Serum Pentraxin 3 as Promising Biomarker for the Long-Lasting Inflammatory Response of COVID-19
Abstract
:1. Introduction
2. Results
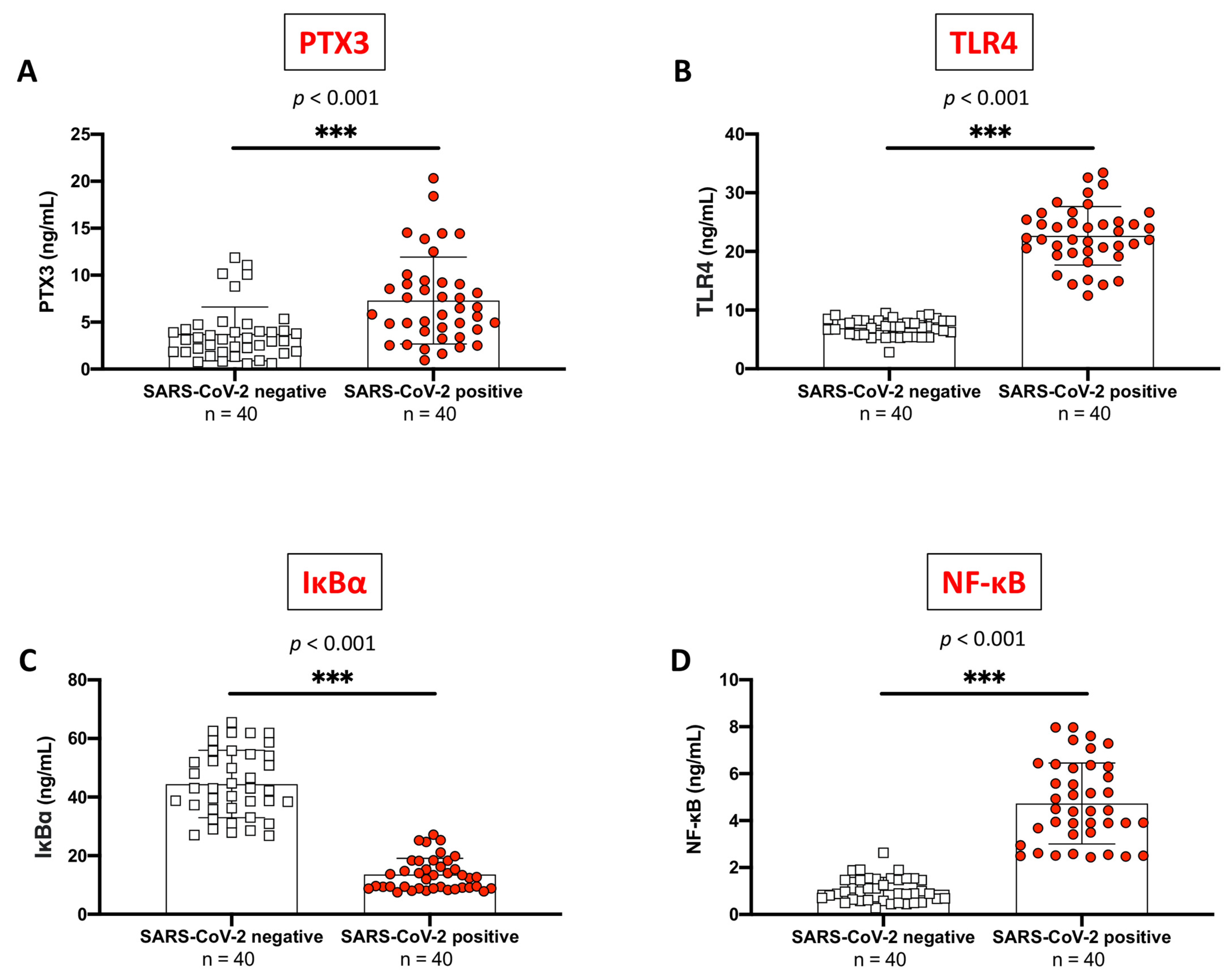
2.1. PTX3 Sustained Inflammation in SARS-CoV-2 Positive Subjects Compared to Never-Infected Individuals
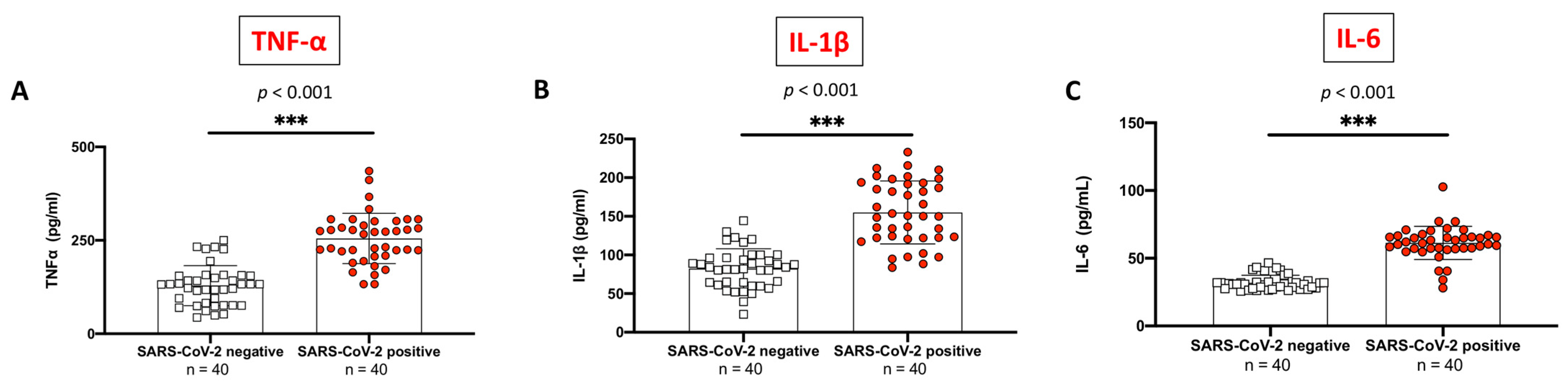
2.2. Evaluation of Pro-Inflammatory Cytokines Profile between SARS-CoV-2 Positive Subjects Compared to Non-Infected Control Individuals
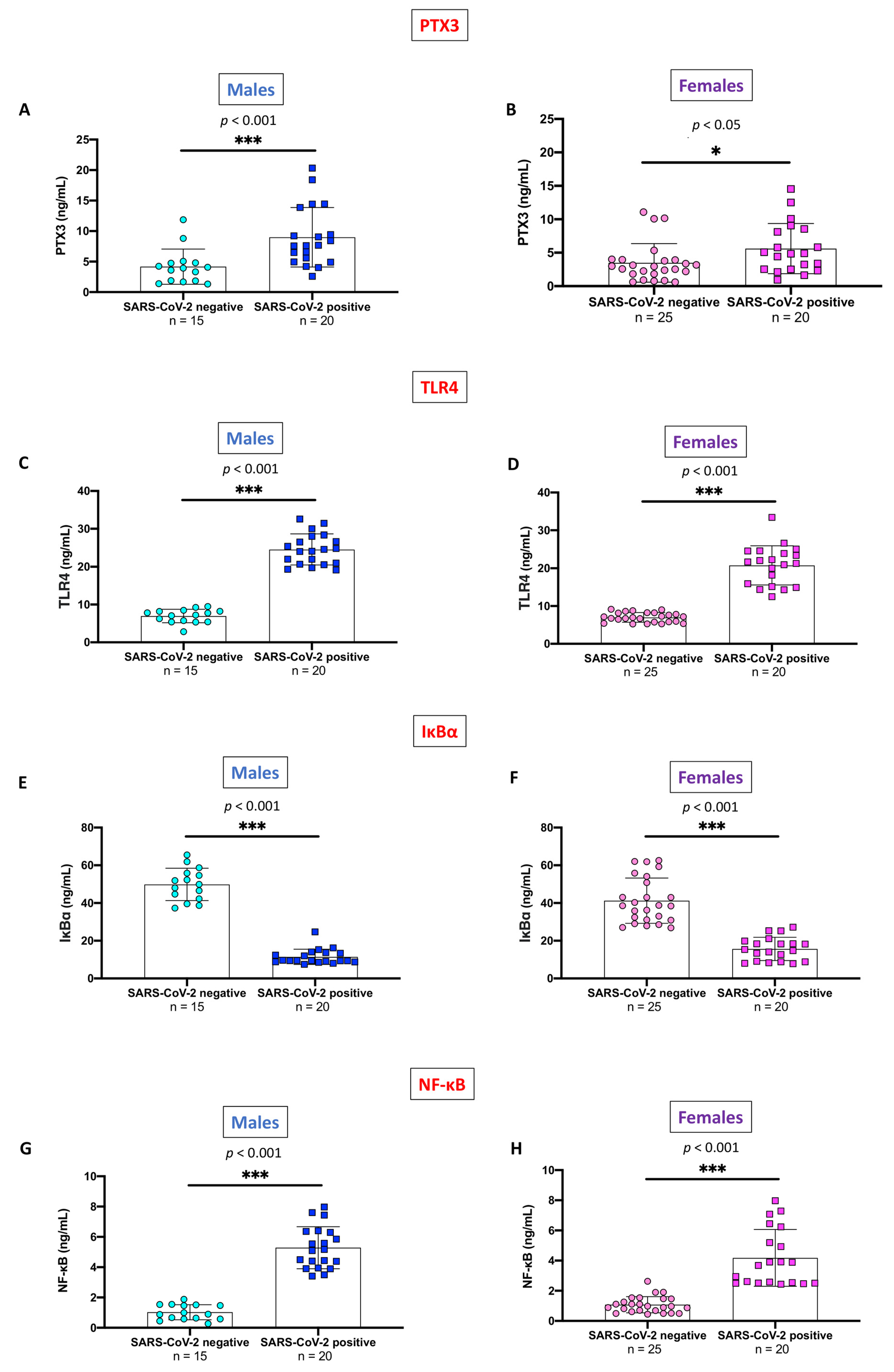
2.3. Evaluation of PTX3 and TLR4/NF-κB Axis, Stratifying SARS-CoV-2 Positive Subjects and Non-Infected Control Individuals by Gender
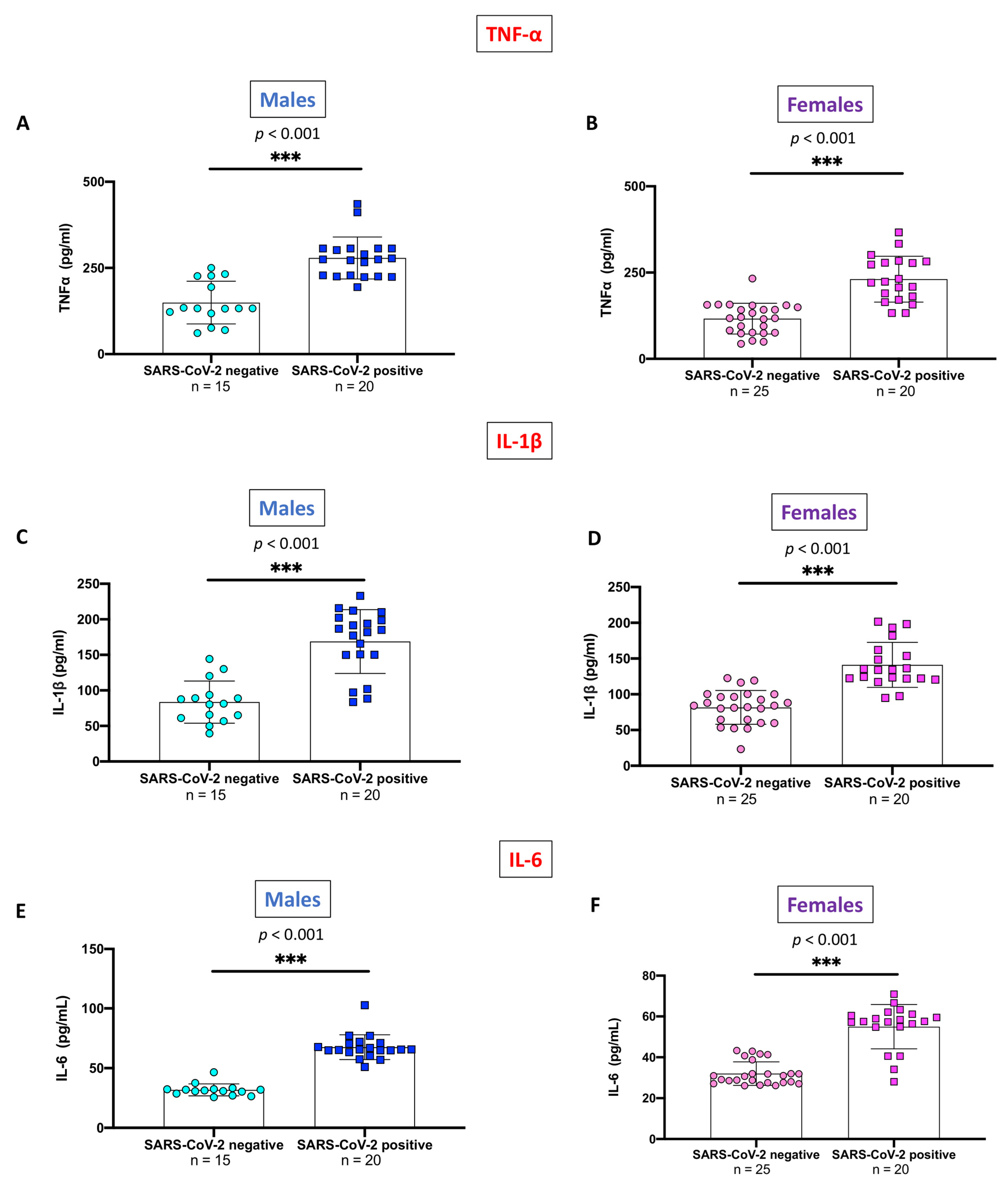
2.4. Evaluation of Pro-Inflammatory Cytokines Stratifying SARS-CoV-2 Positive Subjects and Non-Infected Control Individuals by Gender
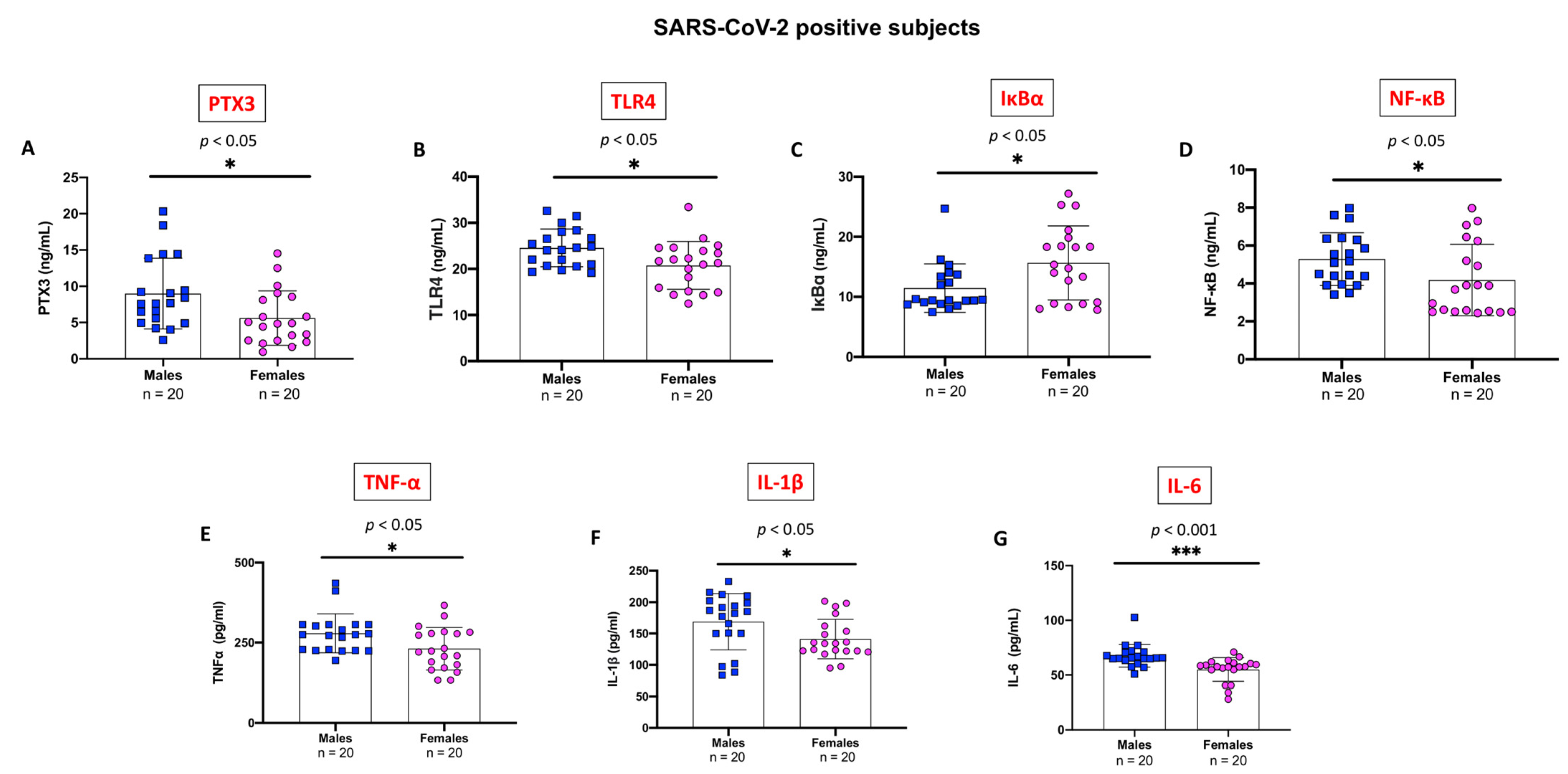
2.5. Evaluation of Pro-Inflammatory Markers Stratifying SARS-CoV-2 Positive Subjects by Gender
3. Discussion
4. Materials and Methods
4.1. Enrolled Subjects
4.2. ELISA Kit
4.3. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pak, A.; Adegboye, O.A.; Adekunle, A.I.; Rahman, K.M.; McBryde, E.S.; Eisen, D.P. Economic Consequences of the COVID-19 Outbreak: The Need for Epidemic Preparedness. Front. Public Health 2020, 8, 241. [Google Scholar] [CrossRef] [PubMed]
- Msemburi, W.; Karlinsky, A.; Knutson, V.; Aleshin-Guendel, S.; Chatterji, S.; Wakefield, J. The WHO estimates of excess mortality associated with the COVID-19 pandemic. Nature 2023, 613, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Kang, L.; Guo, Z.; Liu, J.; Liu, M.; Liang, W. Incubation Period of COVID-19 Caused by Unique SARS-CoV-2 Strains: A Systematic Review and Meta-analysis. JAMA Netw. Open 2022, 5, e2228008. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Liu, J.; Liu, Q.; Kang, L.; Liu, R.; Jing, W.; Wu, Y.; Liu, M. Global Percentage of Asymptomatic SARS-CoV-2 Infections among the Tested Population and Individuals with Confirmed COVID-19 Diagnosis: A Systematic Review and Meta-analysis. JAMA Netw. Open 2021, 4, e2137257. [Google Scholar] [CrossRef] [PubMed]
