T.; Tsuyuki, K.-I.; Ebashi, T.; Nakajima, Y. Isolation and Characterization of Pseudomonas mesoacidophila Producing Trehalulose. Biosci. Biotechnol. Biochem. 1992, 56, 1680–1681. [Google Scholar] [CrossRef]
- Liu, L.; Yu, S.; Zhao, W. A Novel Sucrose Isomerase Producing Isomaltulose from Raoultella terrigena. Appl. Sci. 2021, 11, 5521. [Google Scholar] [CrossRef]
- Aroonnual, A.; Nihira, T.; Seki, T.; Panbangred, W. Role of several key residues in the catalytic activity of sucrose isomerase from Klebsiella pneumoniae NK33-98-8. Enzym. Microb. Technol. 2007, 40, 1221–1227. [Google Scholar] [CrossRef]
- Cha, J.; Jung, J.; Park, S.; Cho, M.; Seo, D.; Ha, S.; Yoon, J.; Lee, O.; Kim, Y.; Park, C. Molecular cloning and functional characterization of a sucrose isomerase (isomaltulose synthase) gene from Enterobacter sp. FMB-1. J. Appl. Microbiol. 2009, 107, 1119–1130. [Google Scholar] [CrossRef]
- Wu, L.; Birch, R.G. Characterization of the Highly Efficient Sucrose Isomerase from Pantoea dispersa UQ68J and Cloning of the Sucrose Isomerase Gene. Appl. Environ. Microbiol. 2005, 71, 1581–1590. [Google Scholar] [CrossRef]
- Watzlawick, H.; Mattes, R. Gene cloning, protein characterization, and alteration of product selectivity for the trehalulose hydrolase and trehalulose synthase from “Pseudomonas mesoacidophila” MX-45. Appl. Environ. Microbiol. 2009, 75, 7026–7036. [Google Scholar] [CrossRef]
- Börnke, F.; Hajirezaei, M.; Sonnewald, U. Cloning and characterization of the gene cluster for palatinose metabolism from the phytopathogenic bacteriumErwinia rhapontici. J. Bacteriol. 2001, 183, 2425–2430. [Google Scholar] [CrossRef]
- Sardiña-Peña, A.J.; Ballinas-Casarrubias, L.; Siqueiros-Cendón, T.S.; Espinoza-Sánchez, E.A.; Flores-Holguín, N.R.; Iglesias-Figueroa, B.F.; Rascón-Cruz, Q. Thermostability improvement of sucrose isomerase PalI NX-5: A comprehensive strategy. Biotechnol. Lett. 2023, 45, 885–904. [Google Scholar] [CrossRef]
- Duan, X.; Cheng, S.; Ai, Y.; Wu, J. Enhancing the thermostability of Serratia plymuthica sucrose isomerase using B-factor-directed mutagenesis. PLoS ONE 2016, 11, e0149208. [Google Scholar] [CrossRef]
- Ravaud, S.; Robert, X.; Watzlawick, H.; Haser, R.; Mattes, R.; Aghajari, N. Trehalulose Synthase Native and Carbohydrate Complexed Structures Provide Insights into Sucrose Isomerization. J. Biol. Chem. 2007, 282, 28126–28136. [Google Scholar] [CrossRef]
- Ravaud, S.; Watzlawick, H.; Haser, R.; Mattes, R.; Aghajari, N. Expression, purification, crystallization and preliminary X-ray crystallographic studies of the trehalulose synthase MutB from Pseudomonas mesoacidophila MX-45. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2005, 61, 100–103. [Google Scholar] [CrossRef]
- Lipski, A.; Watzlawick, H.; Ravaud, S.; Robert, X.; Rhimi, M.; Haser, R.; Mattes, R.; Aghajari, N. Mutations inducing an active-site aperture in Rhizobium sp. sucrose isomerase confer hydrolytic activity. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 298–307. [Google Scholar] [CrossRef]
- Xu, Z.; Li, S.; Li, J.; Li, Y.; Feng, X.; Wang, R.; Xu, H.; Zhou, J. The Structural Basis of Erwinia rhapontici Isomaltulose Synthase. PLoS ONE 2013, 8, e74788. [Google Scholar] [CrossRef]
- Ravaud, S.; Robert, X.; Watzlawick, H.; Haser, R.; Mattes, R.; Aghajari, N. Structural determinants of product specificity of sucrose isomerases. FEBS Lett. 2009, 583, 1964–1968. [Google Scholar] [CrossRef]
- Zhang, D.; Li, N.; Lok, S.-M.; Zhang, L.-H.; Swaminathan, K. Isomaltulose synthase (PalI) of Klebsiella sp. LX3: Crystal structure and implication of mechanism. J. Biol. Chem. 2003, 278, 35428–35434. [Google Scholar] [CrossRef]
- Janeček, Š.; Zámocká, B. A new GH13 subfamily represented by the α-amylase from the halophilic archaeon Haloarcula hispanica. Extremophiles 2020, 24, 207–217. [Google Scholar] [CrossRef]
- Chen, L.; Qu, Z.; Yu, W.; Zheng, L.; Qiao, H.; Wang, D.; Wei, B.; Zhao, Z. Comparative genomic and transcriptome analysis of Bacillus velezensis CL-4 fermented corn germ meal. AMB Express 2023, 13, 1–12. [Google Scholar] [CrossRef]
- Han, S.-R.; Kim, D.W.; Kim, B.; Chi, Y.M.; Kang, S.; Park, H.; Jung, S.-H.; Lee, J.H.; Oh, T.-J. Complete genome sequencing of Shigella sp. PAMC 28760: Identification of CAZyme genes and analysis of their potential role in glycogen metabolism for cold survival adaptation. Microb. Pathog. 2019, 137, 103759. [Google Scholar] [CrossRef]
- Plaza-Vinuesa, L.; Hernandez-Hernandez, O.; Moreno, F.J.; Rivas, B.d.L.; Muñoz, R. Unravelling the diversity of glycoside hydrolase family 13 α-amylases from Lactobacillus plantarum WCFS1. Microb. Cell Factories 2019, 18, 1–11. [Google Scholar] [CrossRef]
- Franceus, J.; Desmet, T. Sucrose Phosphorylase and Related Enzymes in Glycoside Hydrolase Family 13: Discovery, Application and Engineering. Int. J. Mol. Sci. 2020, 21, 2526. [Google Scholar] [CrossRef]
- Yang, W.; Su, L.; Wang, L.; Wu, J.; Chen, S. Alpha-glucanotransferase from the glycoside hydrolase family synthesizes α(1–6)-linked products from starch: Features and synthesis pathways of the products. Trends Food Sci. Technol. 2022, 128, 160–172. [Google Scholar] [CrossRef]
