High Recovery Chromatographic Purification of mRNA at Room Temperature and Neutral pH
Abstract
:1. Introduction
2. Results
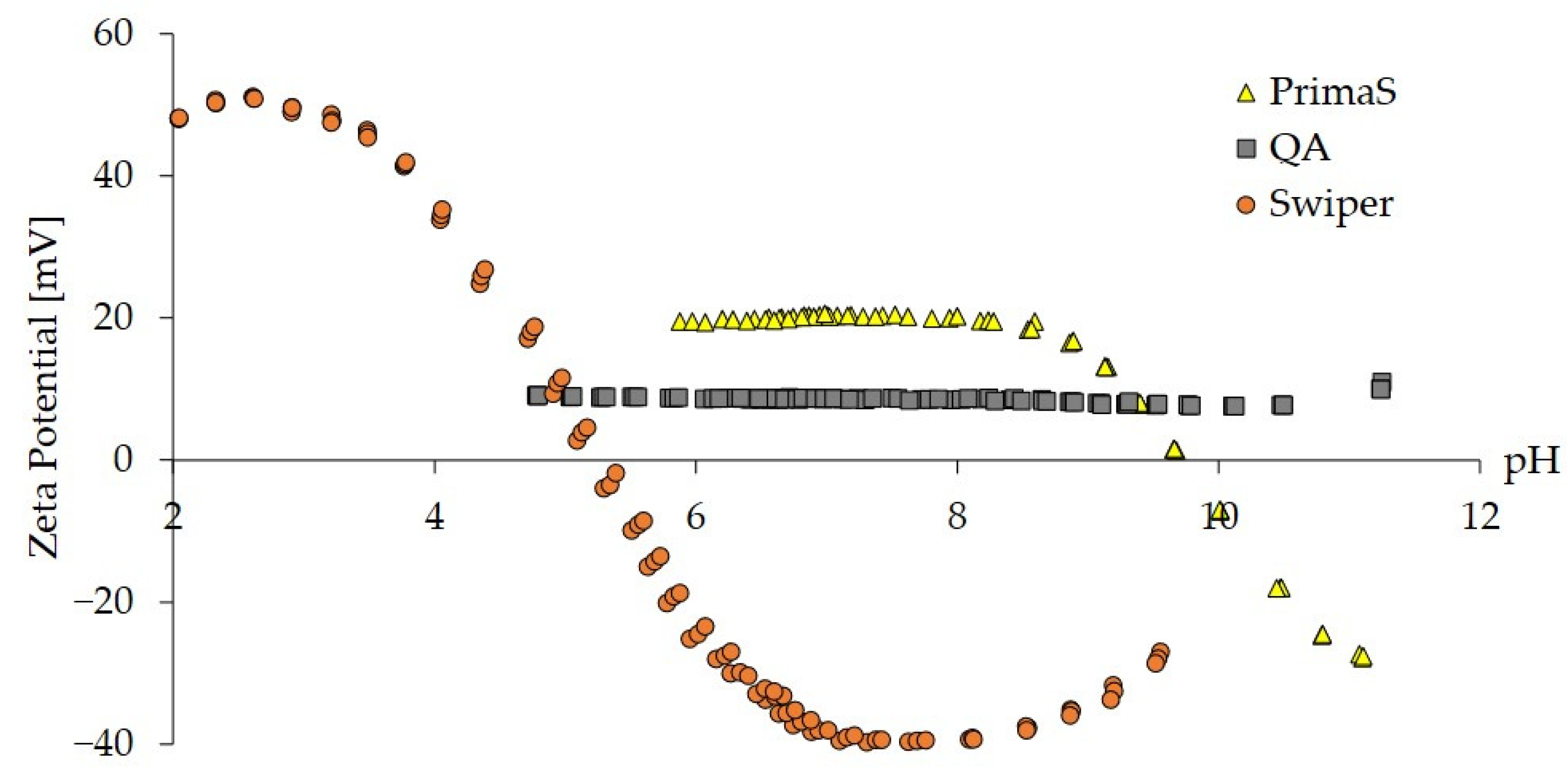
2.1. Zeta Potential Analysis of Monoliths
2.2. Chromatographic Evaluation of CIM Swiper for mRNA Separation
2.3. Optimization of mRNA Elution and mRNA–pDNA Separation
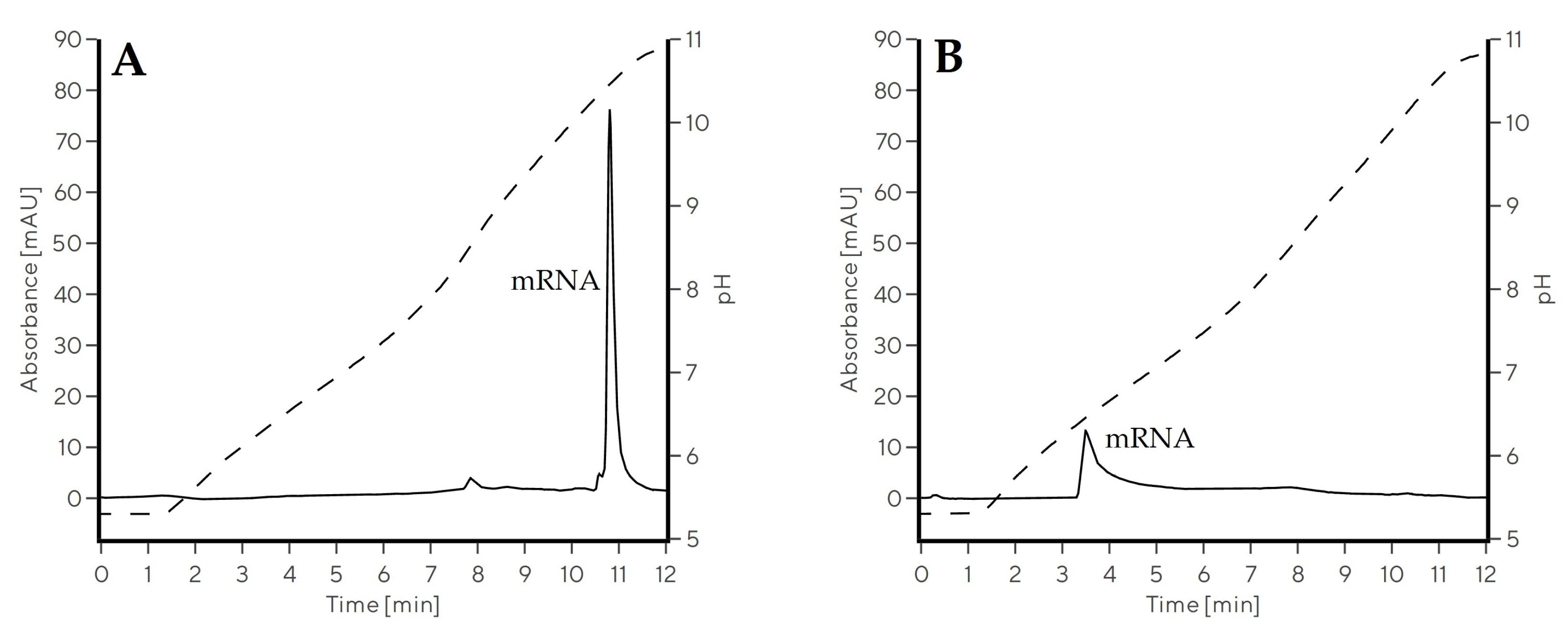
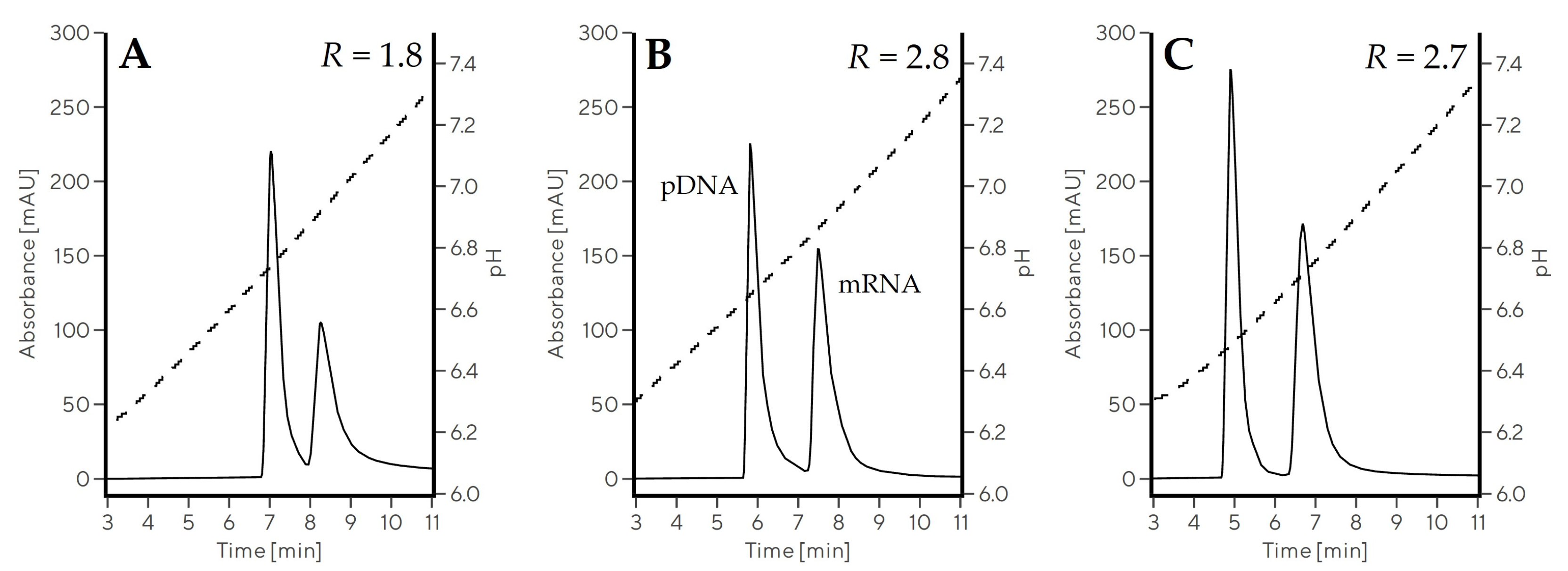
2.3.1. Optimization of the pH Gradient Approach
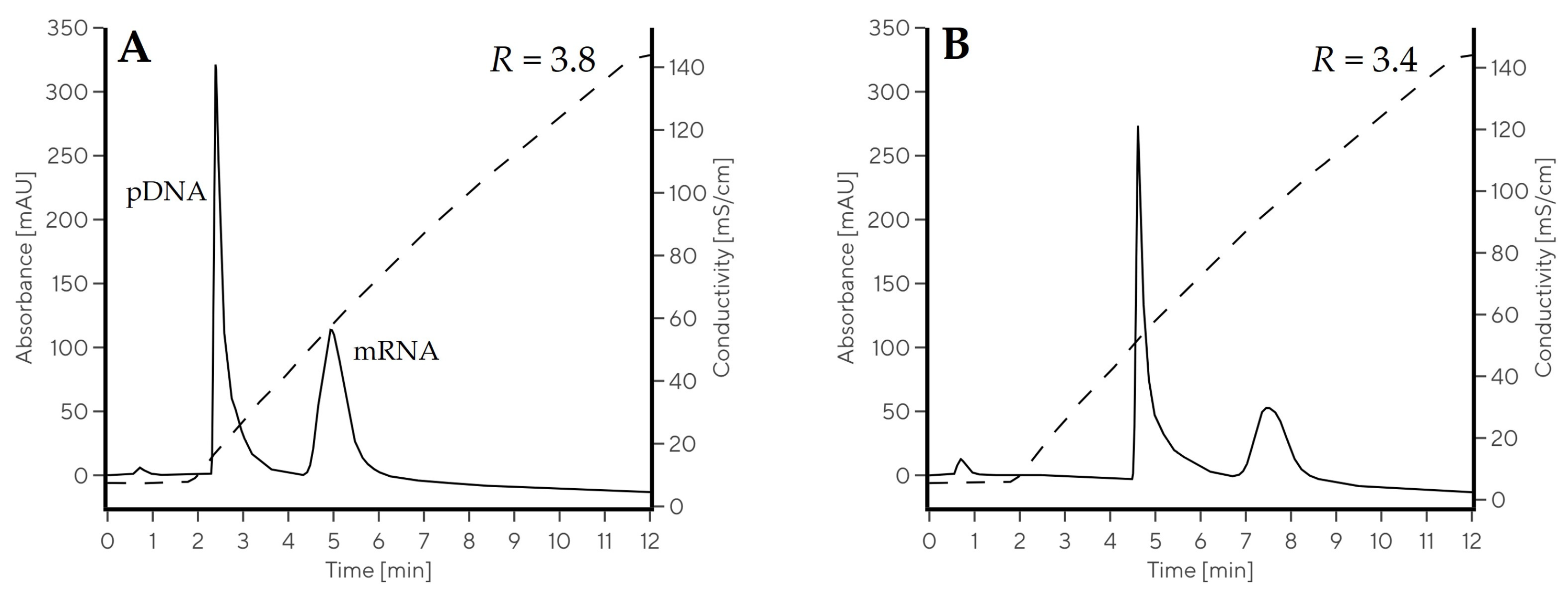
2.3.2. Evaluation of NaCl Gradient for Nucleic Acid Elution
2.3.3. Separation of ssRNAs up to 10,000 nt in Size
2.4. Chromatographic Purification of mRNA from IVT Reaction Mixture at Room Temperature
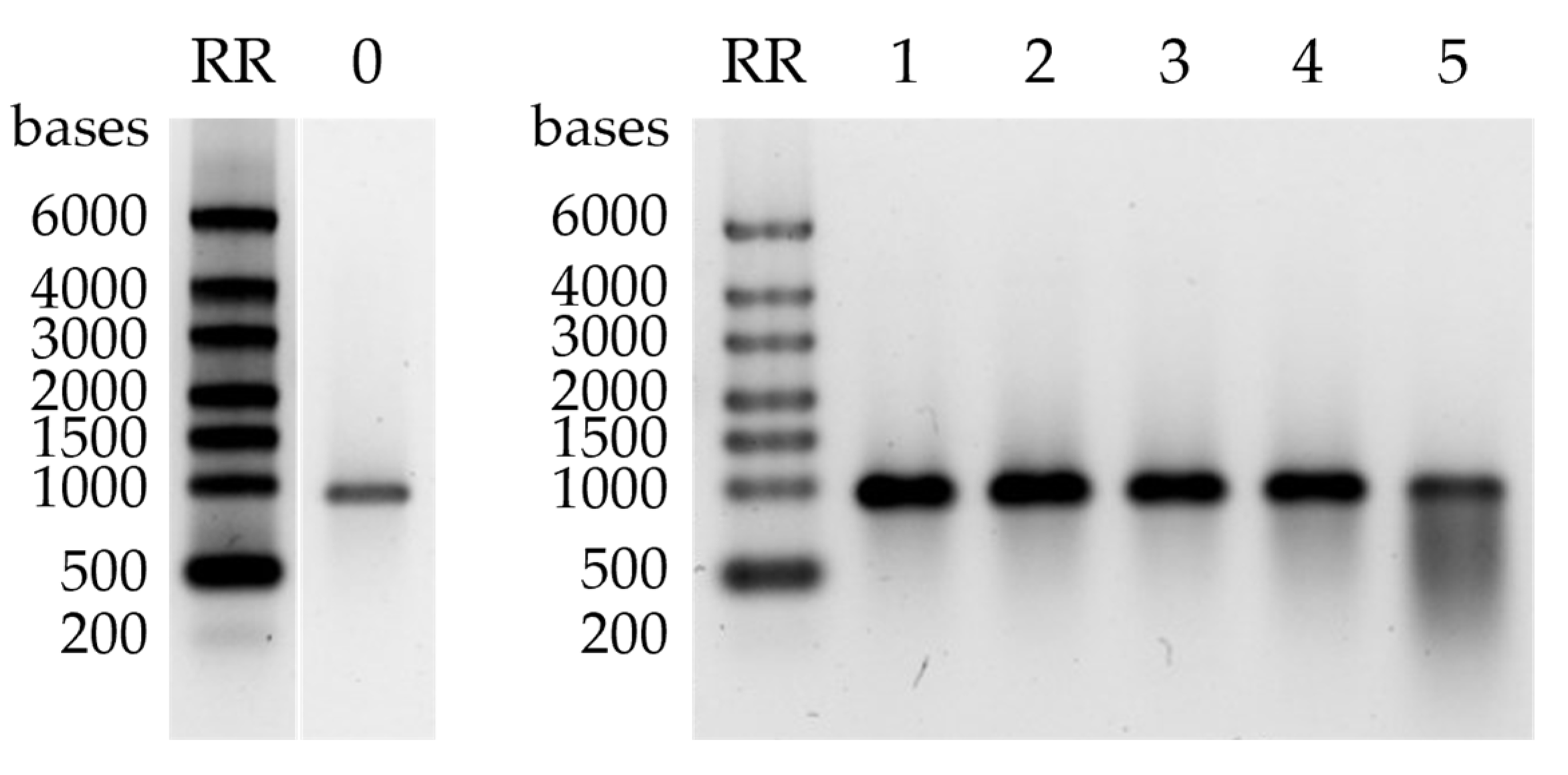
2.5. mRNA Stability Study
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Chromatograhic Columns
4.3. Zeta Potential Analysis of Monoliths
4.4. Preparation of Purified Nucleic Acids
4.5. Chromatography
4.5.1. Evaluation of CIMmic Disks in Ascending pH Gradients
4.5.2. Optimization of mRNA–pDNA Separation in pH and NaCl Gradients
4.5.3. Separation of ssRNAs of Different Sizes
4.5.4. Purification of mRNA from IVT Reaction Mixture
4.6. Nucleic Acids Analytics and Mass Balance Calculation
4.7. mRNA Stability Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AGE | agarose gel electrophoresis |
| BTP | Bis-TRIS propane |
| CAPS | 3-cyclohexyl-3-aminopropanesulfonic acid |
| CIM | convective interaction media |
| CIP | cleaning-in-place |
| CV | column volume |
| dsDNA | double-stranded DNA |
| E | elution |