- Shah, V.K.; Firmal, P.; Alam, A.; Ganguly, D.; Chattopadhyay, S. Overview of Immune Response during SARS-CoV-2 Infection: Lessons from the Past. Front. Immunol. 2020, 11, 1949. [Google Scholar] [CrossRef]
- Wong, R.S.Y. Inflammation in COVID-19: From pathogenesis to treatment. Int. J. Clin. Exp. Pathol. 2021, 14, 831–844. [Google Scholar]
- Mangalmurti, N.; Hunter, C.A. Cytokine Storms: Understanding COVID-19. Immunity 2020, 53, 19–25. [Google Scholar] [CrossRef]
- Raveendran, A.V.; Jayadevan, R.; Sashidharan, S. Long COVID: An overview. Diabetes Metab. Syndr. 2021, 15, 869–875. [Google Scholar] [CrossRef]
- Lopez-Leon, S.; Wegman-Ostrosky, T.; Perelman, C.; Sepulveda, R.; Rebolledo, P.A.; Cuapio, A.; Villapol, S. More than 50 long-term effects of COVID-19: A systematic review and meta-analysis. Sci. Rep. 2021, 11, 16144. [Google Scholar] [CrossRef]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef]
- Low, R.N.; Low, R.J.; Akrami, A. A review of cytokine-based pathophysiology of Long COVID symptoms. Front. Med. 2023, 10, 1011936. [Google Scholar] [CrossRef] [PubMed]
- Schultheiß, C.; Willscher, E.; Paschold, L.; Gottschick, C.; Klee, B.; Henkes, S.-S.; Bosurgi, L.; Dutzmann, J.; Sedding, D.; Frese, T. The IL-1β, IL-6, and TNF cytokine triad is associated with post-acute sequelae of COVID-19. Cell Rep. Med. 2022, 3, 100663. [Google Scholar] [CrossRef] [PubMed]
- Phetsouphanh, C.; Darley, D.R.; Wilson, D.B.; Howe, A.; Munier, C.M.L.; Patel, S.K.; Juno, J.A.; Burrell, L.M.; Kent, S.J.; Dore, G.J.; et al. Immunol.ogical dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat. Immunol. 2022, 23, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Loncharich, M.; Klusewitz, S.; Jones, O. Post-COVID-19 Multisystem Inflammatory Syndrome in Children and Adults: What Happens After Discharge? Cureus 2022, 14, e24438. [Google Scholar] [CrossRef]