- Samanta, S. Structural and Catalytical Features of Different Amylases and their Potential Applications. Jordan J. Biol. Sci. 2022, 15, 311–337. [Google Scholar]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Janeček, Š.; Kuchtová, A.; Petrovičová, S. A novel GH13 subfamily of α-amylases with a pair of tryptophans in the helix α3 of the catalytic TIM-barrel, the LPDlx signature in the conserved sequence region V and a conserved aromatic motif at the C-terminus. Biologia 2015, 70, 1284–1294. [Google Scholar] [CrossRef]
- Sarian, F.D.; Janeček, Š.; Pijning, T.; Ihsanawati; Nurachman, Z.; Radjasa, O.K.; Dijkhuizen, L.; Natalia, D.; van der Maarel, M.J.E.C. A new group of glycoside hydrolase family 13 α-amylases with an aberrant catalytic triad. Sci. Rep. 2017, 7, srep44230. [Google Scholar] [CrossRef] [PubMed]
- Stam, M.R.; Danchin, E.G.; Rancurel, C.; Coutinho, P.M.; Henrissat, B. Dividing the large glycoside hydrolase family 13 into subfamilies: Towards improved functional annotations of α-amylase-related proteins. Protein Eng. Des. Sel. 2006, 19, 555–562. [Google Scholar] [CrossRef]
- Janíčková, Z.; Janeček, Š. Fungal α-amylases from three GH13 subfamilies: Their sequence-structural features and evolutionary relationships. Int. J. Biol. Macromol. 2020, 159, 763–772. [Google Scholar] [CrossRef]
- Marengo, M.; Pezzilli, D.; Gianquinto, E.; Fissore, A.; Oliaro-Bosso, S.; Sgorbini, B.; Spyrakis, F.; Adinolfi, S. Evaluation of Porcine and Aspergillus oryzae α-Amylases as Possible Model for the Human Enzyme. Processes 2022, 10, 780. [Google Scholar] [CrossRef]
- Watanabe, K.; Hata, Y.; Kizaki, H.; Katsube, Y.; Suzuki, Y. The refined crystal structure of Bacillus cereus oligo-1, 6-glucosidase at 2.0 Å resolution: Structural characterization of proline-substitution sites for protein thermostabilization. J. Mol. Biol. 1997, 269, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Møller, M.S.; Fredslund, F.; Majumder, A.; Nakai, H.; Poulsen, J.-C.N.; Lo Leggio, L.; Svensson, B.; Abou Hachem, M. Enzymology and structure of the GH13_31 glucan 1, 6-α-glucosidase that confers isomaltooligosaccharide utilization in the probiotic Lactobacillus acidophilus NCFM. J. Bacteriol. 2012, 194, 4249–4259. [Google Scholar] [CrossRef]
- Lin, M.-G.; Chi, M.-C.; Naveen, V.; Li, Y.-C.; Lin, L.-L.; Hsiao, C.-D. Bacillus licheniformis trehalose-6-phosphate hydrolase structures suggest keys to substrate specificity. Acta Crystallogr. Sect. D Struct. Biol. 2016, 72, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, R.; Arimura, Y.; Ishii, Y.; Kirimura, K. Crystal structure of α-glucosyl transfer enzyme XgtA from Xanthomonas campestris WU-9701. Biochem. Biophys. Res. Commun. 2020, 526, 580–585. [Google Scholar] [CrossRef]
- Auiewiriyanukul, W.; Saburi, W.; Kato, K.; Yao, M.; Mori, H. Function and structure of GH 13_31 α-glucosidase with high α-(1→4)-glucosidic linkage specificity and transglucosylation activity. FEBS Lett. 2018, 592, 2268–2281. [Google Scholar] [CrossRef]
- Cockburn, D.W.; Cerqueira, F.M.; Bahr, C.; Koropatkin, N.M. The structures of the GH13_36 amylases from Eubacterium rectale and Ruminococcus bromii reveal subsite architectures that favor maltose production. Amylase 2020, 4, 24–44. [Google Scholar] [CrossRef]
- Janeček, Š. How many conserved sequence regions are there in the α-amylase family. Biologia 2002, 57 (Suppl. 11), 29–41. [Google Scholar]
- Janeček, Š.; Svensson, B.; MacGregor, E.A. α-Amylase: An enzyme specificity found in various families of glycoside hydrolases. Cell. Mol. Life Sci. 2013, 71, 1149–1170. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Zhang, D.; Li, X.; Zhang, L.-H. Isomaltulose Synthase from Klebsiella sp. Strain LX3: Gene Cloning and Characterization and Engineering of Thermostability. Appl. Environ. Microbiol. 2002, 68, 2676–2682. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Kim, J.H.; Kim, S.Y.; Lee, J.K. Isomaltose Production by Modification of the Fructose-Binding Site on the Basis of the Predicted Structure of Sucrose Isomerase from “Protaminobacter rubrum”. Appl. Environ. Microbiol. 2008, 74, 5183–5194. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Li, S.; Xu, H.; Feng, X.-H.; Cai, H.; Ye, Q. Purification and characterization of a highly selective sucrose isomerase from Erwinia rhapontici NX-5. Bioprocess Biosyst. Eng. 2011, 34, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, P.S.J. The extraction and mechanism of a novel isomaltulose-synthesizing enzyme from Erwinia rhapontici. Biochem. J. 1984, 220, 213–220. [Google Scholar] [CrossRef]
- Zhang, F.; Cheng, F.; Jia, D.-X.; Gu, Y.-H.; Liu, Z.-Q.; Zheng, Y.-G. Characterization of a recombinant sucrose isomerase and its application to enzymatic production of isomaltulose. Biotechnol. Lett. 2021, 43, 261–269. [Google Scholar] [CrossRef]
- Guo, D.; Li, M.; Jiang, M.; Cong, G.; Liu, Y.; Wang, C.; Li, X. Enhanced Extracellular Production and Characterization of Sucrose Isomerase in Bacillus subtilis with Optimized Signal Peptides. Foods 2022, 11, 2468. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, Z.; Ji, X.; Sheng, J. Display of a sucrose isomerase on the cell surface of Yarrowia lipolytica for synthesis of isomaltulose from sugar cane by-products. 3 Biotech 2019, 9, 1–7. [Google Scholar] [CrossRef]
- Xu, Z.; Xue, Y.-P.; Zou, S.-P.; Zheng, Y.-G. Enzyme engineering strategies to confer thermostability. In Biomass, Biofuels, Biochemicals; Elsevier: Amsterdam, The Netherlands, 2020; pp. 67–89. [Google Scholar]