| EDTA | ethylenediaminetetraacetic acid |
| EtOH | ethanol |
| FT | flow-through |
| HEPES | 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid |
| IEP | isoelectric point |
| IVT | in vitro transcription |
| mRNA | messenger ribonucleic acid |
| nt | nucleotides |
| NTP | nucleoside triphosphate |
| Oligo dT | oligo deoxythymidylic acid |
| pKa | acid dissociation constant |
| polyA | polyadenylic acid |
| pDNA | plasmid DNA |
| saRNA | self-amplifying RNA |
| ssRNA | single-stranded RNA |
| W | wash |
References
- Qin, S.; Tang, X.; Chen, Y.; Chen, K.; Fan, N.; Xiao, W.; Zheng, Q.; Li, G.; Teng, Y.; Wu, M.; et al. mRNA-Based Therapeutics: Powerful and Versatile Tools to Combat Diseases. Signal. Transduct. Target. Ther. 2022, 7, 166. [Google Scholar] [CrossRef]
- Rohner, E.; Yang, R.; Foo, K.S.; Goedel, A.; Chien, K.R. Unlocking the Promise of mRNA Therapeutics. Nat. Biotechnol. 2022, 40, 1586–1600. [Google Scholar] [CrossRef]
- Damase, T.R.; Sukhovershin, R.; Boada, C.; Taraballi, F.; Pettigrew, R.I.; Cooke, J.P. The Limitless Future of RNA Therapeutics. Front. Bioeng. Biotechnol. 2021, 9, 628137. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Zhou, S.; Dain, L.; Mei, L.; Zhu, G. Circular RNA: An Emerging Frontier in RNA Therapeutic Targets, RNA Therapeutics, and mRNA Vaccines. J. Control. Release 2022, 348, 84–94. [Google Scholar] [CrossRef]
- Skok, J.; Megušar, P.; Vodopivec, T.; Pregeljc, D.; Mencin, N.; Korenč, M.; Krušič, A.; Celjar, A.M.; Pavlin, N.; Krušič, J.; et al. Gram-Scale mRNA Production Using a 250-mL Single-Use Bioreactor. Chem. Ing. Tech. 2022, 94, 1928–1935. [Google Scholar] [CrossRef]