- Guo, J.; Wang, S.; Xia, H.; Shi, D.; Chen, Y.; Zheng, S.; Chen, Y.; Gao, H.; Guo, F.; Ji, Z.; et al. Cytokine SigNat.ure Associated with Disease Severity in COVID-19. Front. Immunol. 2021, 12, 681516. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Zeng, Z.; Yu, H.; Chen, H.; Qi, W.; Chen, L.; Chen, G.; Yan, W.; Chen, T.; Ning, Q.; Han, M. Longitudinal changes of inflammatory parameters and their correlation with disease severity and outcomes in patients with COVID-19 from Wuhan, China. Crit. Care 2020, 24, 525. [Google Scholar] [CrossRef]
- Brunetta, E.; Folci, M.; Bottazzi, B.; De Santis, M.; Gritti, G.; Protti, A.; Mapelli, S.N.; Bonovas, S.; Piovani, D.; Leone, R.; et al. Macrophage expression and prognostic significance of the long pentraxin PTX3 in COVID-19. Nat. Immunol. 2021, 22, 19–24. [Google Scholar] [CrossRef]
- Lapadula, G.; Leone, R.; Bernasconi, D.P.; Biondi, A.; Rossi, E.; D’Angiò, M.; Bottazzi, B.; Bettini, L.R.; Beretta, I.; Garlanda, C. Long pentraxin 3 (PTX3) levels predict death, intubation and thrombotic events among hospitalized patients with COVID-19. Front. Immunol. 2022, 13, 933960. [Google Scholar] [CrossRef]
- Bottazzi, B.; Doni, A.; Garlanda, C.; Mantovani, A. An integrated view of humoral innate immunity: Pentraxins as a paradigm. Annu. Rev. Immunol. 2009, 28, 157–183. [Google Scholar] [CrossRef]
- Jaillon, S.; Bonavita, E.; Gentile, S.; Rubino, M.; Laface, I.; Garlanda, C.; Mantovani, A. The long pentraxin PTX3 as a key component of humoral inNat.e immunity and a candidate diagnostic for inflammatory diseases. Int. Arch. Allergy Immunol. 2015, 165, 165–178. [Google Scholar] [CrossRef]
- Bottazzi, B.; Inforzato, A.; Messa, M.; Barbagallo, M.; Magrini, E.; Garlanda, C.; Mantovani, A. The pentraxins PTX3 and SAP in inNat.e immunity, regulation of inflammation and tissue remodelling. J. Hepatol. 2016, 64, 1416–1427. [Google Scholar] [CrossRef] [PubMed]
- Porte, R.; Davoudian, S.; Asgari, F.; Parente, R.; Mantovani, A.; Garlanda, C.; Bottazzi, B. The long pentraxin PTX3 as a humoral inNat.e immunity functional player and biomarker of infections and sepsis. Front. Immunol. 2019, 10, 794. [Google Scholar] [CrossRef] [PubMed]