- Bruins, M.E.; Janssen, A.E.M.; Boom, R.M. Thermozymes and Their Applications: A Review of Recent Literature and Patents. Appl. Biochem. Biotechnol. 2001, 90, 155–186. [Google Scholar] [CrossRef]
- Ferdjani, S.; Ionita, M.; Roy, B.; Dion, M.; Djeghaba, Z.; Rabiller, C.; Tellier, C. Correlation between thermostability and stability of glycosidases in ionic liquid. Biotechnol. Lett. 2011, 33, 1215–1219. [Google Scholar] [CrossRef] [PubMed]
- Kazlauskas, R. Engineering more stable proteins. Chem. Soc. Rev. 2018, 47, 9026–9045. [Google Scholar] [CrossRef]
- Cicerone, M.; Giri, J.; Shaked, Z.; Roberts, C. Protein stability—An underappreciated but critical need for drug delivery systems. Adv. Drug Deliv. Rev. 2015, 93, 1. [Google Scholar] [CrossRef]
- Choi, J.-M.; Han, S.-S.; Kim, H.-S. Industrial applications of enzyme biocatalysis: Current status and future aspects. Biotechnol. Adv. 2015, 33, 1443–1454. [Google Scholar] [CrossRef]
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U.T. Biocatalysis: Enzymatic Synthesis for Industrial Applications. Angew. Chem. Int. Ed. 2020, 60, 88–119. [Google Scholar] [CrossRef]
- Turner, P.; Mamo, G.; Karlsson, E.N. Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb. Cell Factories 2007, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Bajaj, B.K. Medium Optimization for Enhanced Production of Protease with Industrially Desirable Attributes from Bacillus subtilis K-1. Chem. Eng. Commun. 2014, 202, 1051–1060. [Google Scholar] [CrossRef]
- Böttcher, D.; Bornscheuer, U.T. Protein engineering of microbial enzymes. Curr. Opin. Microbiol. 2010, 13, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, M.; Signor, G.; Matthews, B.W. Substantial increase of protein stability by multiple disulphide bonds. Nature 1989, 342, 291–293. [Google Scholar] [CrossRef]
- Sapag, A.; Wouters, J.; Lambert, C.; de Ioannes, P.; Eyzaguirre, J.; Depiereux, E. The endoxylanases from family 11: Computer analysis of protein sequences reveals important structural and phylogenetic relationships. J. Biotechnol. 2002, 95, 109–131. [Google Scholar] [CrossRef]
- Warren, G.L.; Petsko, G.A. Composition analysis of α-helices in thermophilic organisms. Protein Eng. Des. Sel. 1995, 8, 905–913. [Google Scholar] [CrossRef]
- Menéndez-Arias, L.; Argosf, P. Engineering protein thermal stability: Sequence statistics point to residue substitutions in α-helices. J. Mol. Biol. 1989, 206, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Chen, S.; Zhao, X.; Nie, Y.; Xu, Y. Computation-aided engineering of starch-debranching pullulanase from Bacillus thermoleovorans for enhanced thermostability. Appl. Microbiol. Biotechnol. 2020, 104, 7551–7562. [Google Scholar] [CrossRef] [PubMed]
- Masakari, Y.; Hara, C.; Araki, Y.; Gomi, K.; Ito, K. Improvement in the thermal stability of Mucor prainii-derived FAD-dependent glucose dehydrogenase via protein chimerization. Enzym. Microb. Technol. 2020, 132, 109387. [Google Scholar] [CrossRef]
- You, S.; Xie, C.; Ma, R.; Huang, H.-Q.; Herman, R.A.; Su, X.-Y.; Ge, Y.; Cai, H.-Y.; Yao, B.; Wang, J.; et al. Improvement in catalytic activity and thermostability of a GH10 xylanase and its synergistic degradation of biomass with cellulase. Biotechnol. Biofuels 2019, 12, 1–15. [Google Scholar] [CrossRef]
- Li, G.; Fang, X.; Su, F.; Chen, Y.; Xu, L.; Yan, Y. Enhancing the Thermostability of Rhizomucor miehei Lipase with a Limited Screening Library by Rational-Design Point Mutations and Disulfide Bonds. Appl. Environ. Microbiol. 2018, 84, e02129-17. [Google Scholar] [CrossRef]
- Pang, B.; Zhou, L.; Cui, W.; Liu, Z.; Zhou, Z. Improvement of the Thermostability and Activity of Pullulanase from Anoxybacillus sp. WB42. Appl. Biochem. Biotechnol. 2020, 191, 942–954. [Google Scholar] [CrossRef]
- Teng, C.; Jiang, Y.; Xu, Y.; Li, Q.; Li, X.; Fan, G.; Xiong, K.; Yang, R.; Zhang, C.; Ma, R.; et al. Improving the thermostability and catalytic efficiency of GH11 xylanase PjxA by adding disulfide bridges. Int. J. Biol. Macromol. 2019, 128, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Bian, F.; Yue, S.; Peng, Z.; Zhang, X.; Chen, G.; Yu, J.; Xuan, N.; Bi, Y. A Comprehensive Alanine-Scanning Mutagenesis Study Reveals Roles for Salt Bridges in the Structure and Activity of Pseudomonas aeruginosa Elastase. PLoS ONE 2015, 10, e0121108. [Google Scholar] [CrossRef][Green Version]
- Wang, R.; Wang, S.; Xu, Y.; Yu, X. Enhancing the thermostability of Rhizopus chinensis lipase by rational design and MD simulations. Int. J. Biol. Macromol. 2020, 160, 1189–1200. [Google Scholar] [CrossRef]
- Kim, T.; Joo, J.C.; Yoo, Y.J. Hydrophobic interaction network analysis for thermostabilization of a mesophilic xylanase. J. Biotechnol. 2012, 161, 49–59. [Google Scholar] [CrossRef]
- Farnoosh, G.; Khajeh, K.; Mohammadi, M.; Hassanpour, K.; Latifi, A.M.; Aghamollaei, H. Catalytic and structural effects of flexible loop deletion in organophosphorus hydrolase enzyme: A thermostability improvement mechanism. J. Biosci. 2020, 45, 1–10. [Google Scholar] [CrossRef]
- Kumar, S.; Ma, B.; Tsai, C.-J.; Nussinov, R. Electrostatic strengths of salt bridges in thermophilic and mesophilic glutamate dehydrogenase monomers. Proteins: Struct. Funct. Bioinform. 2000, 38, 368–383. [Google Scholar] [CrossRef]