- Kavšček, M.; Stražar, M.; Curk, T.; Natter, K.; Petrovič, U. Yeast as a Cell Factory: Current State and Perspectives. Microb. Cell Fact. 2015, 14, 94. [Google Scholar] [CrossRef]
- Li, Y.; Breaker, R.R. Kinetics of RNA Degradation by Specific Base Catalysis of Transesterification Involving the 2′-Hydroxyl Group. J. Am. Chem. Soc. 1999, 121, 5364–5372. [Google Scholar] [CrossRef]
- Pogocki, D.; Schöneich, C. Chemical Stability of Nucleic Acid–Derived Drugs. J. Pharm. Sci. 2000, 89, 443–456. [Google Scholar] [CrossRef]
- Muralidhara, B.K.; Baid, R.; Bishop, S.M.; Huang, M.; Wang, W.; Nema, S. Critical Considerations for Developing Nucleic Acid Macromolecule Based Drug Products. Drug Discov. Today 2016, 21, 430–444. [Google Scholar] [CrossRef]
- Lemire, K.A.; Rodriguez, Y.Y.; McIntosh, M.T. Alkaline Hydrolysis to Remove Potentially Infectious Viral RNA Contaminants from DNA. Virol. J. 2016, 13, 88. [Google Scholar] [CrossRef]
- Wayment-Steele, H.K.; Kim, D.S.; Choe, C.A.; Nicol, J.J.; Wellington-Oguri, R.; Watkins, A.M.; Parra Sperberg, R.A.; Huang, P.S.; Participants, E.; Das, R. Theoretical Basis for Stabilizing Messenger RNA through Secondary Structure Design. Nucleic Acids Res. 2021, 49, 10604–10617. [Google Scholar] [CrossRef]
- Oivanen, M.; Kuusela, S.; Lönnberg, H. Kinetics and Mechanisms for the Cleavage and Isomerization of the Phosphodiester Bonds of RNA by Brønsted Acids and Bases. Chem. Rev. 1998, 98, 961–990. [Google Scholar] [CrossRef]