- Doni, A.; Mantovani, A.; Bottazzi, B.; Russo, R.C. PTX3 Regulation of Inflammation, Hemostatic Response, Tissue Repair, and Resolution of Fibrosis Favors a Role in Limiting Idiopathic Pulmonary Fibrosis. Front. Immunol. 2021, 12, 676702. [Google Scholar] [CrossRef] [PubMed]
- Daigo, K.; Mantovani, A.; Bottazzi, B. The yin-yang of long pentraxin PTX3 in inflammation and immunity. Immunol. Lett 2014, 161, 38–43. [Google Scholar] [CrossRef]
- Doni, A.; Stravalaci, M.; Inforzato, A.; Magrini, E.; Mantovani, A.; Garlanda, C.; Bottazzi, B. The long pentraxin PTX3 as a link between innate immunity, tissue remodeling, and cancer. Front. Immunol. 2019, 10, 712. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.J.; Garred, P. Pentraxins in Complement Activation and Regulation. Front. Immunol. 2018, 9, 3046. [Google Scholar] [CrossRef]
- Rathore, M.; Girard, C.; Ohanna, M.; Tichet, M.; Ben Jouira, R.; Garcia, E.; Larbret, F.; Gesson, M.; Audebert, S.; Lacour, J.P.; et al. Cancer cell-derived long pentraxin 3 (PTX3) promotes melanoma migration through a toll-like receptor 4 (TLR4)/NF-kappaB signaling pathway. Oncogene 2019, 38, 5873–5889. [Google Scholar] [CrossRef]
- Garlanda, C.; Bottazzi, B.; Magrini, E.; Inforzato, A.; Mantovani, A. PTX3, a Humoral Pattern Recognition Molecule, in InNat.e Immunity, Tissue Repair, and Cancer. Physio.l Rev. 2018, 98, 623–639. [Google Scholar] [CrossRef]
- Genc, A.B.; Yaylaci, S.; Dheir, H.; Genc, A.C.; Issever, K.; Cekic, D.; Kocayigit, H.; Cokluk, E.; Karacan, A.; Sekeroglu, M.R.; et al. The predictive and diagnostic accuracy of long pentraxin-3 in COVID-19 pneumonia. Turk. J. Med. Sci. 2021, 51, 448–453. [Google Scholar] [CrossRef]
- Capra, A.P.; Ardizzone, A.; Panto, G.; Paterniti, I.; Campolo, M.; Crupi, L.; Squeri, R.; Esposito, E. The Prognostic Value of Pentraxin-3 in COVID-19 Patients: A Systematic Review and Meta-Analysis of Mortality Incidence. Int. J. Mol. Sci. 2023, 24, 3537. [Google Scholar] [CrossRef]
- Mukherjee, S. Toll-like receptor 4 in COVID-19: Friend or foe? Future Virol. 2022, 17, 415–417. [Google Scholar] [CrossRef]