- Xue, H.; Zhou, J.; You, C.; Huang, Q.; Lu, H. Amino acid substitutions in the N-terminus, cord and α-helix domains improved the thermostability of a family 11 xylanase XynR8. J. Ind. Microbiol. Biotechnol. 2012, 39, 1279–1288. [Google Scholar] [CrossRef]
- McCarthy, A.A.; Morris, D.D.; Bergquist, P.L.; Baker, E.N. Structure of XynB, a highly thermostable β-1, 4-xylanase from Dictyoglomus thermophilum Rt46B. 1, at 1.8 Å resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2000, 56, 1367–1375. [Google Scholar] [CrossRef] [PubMed]
- Maenpuen, S.; Pongsupasa, V.; Pensook, W.; Anuwan, P.; Kraivisitkul, N.; Pinthong, C.; Phonbuppha, J.; Luanloet, T.; Wijma, H.J.; Fraaije, M.W.; et al. Creating Flavin Reductase Variants with Thermostable and Solvent-Tolerant Properties by Rational-Design Engineering. ChemBioChem 2020, 21, 1481–1491. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Vermaas, J.V.; Zheng, J.; Wang, Y.; Tu, T.; Wang, X.; Xie, X.; Yao, B.; Beckham, G.T.; Luo, H. Activity and Thermostability of GH5 Endoglucanase Chimeras from Mesophilic and Thermophilic Parents. Appl. Environ. Microbiol. 2019, 85, e02079-18. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, S.; Murthy, M. Protein thermal stability: Insights from atomic displacement parameters (B values). Protein Eng. Des. Sel. 2000, 13, 9–13. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Pereira, P.C. Biocatalysis engineering: The big picture. Chem. Soc. Rev. 2017, 46, 2678–2691. [Google Scholar] [CrossRef] [PubMed]
- Estell, D.A.; Graycar, T.P.; Wells, J.A. Engineering an enzyme by site-directed mutagenesis to be resistant to chemical oxidation. Pediatrics 1985, 260, 6518–6521. [Google Scholar] [CrossRef]
- Zhang, H.; Sang, J.; Zhang, Y.; Sun, T.; Liu, H.; Yue, R.; Zhang, J.; Wang, H.; Dai, Y.; Lu, F.; et al. Rational design of a Yarrowia lipolytica derived lipase for improved thermostability. Int. J. Biol. Macromol. 2019, 137, 1190–1198. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, H.; Xu, L.; Zhu, X.; Li, X. Site-directed mutagenesis of a hyperthermophilic endoglucanase Cel12B from Thermotoga maritima based on rational design. PLoS ONE 2015, 10, e0133824. [Google Scholar] [CrossRef]
- Basheer, S.M.; Chellappan, S. Enzyme engineering. In Bioresources and Bioprocess in Biotechnology; Springer: Berlin/Heidelberg, Germany, 2017; pp. 151–168. [Google Scholar]
- Lutz, S. Beyond directed evolution—Semi-rational protein engineering and design. Curr. Opin. Biotechnol. 2010, 21, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Kazlauskas, R.J.; Bornscheuer, U.T. Finding better protein engineering strategies. Nat. Chem. Biol. 2009, 5, 526–529. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.C.; Jin, H.-M.; Kuchner, O.; Arnold, F.H. Strategies for the in vitro evolution of protein function: Enzyme evolution by random recombination of improved sequences. J. Mol. Biol. 1997, 272, 336–347. [Google Scholar] [CrossRef]
- Chen, K.; Arnold, F.H. Enzyme Engineering for Nonaqueous Solvents: Random Mutagenesis to Enhance Activity of Subtilisin E in Polar Organic Media. Bio/Technology 1991, 9, 1073–1077. [Google Scholar] [CrossRef]
- Tizei, P.A.; Csibra, E.; Torres, L.; Pinheiro, V.B. Selection platforms for directed evolution in synthetic biology. Biochem. Soc. Trans. 2016, 44, 1165–1175. [Google Scholar] [CrossRef]
- Cirino, P.C.; Mayer, K.M.; Umeno, D. Generating mutant libraries using error-prone PCR. In Directed Evolution Library Creation; Springer: Berlin/Heidelberg, Germany, 2003; pp. 3–9. [Google Scholar]
- Sharma, A.; Gupta, G.; Ahmad, T.; Mansoor, S.; Kaur, B. Enzyme Engineering: Current Trends and Future Perspectives. Food Rev. Int. 2019, 37, 121–154. [Google Scholar] [CrossRef]
- Khan, M.F.; Kundu, D.; Hazra, C.; Patra, S. A strategic approach of enzyme engineering by attribute ranking and enzyme immobilization on zinc oxide nanoparticles to attain thermostability in mesophilic Bacillus subtilis lipase for detergent formulation. Int. J. Biol. Macromol. 2019, 136, 66–82. [Google Scholar] [CrossRef]
- Packer, M.S.; Liu, D.R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015, 16, 379–394. [Google Scholar] [CrossRef]
- Zhou, C.; Xue, Y.; Ma, Y. Evaluation and directed evolution for thermostability improvement of a GH 13 thermostable α-glucosidase from Thermus thermophilus TC11. BMC Biotechnol. 2015, 15, 97. [Google Scholar] [CrossRef]
- Huang, L.; Shan, M.; Ma, J.; Li, Y.; Xu, Z.; Shao, S.; Wang, X.; Wang, K.; Xiao, D.; Lu, F.; et al. Directed evolution of α-amylase from Bacillus licheniformis to enhance its acid-stable performance. Biologia 2019, 74, 1363–1372. [Google Scholar] [CrossRef]
- Chen, C.; Su, L.; Xu, F.; Xia, Y.; Wu, J. Improved Thermostability of Maltooligosyltrehalose Synthase from Arthrobacter ramosus by Directed Evolution and Site-Directed Mutagenesis. J. Agric. Food Chem. 2019, 67, 5587–5595. [Google Scholar] [CrossRef] [PubMed]
- Steiner, K.; Schwab, H. Recent advances in rational approaches for enzyme engineering. Comput. Struct. Biotechnol. J. 2012, 2, e201209010. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Qu, G.; Li, J.K.; Fan, W.; Ma, J.A.; Xu, Y.; Nie, Y.; Sun, Z. Conformational dynamics-guided loop engineering of an alcohol dehydrogenase: Capture, turnover and enantioselective transformation of difficult-to-reduce ketones. Adv. Synth. Catal. 2019, 361, 3182–3190. [Google Scholar] [CrossRef]