- Tanaka, M.; Chock, P.B. Oxidative Modifications of RNA and Its Potential Roles in Biosystem. Front. Mol. Biosci. 2021, 8, 685331. [Google Scholar] [CrossRef]
- Abdulrahman, A.; Ghanem, A. Recent Advances in Chromatographic Purification of Plasmid DNA for Gene Therapy and DNA Vaccines: A Review. Anal. Chim. Acta 2018, 1025, 41–57. [Google Scholar] [CrossRef]
- Korenč, M.; Mencin, N.; Puc, J.; Skok, J.; Nemec, K.Š.; Celjar, A.M.; Gagnon, P.; Štrancar, A.; Sekirnik, R. Chromatographic Purification with CIMmultusTM Oligo DT Increases mRNA Stability. Cell Gene Ther. Insights 2021, 7, 1207–1216. [Google Scholar] [CrossRef]
- Svec, F. Monolithic Columns: A Historical Overview. Electrophoresis 2017, 38, 2810–2820. [Google Scholar] [CrossRef]
- Gagnon, P.; Goričar, B.; Peršič, Š.; Černigoj, U.; Štrancar, A. Two New Capture Options for Improved Purification of Large mRNA. Cell Gene Ther. Insights 2020, 6, 1035–1046. [Google Scholar] [CrossRef]
- Mencin, N.; Štepec, D.; Margon, A.; Vidič, J.; Dolenc, D.; Simčič, T.; Rotar, S.; Sekirnik, R.; Štrancar, A.; Černigoj, U. Development and Scale-up of Oligo-DT Monolithic Chromatographic Column for mRNA Capture through Understanding of Base-Pairing Interactions. Sep. Purif. Technol. 2023, 304, 122320. [Google Scholar] [CrossRef]
- Megušar, P.; Miklavčič, R.; Korenč, M.; Ličen, J.; Vodopivec, T.; Černigoj, U.; Štrancar, A.; Sekirnik, R. Scalable Multimodal Weak Anion Exchange Chromatographic Purification for Stable mRNA Drug Substance. Electrophoresis 2023, in press. [Google Scholar]