- Brandao, S.C.S.; Ramos, J.d.O.X.; Dompieri, L.T.; Godoi, E.T.A.M.; Figueiredo, J.L.; Sarinho, E.S.C.; Chelvanambi, S.; Aikawa, M. Is Toll-like receptor 4 involved in the severity of COVID-19 pathology in patients with cardiometabolic comorbidities? Cytokine Growth Factor Rev. 2021, 58, 102–110. [Google Scholar] [CrossRef]
- Ciarambino, T.; Para, O.; Giordano, M. Immune system and COVID-19 by sex differences and age. Womens Health 2021, 17, 17455065211022262. [Google Scholar] [CrossRef]
- Pela, G.; Goldoni, M.; Solinas, E.; Cavalli, C.; Tagliaferri, S.; Ranzieri, S.; Frizzelli, A.; Marchi, L.; Mori, P.A.; Majori, M.; et al. Sex-Related Differences in Long-COVID-19 Syndrome. J. Women’s Health 2022, 31, 620–630. [Google Scholar] [CrossRef]
- Qi, S.; Ngwa, C.; Morales Scheihing, D.A.; Al Mamun, A.; Ahnstedt, H.W.; Finger, C.E.; Colpo, G.D.; Sharmeen, R.; Kim, Y.; Choi, H.A.; et al. Sex differences in the immune response to acute COVID-19 respiratory tract infection. Biol. Sex Differ. 2021, 12, 66. [Google Scholar] [CrossRef]
- Yang, H.; Rao, Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021, 19, 685–700. [Google Scholar] [CrossRef]
- Ardizzone, A.; Capra, A.P.; Campolo, M.; Filippone, A.; Esposito, E.; Briuglia, S. Neurofibromatosis: New Clinical Challenges in the Era of COVID-19. Biomedicines 2022, 10, 940. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Huang, S.; Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 2021, 93, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Montero, M.T.V.; Rowe, K.; Kirton, R.; Kunik, F., Jr. Epidemiology, pathogenesis, clinical presentations, diagnosis and treatment of COVID-19: A review of current evidence. Expert. Rev. Clin. Pharmacol. 2021, 14, 601–621. [Google Scholar] [CrossRef] [PubMed]
- Castanares-Zapatero, D.; Chalon, P.; Kohn, L.; Dauvrin, M.; Detollenaere, J.; Maertens de Noordhout, C.; Primus-de Jong, C.; Cleemput, I.; Van den Heede, K. Pathophysiology and mechanism of long COVID: A comprehensive review. Ann. Med. 2022, 54, 1473–1487. [Google Scholar] [CrossRef] [PubMed]