- Li, G.; Chen, Y.; Fang, X.; Su, F.; Xu, L.; Yan, Y. Identification of a hot-spot to enhance Candida rugosa lipase thermostability by rational design methods. RSC Adv. 2018, 8, 1948–1957. [Google Scholar] [CrossRef]
- Bashirova, A.; Pramanik, S.; Volkov, P.; Rozhkova, A.; Nemashkalov, V.; Zorov, I.; Gusakov, A.; Sinitsyn, A.; Schwaneberg, U.; Davari, M.D. Disulfide Bond Engineering of an Endoglucanase from Penicillium verruculosum to Improve Its Thermostability. Int. J. Mol. Sci. 2019, 20, 1602. [Google Scholar] [CrossRef]
- Cui, X.; Yuan, X.; Li, S.; Hu, X.; Zhao, J.; Zhang, G. Simultaneously improving the specific activity and thermostability of α-amylase BLA by rational design. Bioprocess Biosyst. Eng. 2022, 45, 1839–1848. [Google Scholar] [CrossRef]
- He, J.; Tang, F.; Chen, D.; Yu, B.; Luo, Y.; Zheng, P.; Mao, X.; Yu, J.; Yu, F. Design, expression and functional characterization of a thermostable xylanase from Trichoderma reesei. PLoS ONE 2019, 14, e0210548. [Google Scholar] [CrossRef]
- Chen, A.; Li, Y.; Nie, J.; McNeil, B.; Jeffrey, L.; Yang, Y.; Bai, Z. Protein engineering of Bacillus acidopullulyticus pullulanase for enhanced thermostability using in silico data driven rational design methods. Enzym. Microb. Technol. 2015, 78, 74–83. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, Y.; Zhou, X.; Liu, N.; Ming, D.; Zhu, L.; Jiang, L. Improving the thermostability of trehalose synthase from Thermomonospora curvata by covalent cyclization using peptide tags and investigation of the underlying molecular mechanism. Int. J. Biol. Macromol. 2020, 168, 13–21. [Google Scholar] [CrossRef]
- Krüger, D.M.; Rathi, P.C.; Pfleger, C.; Gohlke, H. CNA web server: Rigidity theory-based thermal unfolding simulations of proteins for linking structure, (thermo-)stability, and function. Nucleic Acids Res. 2013, 41, W340–W348. [Google Scholar] [CrossRef]
- Ban, X.; Wang, T.; Fan, W.; Li, C.; Gu, Z.; Cheng, L.; Hong, Y.; Li, Z. Thermostability and catalytic ability enhancements of 1, 4-α-glucan branching enzyme by introducing salt bridges at flexible amino acid sites. Int. J. Biol. Macromol. 2023, 224, 1276–1282. [Google Scholar] [CrossRef]
- Zhang, H.; Zhai, W.; Lin, L.; Wang, P.; Xu, X.; Wei, W.; Wei, D. In Silico Rational Design and Protein Engineering of Disulfide Bridges of an α-Amylase from Geobacillus sp. to Improve Thermostability. Starch-Stärke 2021, 73, 2000274. [Google Scholar] [CrossRef]
- Han, C.; Wang, Q.; Sun, Y.; Yang, R.; Liu, M.; Wang, S.; Liu, Y.; Zhou, L.; Li, D. Improvement of the catalytic activity and thermostability of a hyperthermostable endoglucanase by optimizing N-glycosylation sites. Biotechnol. Biofuels 2020, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Vicente, A.I.; Viña-Gonzalez, J.; Mateljak, I.; Monza, E.; Lucas, F.; Guallar, V.; Alcalde, M. Enhancing thermostability by modifying flexible surface loops in an evolved high-redox potential laccase. AIChE J. 2019, 66, e16747. [Google Scholar] [CrossRef]
- Modarres, H.P.; Mofrad, M.R.; Sanati-Nezhad, A. Protein thermostability engineering. RSC Adv. 2016, 6, 115252–115270. [Google Scholar] [CrossRef]
- Ren, X.; Wang, J.; Li, Y.; Wang, F.; Wang, R.; Li, P.; Ma, C.; Su, J. Computational and Enzymatic Analyses Unveil the Catalytic Mechanism of Thermostable Trehalose Synthase and Suggest Strategies for Improved Bioconversion. J. Agric. Food Chem. 2019, 67, 8177–8185. [Google Scholar] [CrossRef]
- Peng, M.; Maier, M.; Esch, J.; Schug, A.; Rabe, K.S. Direct coupling analysis improves the identification of beneficial amino acid mutations for the functional thermostabilization of a delicate decarboxylase. Biol. Chem. 2019, 400, 1519–1527. [Google Scholar] [CrossRef]
- Ali, M.; Ishqi, H.M.; Husain, Q. Enzyme engineering: Reshaping the biocatalytic functions. Biotechnol. Bioeng. 2020, 117, 1877–1894. [Google Scholar] [CrossRef]
- Ban, X.; Wu, J.; Kaustubh, B.; Lahiri, P.; Dhoble, A.S.; Gu, Z.; Li, C.; Cheng, L.; Hong, Y.; Tong, Y.; et al. Additional salt bridges improve the thermostability of 1,4-α-glucan branching enzyme. Food Chem. 2020, 316, 126348. [Google Scholar] [CrossRef]
- Xu, Z.; Cen, Y.-K.; Zou, S.-P.; Xue, Y.-P.; Zheng, Y.-G. Recent advances in the improvement of enzyme thermostability by structure modification. Crit. Rev. Biotechnol. 2020, 40, 83–98. [Google Scholar] [CrossRef]
- Suzuki, Y. A general principle of increasing protein thermostability. Proc. Jpn. Acad. Ser. B 1989, 65, 146–148. [Google Scholar] [CrossRef]
- Suzuki, Y.; Oishi, K.; Nakano, H.; Nagayama, T. A strong correlation between the increase in number of proline residues and the rise in thermostability of five Bacillus oligo-1,6-glucosidases. Appl. Microbiol. Biotechnol. 1987, 26, 546–551. [Google Scholar] [CrossRef]
- Igarashi, K.; Ozawa, T.; Ikawa-Kitayama, K.; Hayashi, Y.; Araki, H.; ENDo, K.; Hagihara, H.; Ozaki, K.; Kawai, S.; Ito, S. Thermostabilization by proline substitution in an alkaline, liquefying α-amylase from Bacillus sp. strain KSM-1378. Biosci. Biotechnol. Biochem. 1999, 63, 1535–1540. [Google Scholar] [PubMed]
- Carugo, O. How large B-factors can be in protein crystal structures. BMC Bioinform. 2018, 19, 61. [Google Scholar] [CrossRef] [PubMed]