- Feng, X.; Su, Z.; Cheng, Y.; Ma, G.; Zhang, S. Messenger RNA Chromatographic Purification: Advances and Challenges. J. Chromatogr. A 2023, 1707, 464321. [Google Scholar] [CrossRef]
- Issa, W.J.; Barberio, J.L.; Aunins, J.G.; Afeyan, N.B. Ion Exchange Purification of mRNA. U.S. Patent 10,590,161 B2, 17 March 2014. [Google Scholar]
- Kanavarioti, A. HPLC Methods for Purity Evaluation of Man-Made Single-Stranded RNAs. Sci. Rep. 2019, 9, 1019. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, P. Purification of Nucleic Acids: A Handbook for Purification of Plasmid DNA and mRNA for Gene Therapy and Vaccines; BIA Separations: Ajdovščina, Slovenia, 2020. [Google Scholar]
- Karikó, K.; Muramatsu, H.; Ludwig, J.; Weissman, D. Generating the Optimal mRNA for Therapy: HPLC Purification Eliminates Immune Activation and Improves Translation of Nucleoside-Modified, Protein-Encoding mRNA. Nucleic Acids Res. 2011, 39, e142. [Google Scholar] [CrossRef] [PubMed]
- Weissman, D.; Pardi, N.; Muramatsu, H.; Karikó, K. HPLC Purification of in Vitro Transcribed Long RNA. Methods Mol. Biol. 2013, 969, 43–54. [Google Scholar] [PubMed]
- Grinsted, J.; Liddell, J.; Bouleghlimat, E.; Kwok, K.Y.; Taylor, G.; Marques, M.P.C.; Bracewell, D.G. Purification of Therapeutic & Prophylactic mRNA by Affinity Chromatography. Cell Gene Ther. Insights 2022, 8, 335–349. [Google Scholar]
- Pregeljc, D.; Skok, J.; Vodopivec, T.; Mencin, N.; Krušič, A.; Ličen, J.; Nemec, K.Š.; Štrancar, A.; Sekirnik, R.; Bia, S. Increasing Yield of in Vitro Transcription Reaction with At-Line High Pressure Liquid Chromatography Monitoring. Biotechnol. Bioeng. 2023, 120, 737–747. [Google Scholar] [CrossRef]