- Bozza, S.; Bistoni, F.; Gaziano, R.; Pitzurra, L.; Zelante, T.; Bonifazi, P.; Perruccio, K.; Bellocchio, S.; Neri, M.; Iorio, A.M.; et al. Pentraxin 3 protects from MCMV infection and reactivation through TLR sensing pathways leading to IRF3 activation. Blood 2006, 108, 3387–3396. [Google Scholar] [CrossRef]
- Reading, P.C.; Bozza, S.; Gilbertson, B.; Tate, M.; Moretti, S.; Job, E.R.; Crouch, E.C.; Brooks, A.G.; Brown, L.E.; Bottazzi, B.; et al. Antiviral activity of the long chain pentraxin PTX3 against influenza viruses. J. Immunol. 2008, 180, 3391–3398. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Ma, X.; Zhang, J.; Zhang, Y.; Bai, X.; Hwang, D.M.; Keshavjee, S.; Levy, G.A.; McGilvray, I.; Liu, M. Protective effects of long pentraxin PTX3 on lung injury in a severe acute respiratory syndrome model in mice. Lab. Investig. 2012, 92, 1285–1296. [Google Scholar] [CrossRef]
- Stravalaci, M.; Pagani, I.; Paraboschi, E.M.; Pedotti, M.; Doni, A.; Scavello, F.; Mapelli, S.N.; Sironi, M.; Perucchini, C.; Varani, L.; et al. Recognition and inhibition of SARS-CoV-2 by humoral inNat.e immunity pattern recognition molecules. Nat. Immunol. 2022, 23, 275–286. [Google Scholar] [CrossRef]
- Gutmann, C.; Takov, K.; Burnap, S.A.; Singh, B.; Ali, H.; Theofilatos, K.; Reed, E.; Hasman, M.; Nabeebaccus, A.; Fish, M.; et al. SARS-CoV-2 RNAemia and proteomic trajectories inform prognostication in COVID-19 patients admitted to intensive care. Nat. Commun. 2021, 12, 3406. [Google Scholar] [CrossRef]
- Hansen, C.B.; Sandholdt, H.; Moller, M.E.E.; Perez-Alos, L.; Pedersen, L.; Bastrup Israelsen, S.; Garred, P.; Benfield, T. Prediction of Respiratory Failure and Mortality in COVID-19 Patients Using Long Pentraxin PTX3. J. Innate Immun. 2022, 14, 493–501. [Google Scholar] [CrossRef]
- Chaturvedi, R.; Lui, B.; Aaronson, J.A.; White, R.S.; Samuels, J.D. COVID-19 complications in males and females: Recent developments. J. Comp. Eff. Res. 2022, 11, 689–698. [Google Scholar] [CrossRef]
- Jin, J.M.; Bai, P.; He, W.; Wu, F.; Liu, X.F.; Han, D.M.; Liu, S.; Yang, J.K. Gender Differences in Patients with COVID-19: Focus on Severity and Mortality. Front. Public Health 2020, 8, 152. [Google Scholar] [CrossRef]