- Merritt, E.A. To B or not to B: A question of resolution? Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 468–477. [Google Scholar] [CrossRef]
- Kuriyan, J.; Karplus, M.; Petsko, G.A. Estimation of uncertainties in X-ray refinement results by use of perturbed structures. Proteins Struct. Funct. Bioinform. 1987, 2, 1–12. [Google Scholar] [CrossRef]
- Bendl, J.; Stourac, J.; Sebestova, E.; Vavra, O.; Musil, M.; Brezovsky, J.; Damborsky, J. HotSpot Wizard 2.0: Automated design of site-specific mutations and smart libraries in protein engineering. Nucleic Acids Res. 2016, 44, W479–W487. [Google Scholar] [CrossRef]
- Helenius, A.; Aebi, M. Roles of N-Linked Glycans in the Endoplasmic Reticulum. Annu. Rev. Biochem. 2004, 73, 1019–1049. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.; Burnina, I.; Lynaugh, H.; Li, H. O-linked glycosylation analysis of recombinant human granulocyte colony-stimulating factor produced in glycoengineered Pichia pastoris by liquid chromatography and mass spectrometry. J. Chromatogr. B 2014, 945-946, 135–140. [Google Scholar] [CrossRef]
- Knauer, R.; Lehle, L. The oligosaccharyltransferase complex from yeast. Biochim. Biophys. Acta (BBA)-Gen. Subj. 1999, 1426, 259–273. [Google Scholar] [CrossRef]
- Ryu, K.-S.; Lee, J.-O.; Kwon, T.H.; Choi, H.-H.; Park, H.-S.; Hwang, S.K.; Lee, Z.-W.; Lee, K.-B.; Han, Y.H.; Choi, Y.-S.; et al. The presence of monoglucosylated N196-glycan is important for the structural stability of storage protein, arylphorin. Biochem. J. 2009, 421, 87–96. [Google Scholar] [CrossRef]
- Aebi, M.; Bernasconi, R.; Clerc, S.; Molinari, M. N-glycan structures: Recognition and processing in the ER. Trends Biochem. Sci. 2010, 35, 74–82. [Google Scholar] [CrossRef]
- Haltiwanger, R.S.; Lowe, J.B. Role of Glycosylation in Development. Annu. Rev. Biochem. 2004, 73, 491–537. [Google Scholar] [CrossRef] [PubMed]
- Colley, K.J.; Varki, A.; Kinoshita, T. Cellular Organization of Glycosylation, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
- Katla, S.; Yoganand, K.; Hingane, S.; Kumar, C.R.; Anand, B.; Sivaprakasam, S. Novel glycosylated human interferon alpha 2b expressed in glycoengineered Pichia pastoris and its biological activity: N-linked glycoengineering approach. Enzym. Microb. Technol. 2019, 128, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Benoit, I.; Asther, M.; Sulzenbacher, G.; Record, E.; Marmuse, L.; Parsiegla, G.; Gimbert, I.; Asther, M.; Bignon, C. Respective importance of protein folding and glycosylation in the thermal stability of recombinant feruloyl esterase A. FEBS Lett. 2006, 580, 5815–5821. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Huang, S.; Kaleem, I.; Li, C. N-glycosylation enhances functional and structural stability of recombinant β-glucuronidase expressed in Pichia pastoris. J. Biotechnol. 2013, 164, 75–81. [Google Scholar] [CrossRef]
- Hu, X.; Yuan, X.; He, N.; Zhuang, T.Z.; Wu, P.; Zhang, G. Expression of Bacillus licheniformis α-amylase in Pichia pastoris without antibiotics-resistant gene and effects of glycosylation on the enzymic thermostability. 3 Biotech 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Maksimainen, M.; Hakulinen, N.; Kallio, J.M.; Timoharju, T.; Turunen, O.; Rouvinen, J. Crystal structures of Trichoderma reesei β-galactosidase reveal conformational changes in the active site. J. Struct. Biol. 2011, 174, 156–163. [Google Scholar] [CrossRef]
- Nagae, M.; Yamaguchi, Y. Function and 3D Structure of the N-Glycans on Glycoproteins. Int. J. Mol. Sci. 2012, 13, 8398–8429. [Google Scholar] [CrossRef]
- Joao, H.C.; Scragg, I.G.; Dwek, R.A. Effects of glycosylation on protein conformation and amide proton exchange rates in RNase B. FEBS Lett. 1992, 307, 343–346. [Google Scholar] [CrossRef]
- Gavel, Y.; Heijne, G.V. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: Implications for protein engineering. Protein Eng. Des. Sel. 1990, 3, 433–442. [Google Scholar] [CrossRef]
- Trimble, R.B.; Lubowski, C.; Hauer III, C.R.; Stack, R.; McNaughton, L.; Gemmill, T.R.; Kumar, S.A. Characterization of N-and O-linked glycosylation of recombinant human bile salt–stimulated lipase secreted by Pichia pastoris. Glycobiology 2004, 14, 265–274. [Google Scholar] [CrossRef]
- Wang, Z.; Guo, C.; Liu, L.; Huang, H. Effects of N-glycosylation on the biochemical properties of recombinant bEKL expressed in Pichia pastoris. Enzym. Microb. Technol. 2018, 114, 40–47. [Google Scholar] [CrossRef]
- Villiger, T.K.; Scibona, E.; Stettler, M.; Broly, H.; Morbidelli, M.; Soos, M. Controlling the time evolution of mAb N-linked glycosylation - Part II: Model-based predictions. Biotechnol. Prog. 2016, 32, 1135–1148. [Google Scholar] [CrossRef] [PubMed]
- Hang, I.; Lin, C.-W.; Grant, O.C.; Fleurkens, S.; Villiger, T.K.; Soos, M.; Morbidelli, M.; Woods, R.J.; Gauss, R.; Aebi, M. Analysis of site-specificN-glycan remodeling in the endoplasmic reticulum and the Golgi. Glycobiology 2015, 25, 1335–1349. [Google Scholar] [CrossRef] [PubMed]
- Ravaud, S.; Watzlawick, H.; Mattes, R.; Haser, R.; Aghajari, N. Towards the three-dimensional structure of a sucrose isomerase from Pseudomonas mesoacidophila MX-45. Biol. Bratisl. 2005, 60, 89–95. [Google Scholar]