- Park, J.-W.; Choi, K.H.; Koh Park, K.-H. Acid-Base Equilibria and Related Properites of Chitosan. Bull. Korean Chem. Soc. 1983, 4, 68–72. [Google Scholar]
- Wang, Q.Z.; Chen, X.G.; Liu, N.; Wang, S.X.; Liu, C.S.; Meng, X.H.; Liu, C.G. Protonation Constants of Chitosan with Different Molecular Weight and Degree of Deacetylation. Carbohydr. Polym. 2006, 65, 194–201. [Google Scholar] [CrossRef]
- Cao, W.; Easley, C.J.; Ferrance, J.P.; Landers, J.P. Chitosan as a Polymer for PH-Induced DNA Capture in a Totally Aqueous System. Anal. Chem. 2006, 78, 7222–7228. [Google Scholar] [CrossRef]
- Hagan, K.A.; Meier, W.L.; Ferrance, J.P.; Landers, J.P. Chitosan-Coated Silica as a Solid Phase for RNA Purification in a Microfluidic Device. Anal. Chem. 2009, 81, 5249–5256. [Google Scholar] [CrossRef]
- Baker, M.J. Isolation of Nucleic Acids. U.S. Patent 6914137B2, 5 July 2005. [Google Scholar]
- Luxbacher, T. The Zeta Potential for Solid Surface Analysis, 1st ed.; Anton Paar GmbH: Graz, Austria, 2014. [Google Scholar]
- Fievet, P.; Szymczyk, A.; Labbez, C.; Aoubiza, B.; Simon, C.; Foissy, A.; Pagetti, J. Determining the Zeta Potential of Porous Membranes Using Electrolyte Conductivity inside Pores. J. Colloid. Interface Sci. 2001, 235, 383–390. [Google Scholar] [CrossRef]