- Muñoz-Wolf, N.; Lavelle, E.C. Innate immune receptors. In NLR Proteins: Methods and Protocols: Methods in Molecular Biology; Di Virgilio, F., Pelegrín, P., Eds.; Humana Press: New York, NY, USA, 2016; Volume 1417, pp. 1–43. [Google Scholar]
- Ardizzone, A.; Filippone, A.; Mannino, D.; Scuderi, S.A.; Casili, G.; Lanza, M.; Cucinotta, L.; Campolo, M.; Esposito, E. Ulva pertusa, a Marine Green Alga, Attenuates DNBS-Induced Colitis Damage via NF-kappaB/Nrf2/SIRT1 Signaling Pathways. J. Clin. Med. 2022, 11, 4301. [Google Scholar] [CrossRef]
- Campolo, M.; Lanza, M.; Paterniti, I.; Filippone, A.; Ardizzone, A.; Casili, G.; Scuderi, S.A.; Puglisi, C.; Mare, M.; Memeo, L.; et al. PEA-OXA Mitigates Oxaliplatin-Induced Painful Neuropathy through NF-kappaB/Nrf-2 Axis. Int. J. Mol. Sci. 2021, 22, 3927. [Google Scholar] [CrossRef]
- Qi, S.; Zhao, F.; Li, Z.; Liang, F.; Yu, S. Silencing of PTX3 alleviates LPS-induced inflammatory pain by regulating TLR4/NF-kappaB signaling pathway in mice. Biosci. Rep. 2020, 40, BSR20194208. [Google Scholar] [CrossRef] [PubMed]
- Ardizzone, A.; Repici, A.; Capra, A.P.; De Gaetano, F.; Bova, V.; Casili, G.; Campolo, M.; Esposito, E. Efficacy of the Radical Scavenger, Tempol, to Reduce Inflammation and Oxidative Stress in a Murine Model of Atopic Dermatitis. Antioxidants 2023, 12, 1278. [Google Scholar] [CrossRef] [PubMed]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine sigNat.ure predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Pere, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Sulicka-Grodzicka, J.; Surdacki, A.; Surmiak, M.; Sanak, M.; Wizner, B.; Sydor, W.; Bociąga-Jasik, M.; Strach, M.; Korkosz, M.; Skladany, L. Chemerin as a Potential Marker of Resolution of Inflammation in COVID-19 Infection. Biomedicines 2022, 10, 2462. [Google Scholar] [CrossRef]
- Ardizzone, A.; Mannino, D.; Capra, A.P.; Repici, A.; Filippone, A.; Esposito, E.; Campolo, M. New Insights into the Mechanism of Ulva pertusa on Colitis in Mice: Modulation of the Pain and Immune System. Mar. Drugs 2023, 21, 298. [Google Scholar] [CrossRef]