- Wu, X.; Wang, Y.; Tong, B.; Chen, X.; Chen, J. Purification and biochemical characterization of a thermostable and acid-stable alpha-amylase from Bacillus licheniformis B4-423. Int. J. Biol. Macromol. 2018, 109, 329–337. [Google Scholar] [CrossRef]
- Laderman, K.; Davis, B.; Krutzsch, H.; Lewis, M.; Griko, Y.; Privalov, P.; Anfinsen, C. The purification and characterization of an extremely thermostable alpha-amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus. Pediatrics 1993, 268, 24394–24401. [Google Scholar] [CrossRef]
- Kikani, B.; Singh, S. Single step purification and characterization of a thermostable and calcium independent α-amylase from Bacillus amyloliquifaciens TSWK1-1 isolated from Tulsi Shyam hot spring reservoir, Gujarat (India). Int. J. Biol. Macromol. 2011, 48, 676–681. [Google Scholar] [CrossRef]
- Wang, J.; Ren, X.; Wang, R.; Su, J.; Wang, F. Structural Characteristics and Function of a New Kind of Thermostable Trehalose Synthase from Thermobaculum terrenum. J. Agric. Food Chem. 2017, 65, 7726–7735. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, M.; Wyss, M. Engineering proteins for thermostability: The use of sequence alignments versus rational design and directed evolution. Curr. Opin. Biotechnol. 2001, 12, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Dagan, S.; Hagai, T.; Gavrilov, Y.; Kapon, R.; Levy, Y.; Reich, Z. Stabilization of a protein conferred by an increase in folded state entropy. Proc. Natl. Acad. Sci. USA 2013, 110, 10628–10633. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.J.; Eisenberg, D. Transproteomic evidence of a loop-deletion mechanism for enhancing protein thermostability. J. Mol. Biol. 1999, 290, 595–604. [Google Scholar] [CrossRef]
- Lu, Z.; Wang, Q.; Jiang, S.; Zhang, G.; Ma, Y. Truncation of the unique N-terminal domain improved the thermos-stability and specific activity of alkaline α-amylase Amy703. Sci. Rep. 2016, 6, 22465. [Google Scholar] [CrossRef]
- Feller, G.; Bonneau, M.; Da Lage, J.L. Amyrel, a novel glucose-forming α-amylase from Drosophila with 4-α-glucanotransferase activity by disproportionation and hydrolysis of maltooligosaccharides. Glycobiology 2021, 31, 1134–1144. [Google Scholar] [CrossRef]
- Rhimi, M.; Da Lage, J.-L.; Haser, R.; Feller, G.; Aghajari, N. Structural and Functional Characterization of Drosophila melanogaster α-Amylase. Molecules 2023, 28, 5327. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Wang, X.; Zhang, D.; Wu, S.; Zhang, G. Enhanced thermal stability of lichenase from Bacillus subtilis 168 by SpyTag/SpyCatcher-mediated spontaneous cyclization. Biotechnol. Biofuels 2016, 9, 1–9. [Google Scholar] [CrossRef]
- Sun, X.-B.; Cao, J.-W.; Wang, J.-K.; Lin, H.-Z.; Gao, D.-Y.; Qian, G.-Y.; Park, Y.-D.; Chen, Z.-F.; Wang, Q. SpyTag/SpyCatcher molecular cyclization confers protein stability and resilience to aggregation. New Biotechnol. 2019, 49, 28–36. [Google Scholar] [CrossRef]
- Wang, Y.; Tian, J.; Xiao, Y.; Wang, Y.; Sun, H.; Chang, Y.; Luo, H. SpyTag/SpyCatcher cyclization enhances the thermostability and organic solvent tolerance of l-phenylalanine aldolase. Biotechnol. Lett. 2020, 41, 987–994. [Google Scholar] [CrossRef]
- Dani, V.S.; Ramakrishnan, C.; Varadarajan, R. MODIP revisited: Re-evaluation and refinement of an automated procedure for modeling of disulfide bonds in proteins. Protein Eng. Des. Sel. 2003, 16, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Du, M.; Cheng, B.; Wang, L.; Liu, X.; Ma, C.; Yang, C.; Xu, P. Close relationship of a novel Flavobacteriaceaeα-amylase with archaeal α-amylases and good potentials for industrial applications. Biotechnol. Biofuels 2014, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Ismaya, W.T.; Hasan, K.; Kardi, I.; Zainuri, A.; Rahmawaty, R.I.; Permanahadi, S.; El Viera, B.V.; Harinanto, G.; Gaffar, S.; Natalia, D.; et al. Chemical modification of Saccharomycopsis fibuligera R64 α-amylase to improve its stability against thermal, chelator, and proteolytic inactivation. Appl. Biochem. Biotechnol. 2013, 170, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.A.K. Studies on stabilization of amylase by covalent coupling to soluble polysaccharides. Enzym. Microb. Technol. 1991, 13, 164–170. [Google Scholar] [CrossRef]
- Klibanov, A.M. Immobilized enzymes and cells as practical catalysts. Science 1983, 219, 722–727. [Google Scholar] [CrossRef]
- Villalonga, R.; Gómez, L.; Ramírez, H.L.; Villalonga, M.L. Stabilization of α-amylase by chemical modification with carboxymethylcellulose. J. Chem. Technol. Biotechnol. 1999, 74, 635–638. [Google Scholar] [CrossRef]
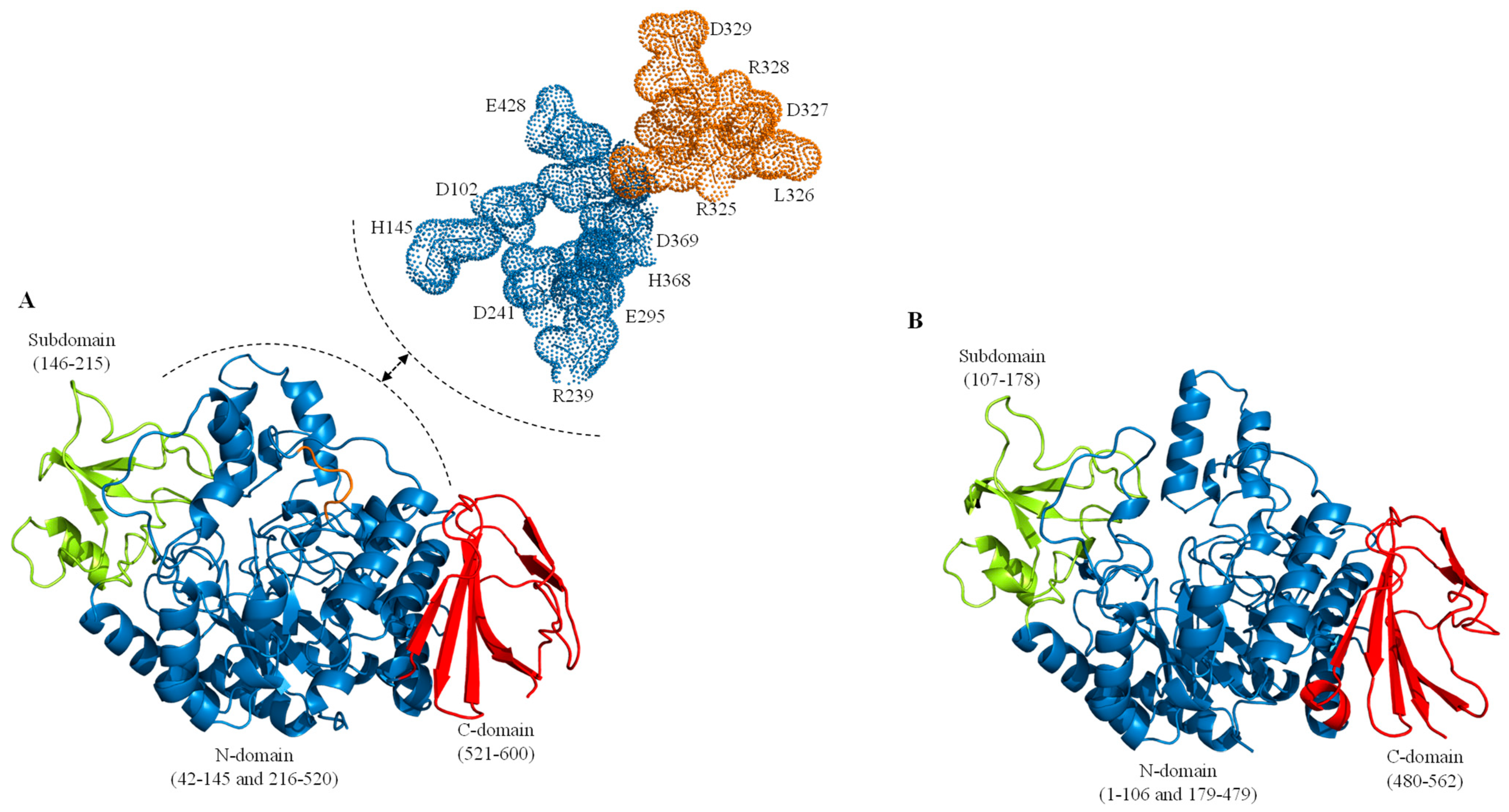
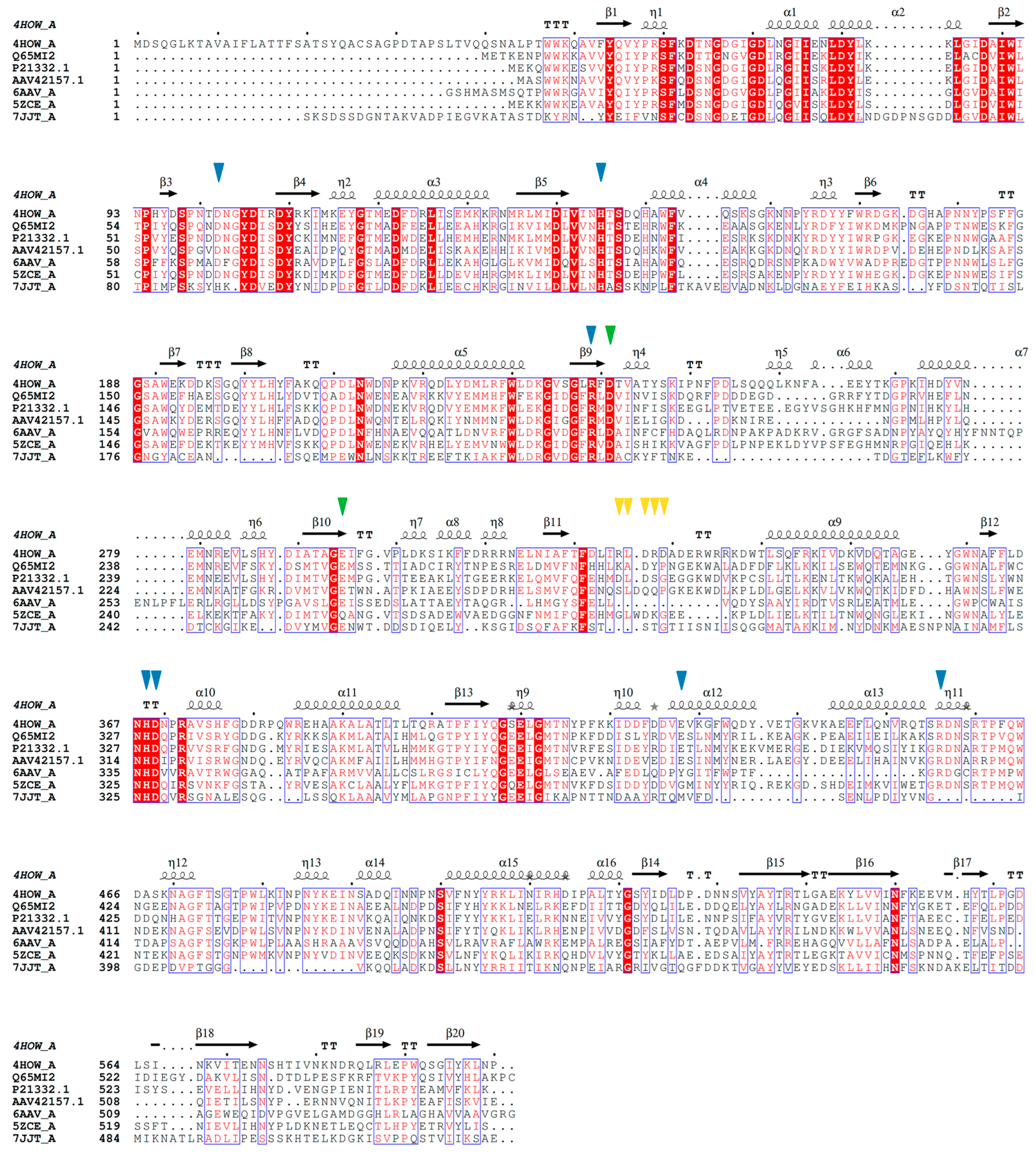
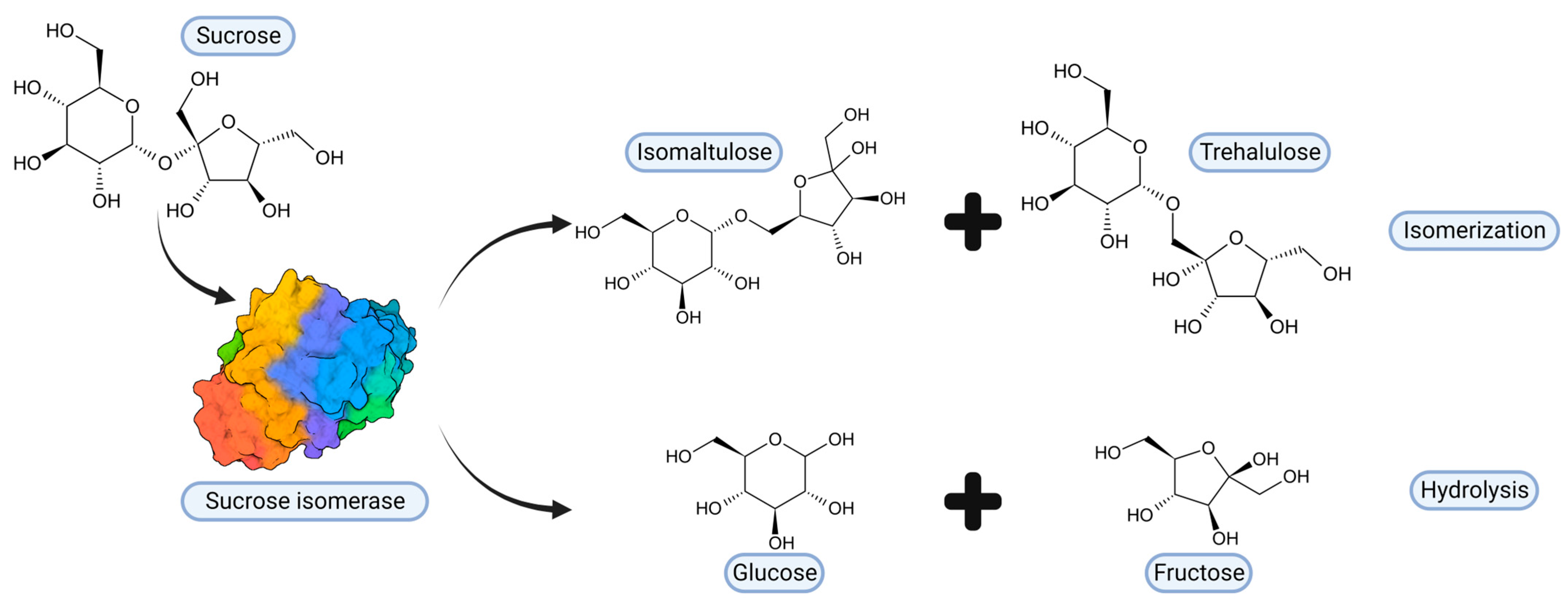

| Microbial Source | Sucrose Isomerase | Mutation | PDB ID | Interacted Chemical | Resolution (Å) | References |
|---|---|---|---|---|---|---|
| Pseudomonas mesoacidophila MX-45 (Rhizobium sp. MX-45) | MutB | Non-mutant | 2PWH | native | 2.0 | [56] |
| Non-mutant | 1ZJA | MutB-Tris | 1.6 | |||
| Non-mutant | 2PWD | MutB-deoxynojirimycin | 1.8 | |||
| Non-mutant | 2PWG | MutB-castanospermine | 2.2 | |||
| D200A | 2PWF | MutB-glucose | 1.8 | |||
| E254Q | 2PWE | MutB-sucrose | 2.0 | |||
| Non-mutant | 1ZJB | MutB-Tris | 1.8 | [57] | ||
| A258V | 4GO8 | MutB-Tris | 2.15 | |||
| D415N | 4GO9 | MutB-Tris | 2.2 | |||
| D200A-D415N (inactive enzyme) | 4HA1 | MutB-isomaltulose-glucose-Ca2+ | 2.2 | [58] | ||