- Buszewski, B.; Jaćkowska, M.; Bocian, S.; Dziubakiewicz, E. Application of the Zeta Potential for Stationary Phase Characterization in Ion Chromatography. J. Sep. Sci. 2013, 36, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Urthaler, J.; Schlegl, R.; Podgornik, A.; Strancar, A.; Jungbauer, A.; Necina, R. Application of Monoliths for Plasmid DNA Purification Development and Transfer to Production. J. Chromatogr. A 2005, 1065, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Černigoj, U.; Štrancar, A. Scale-Up of Plasmid DNA Downstream Process Based on Chromatographic Monoliths. Methods Mol. Biol. 2021, 2197, 167–192. [Google Scholar] [PubMed]
- Tan, Z.J.; Chen, S.J. Salt Contribution to RNA Tertiary Structure Folding Stability. Biophys. J. 2011, 101, 176. [Google Scholar] [CrossRef]
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Iviglia, G.; Morra, M.; Cassinelli, C.; Torre, E.; Rodriguez, Y.; Baena, R. New Collagen-Coated Calcium Phosphate Synthetic Bone Filler (Synergoss®): A Comparative Surface Analysis. Int. J. Appl. Ceram. Technol. 2018, 15, 910–920. [Google Scholar] [CrossRef]

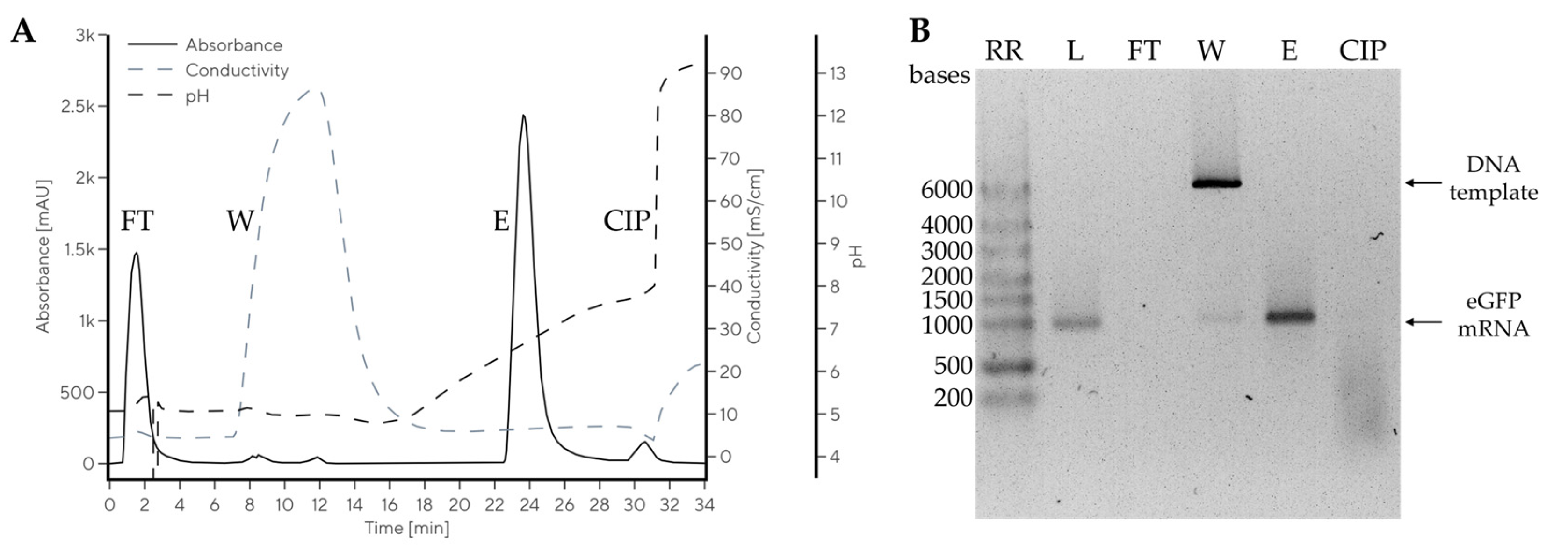
| Gradient | Additive | Conductivity (mS/cm) | Elution pH of mRNA | ΔpHmRNA-pDNA | RmRNA-pDNA |
|---|---|---|---|---|---|
| Gradient 2 | / | 1 | 7.03 | 0.14 | 0.4 |
| Gradient 3 | NaCl | 6 | 6.74 | 0.24 | 2.3 |
| Gradient 4 | Na-phosphate | 8 | 6.37 | 0.29 | 3.1 |
| Gradient 5 | Na-citrate | 11 | 6.56 | 0.37 | 5.9 |
| Gradient 6 | Na-EDTA | 11 | 6.65 | 0.29 | 5.8 |
| Gradient | Elution Type | w0.5 pDNA (min) | w0.5 mRNA (min) | RmRNA-pDNA |
|---|---|---|---|---|
| Gradient 7 | pH | 0.32 | 0.50 | 1.8 |
| Gradient 8 | pH | 0.30 | 0.41 | 2.8 |
| Gradient 9 | pH | 0.31 | 0.47 | 2.7 |