- Karimi, A.; Shobeiri, P.; Kulasinghe, A.; Rezaei, N. Novel Systemic Inflammation Markers to Predict COVID-19 Prognosis. Front. Immunol. 2021, 12, 741061. [Google Scholar] [CrossRef]
| Total Subjects | SARS-CoV-2 Positive | Never-Infected Subjects | ||||
|---|---|---|---|---|---|---|
| Sex | 35 (M) 45 (F) | 20 (M) 20 (F) | 15 (M) 25 (F) | |||
| Age | 40.9 ± 11.6 | 39.5 ± 10.8 | 43.0 ± 13.1 | |||
| Males | Females | Males | Females | Males | Females | |
| 40.2 ± 12.0 | 41.5 ± 11.7 | 39.7 ± 13.0 | 39.4 ± 8.5 | 41.5 ± 11.0 | 43.9 ± 14.9 | |
| Comorbidities | None | None | None | |||
| Months from COVID-19 positivity | 6.24 ± 3.51 | Males | Females | / | ||
| 5.00 ± 2.55 | 7.63 ± 4.07 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Capra, A.P.; Crupi, L.; Pantò, G.; Repici, A.; Calapai, F.; Squeri, R.; Ardizzone, A.; Esposito, E. Serum Pentraxin 3 as Promising Biomarker for the Long-Lasting Inflammatory Response of COVID-19. Int. J. Mol. Sci. 2023, 24, 14195. https://doi.org/10.3390/ijms241814195
Capra AP, Crupi L, Pantò G, Repici A, Calapai F, Squeri R, Ardizzone A, Esposito E. Serum Pentraxin 3 as Promising Biomarker for the Long-Lasting Inflammatory Response of COVID-19. International Journal of Molecular Sciences. 2023; 24(18):14195. https://doi.org/10.3390/ijms241814195
Chicago/Turabian StyleCapra, Anna Paola, Lelio Crupi, Giuseppe Pantò, Alberto Repici, Fabrizio Calapai, Raffaele Squeri, Alessio Ardizzone, and Emanuela Esposito. 2023. "Serum Pentraxin 3 as Promising Biomarker for the Long-Lasting Inflammatory Response of COVID-19" International Journal of Molecular Sciences 24, no. 18: 14195. https://doi.org/10.3390/ijms241814195
APA StyleCapra, A. P., Crupi, L., Pantò, G., Repici, A., Calapai, F., Squeri, R., Ardizzone, A., & Esposito, E. (2023). Serum Pentraxin 3 as Promising Biomarker for the Long-Lasting Inflammatory Response of COVID-19. International Journal of Molecular Sciences, 24(18), 14195. https://doi.org/10.3390/ijms241814195