| Non-mutant | 4H8V | MutB-trehalulose-Ca2+ | 1.95 | |||
| D200A-D415N (inactive enzyme) | 4H8U | MutB-trehalulose-glycerol-Ca2+ | 2.0 | |||
| E254Q-D415N (inactive enzyme) | 4H8H | MutB-SO42−-glycerol-Ca2+ | 2.0 | |||
| D200A-D415N (inactive enzyme) | 4H7V | MutB-glycerol-glucose-Ca2+ | 1.8 | |||
| R284C | 4H2C | MutB-glycerol-Ca2+ | 1.7 | |||
| R284C | 4GIN | MutB-glycerol-Ca2+ | 1.9 | |||
| F164L | 4GIA | MutB-Tris-glycerol-Ca2+ | 2.01 | |||
| F164L | 4GI9 | MutB-Tris-glycerol-Ca2+ | 2.15 | |||
| F164L | 4GI8 | MutB-Tris-glycerol-Ca2+ | 1.95 | |||
| F164L | 4GI6 | MutB-Tris-glycerol-glucose-Ca2+ | 2.15 | |||
| Erwinia rhapontici NX-5 | NX-5 | Non-mutant | 4HOW | (NX-5)-glycerol-Ca2+ | 1.7 | [59] |
| Non-mutant | 4HOX | (NX-5)-Tris-glycerol-Ca2+ | 2.0 | |||
| D241A | 4HOZ | (NX-5)-glucose-glycerol-Ca2+ | 2.0 | |||
| E295A | 4HP5 | (NX-5)-glucose-glycerol-Ca2+ | 2.0 | |||
| E295Q | 4HPH | (NX-5)-sucrose-glycerol-Ca2+ | 1.7 | |||
| Protaminobacter rubrum CBS574.77 | SmuA | Non-mutant | 3GBD | SmuA-C6H5O73−-ethylene glycol | 1.95 | [60] |
| Non-mutant | 3GBE | SmuA-C6H5O73−-ethylene glycol-deoxynojirimycin | 1.7 | |||
| Klebsiella sp. LX3 | PalI | Non-mutant | 1M53 | No information | 2.2 | [61] |
| Isoform | Top (°C) | pH | Specific Activity (U/mg) | Km (mM) | kcat/Km (mM−1·s−1) | Isomaltulose Ratio (%) | References |
|---|---|---|---|---|---|---|---|
| Wild-type PalI NX-5 | 30 | 6 | 423 | 222 | NR | 83 | [87] |
| Recombinant PalI NX-5 | 30 | 5 | NR | 257 | NR | 87 | [39] |
| Recombinant PalI NX-5 | 30 | 6 | 483.8 | 255.1 | 2.2 | 78 | [54] |
| Wild-type PalI D12 | 40 | 6 | 19.8 | 138 | NR | 65.7 | [40] |
| Wild-type NCPPB 1578 | 30 | NR | 4.11 a | 280 | NR | 85 | [88] |
| Recombinant CBS 574.77 | 35 | NR | NR | 32.4 | 1301 | 88.5 | [86] |
| Recombinant PalI NK33 | 35 | 6 | 2362 | 42.7 | NR | 76.8 | [49] |
| Recombinant UQ68J | 35 | 5 | 562 | 39.9 | 17.9 | 91 | [51] |
| Recombinant UQ14S | 35 | 6 | 351 | 76 | 6.2 | 66 | [51] |
| Recombinant PalI LX3 | 35 | 6 | 328 | 54.6 | 0.27 | 83 | [85] |
| Wild-type ATCC15928 | 30 | 6.2 | 120 | 65 | NR | 72.6 | [41] |
| Recombinant AS9 | 30 | 6 | 957.5 | 30.1 | 33 | 76.3 | [55] |
| Recombinant Ejp617 | 40 | 6 | 118.87 | 69.28 | NR | 80.5 | [89] |
| Recombinant FMB-1 | 50 | 5–6 | 49 | NR | NR | 78 | [50] |
| Recombinant Pal-2 | 40 | 5.5 | 286.4 | 62.9 | NR | 81.7 | [48] |
| Isoform | Modification | Strategy for Thermostabilization | Stabilizing Interaction | Top (°C) | Half-Life (min) | Specific Activity (U/mg) | Km (mM) | kcat/Km (mM−1·s−1) | References |
|---|---|---|---|---|---|---|---|---|---|
| PalI NX-5 | Glycosylation | B-factor analysis and glycosylation site engineering. | Strengthening of the hydrogen-bonding network and glycosylation of the flexible terminal C region. | 30 | 10.1 a | 483.8 | 255.1 | 2.21 | [54] |
| PalI NX-5 | Glycosylation—K174Q | 35 | 22.3 a | 529.9 | 241.9 | 2.55 | |||
| PalI NX-5 | Glycosylation—L202E | 35 | 17.5 a | 509.1 | 234.9 | 2.52 | |||
| PalI NX-5 | Glycosylation—K174Q/L202E | 35 | 29.2 a | 509.3 | 231.2 | 2.57 | |||
| PalI AS9 | Native | B-factor analysis | Strengthening of the hydrogen bridge network | 30 | 39.2 b | 957.5 | 30.1 | 33 | [55] |
| PalI AS9 | E175N | 35 | 90.2 b | 1017.6 | 28.1 | 45.6 | |||
| PalI AS9 | K576D | 35 | 69.8 b | 1045.7 | 29.5 | 34.4 | |||
| PalI AS9 | E175N/K576D | 35 | 300 b | 1218.9 | 26.8 | 39.4 | |||
| PalI LX3 | Native | Proline theory | Loop stabilization | 35 | 1.81 c | 328 | 54.6 | 0.27 | [85] |
| PalI LX3 | E498P | 40 | 9.45 c | 350 | NR | 0.29 | |||
| PalI LX3 | E498P/R310P | 40 | 13.61 c | 340 | NR | 0.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sardiña-Peña, A.J.; Mesa-Ramos, L.; Iglesias-Figueroa, B.F.; Ballinas-Casarrubias, L.; Siqueiros-Cendón, T.S.; Espinoza-Sánchez, E.A.; Flores-Holguín, N.R.; Arévalo-Gallegos, S.; Rascón-Cruz, Q. Analyzing Current Trends and Possible Strategies to Improve Sucrose Isomerases’ Thermostability. Int. J. Mol. Sci. 2023, 24, 14513. https://doi.org/10.3390/ijms241914513
Sardiña-Peña AJ, Mesa-Ramos L, Iglesias-Figueroa BF, Ballinas-Casarrubias L, Siqueiros-Cendón TS, Espinoza-Sánchez EA, Flores-Holguín NR, Arévalo-Gallegos S, Rascón-Cruz Q. Analyzing Current Trends and Possible Strategies to Improve Sucrose Isomerases’ Thermostability. International Journal of Molecular Sciences. 2023; 24(19):14513. https://doi.org/10.3390/ijms241914513
Chicago/Turabian StyleSardiña-Peña, Amado Javier, Liber Mesa-Ramos, Blanca Flor Iglesias-Figueroa, Lourdes Ballinas-Casarrubias, Tania Samanta Siqueiros-Cendón, Edward Alexander Espinoza-Sánchez, Norma Rosario Flores-Holguín, Sigifredo Arévalo-Gallegos, and Quintín Rascón-Cruz. 2023. "Analyzing Current Trends and Possible Strategies to Improve Sucrose Isomerases’ Thermostability" International Journal of Molecular Sciences 24, no. 19: 14513. https://doi.org/10.3390/ijms241914513
APA StyleSardiña-Peña, A. J., Mesa-Ramos, L., Iglesias-Figueroa, B. F., Ballinas-Casarrubias, L., Siqueiros-Cendón, T. S., Espinoza-Sánchez, E. A., Flores-Holguín, N. R., Arévalo-Gallegos, S., & Rascón-Cruz, Q. (2023). Analyzing Current Trends and Possible Strategies to Improve Sucrose Isomerases’ Thermostability. International Journal of Molecular Sciences, 24(19), 14513. https://doi.org/10.3390/ijms241914513