| Gradient 10 | salt | 0.17 | 0.62 | 3.8 |
| Gradient 11 | salt | 0.17 | 0.83 | 3.4 |
| Sample | ssRNA Size (nt) | DNA Size (kbp) | RssRNA-DNA | ssRNA Recovery (%) |
|---|---|---|---|---|
| eGFP mRNA | 995 nt | 3.3 | 2.5 | 95 |
| mFIX mRNA | 4000 nt | 6.6 | 2.8 | 91 |
| saRNA | 10,000 nt | 12.0 | 2.8 | 80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miklavčič, R.; Megušar, P.; Kodermac, Š.M.; Bakalar, B.; Dolenc, D.; Sekirnik, R.; Štrancar, A.; Černigoj, U. High Recovery Chromatographic Purification of mRNA at Room Temperature and Neutral pH. Int. J. Mol. Sci. 2023, 24, 14267. https://doi.org/10.3390/ijms241814267
Miklavčič R, Megušar P, Kodermac ŠM, Bakalar B, Dolenc D, Sekirnik R, Štrancar A, Černigoj U. High Recovery Chromatographic Purification of mRNA at Room Temperature and Neutral pH. International Journal of Molecular Sciences. 2023; 24(18):14267. https://doi.org/10.3390/ijms241814267
Chicago/Turabian StyleMiklavčič, Rok, Polona Megušar, Špela Meta Kodermac, Blaž Bakalar, Darko Dolenc, Rok Sekirnik, Aleš Štrancar, and Urh Černigoj. 2023. "High Recovery Chromatographic Purification of mRNA at Room Temperature and Neutral pH" International Journal of Molecular Sciences 24, no. 18: 14267. https://doi.org/10.3390/ijms241814267
APA StyleMiklavčič, R., Megušar, P., Kodermac, Š. M., Bakalar, B., Dolenc, D., Sekirnik, R., Štrancar, A., & Černigoj, U. (2023). High Recovery Chromatographic Purification of mRNA at Room Temperature and Neutral pH. International Journal of Molecular Sciences, 24(18), 14267. https://doi.org/10.3390/ijms241814267






