Epigallocatechin Gallate Affects the Structure of Chromatosomes, Nucleosomes and Their Complexes with PARP1
Abstract
1. Introduction
2. Results
2.1. Binding of EGCG to DNA in Living Cells
2.2. Experimental Approaches for Analysis of EGCG–Nucleosome Interaction
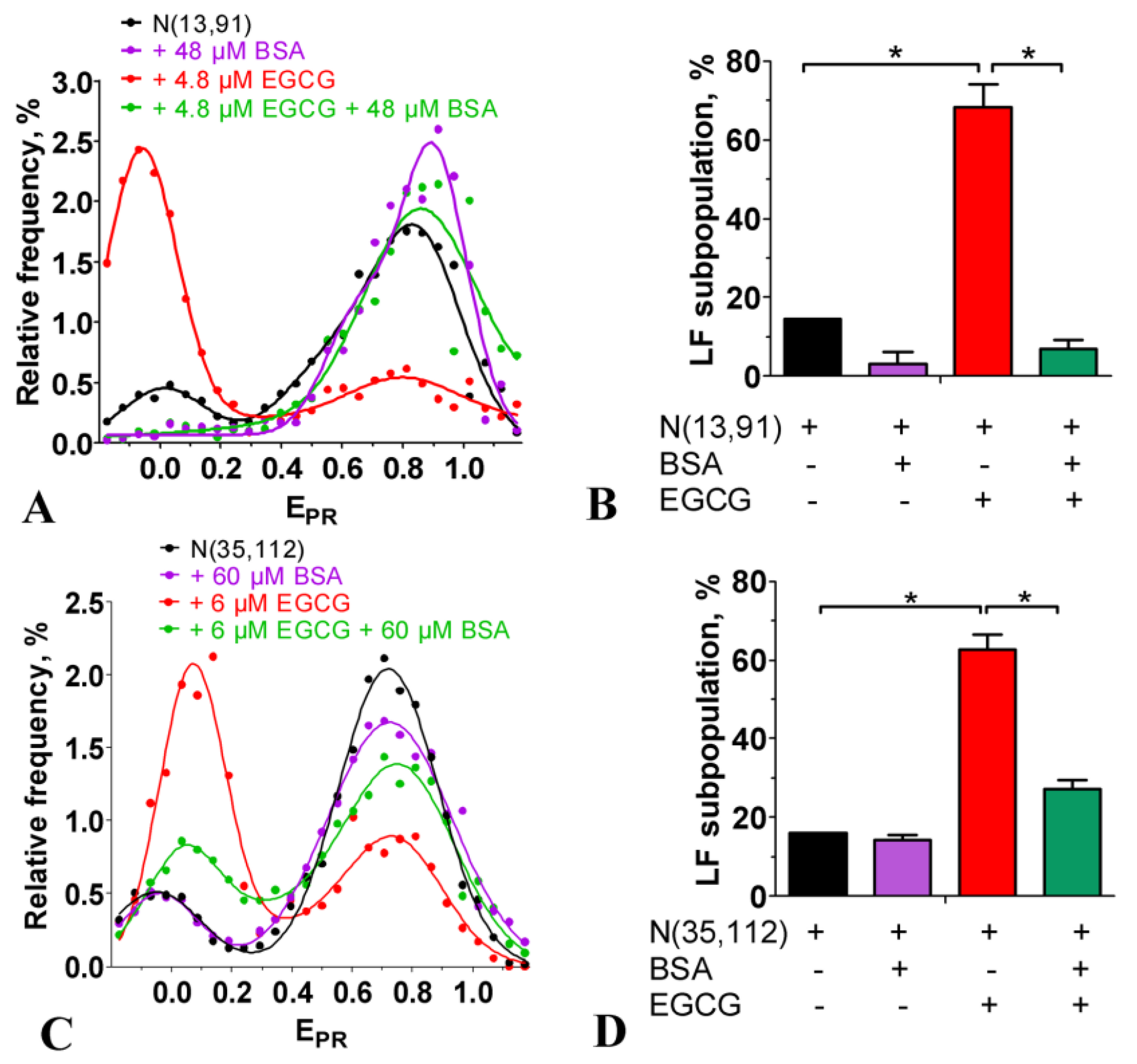
2.3. Effect of EGCG on the Structure of Nucleosomes
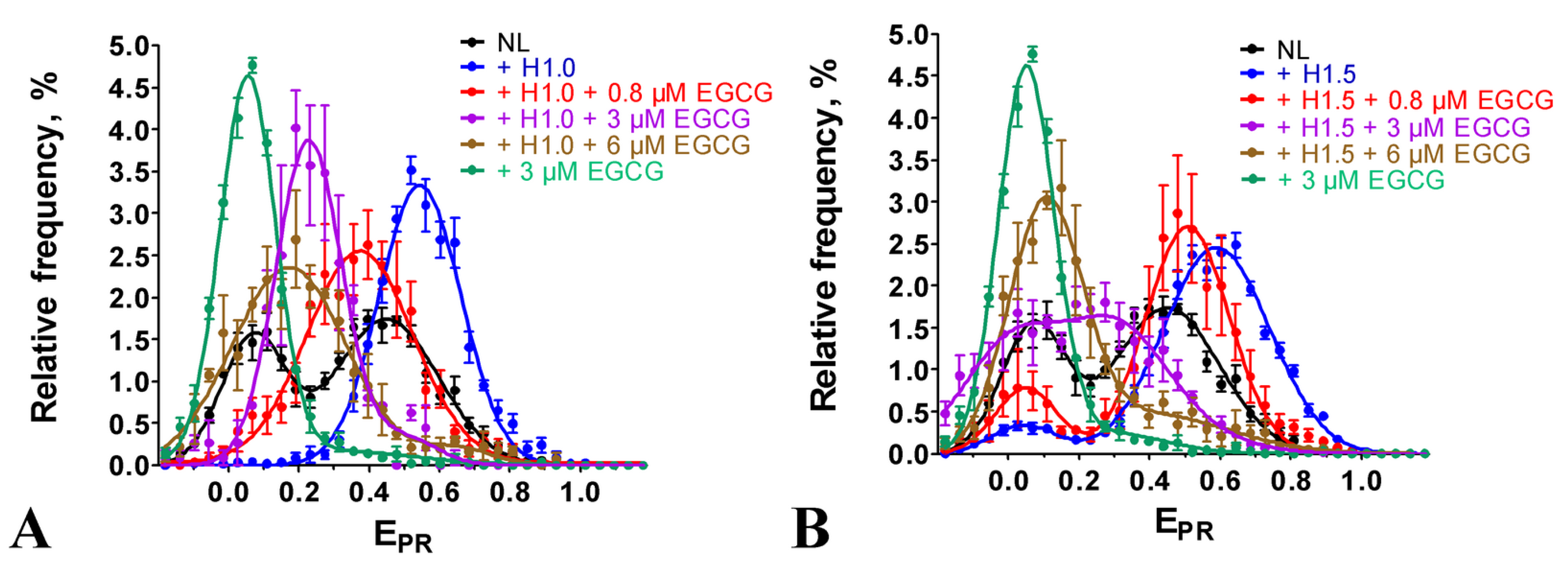
2.4. Effect of EGCG on the Structure of Chromatosomes
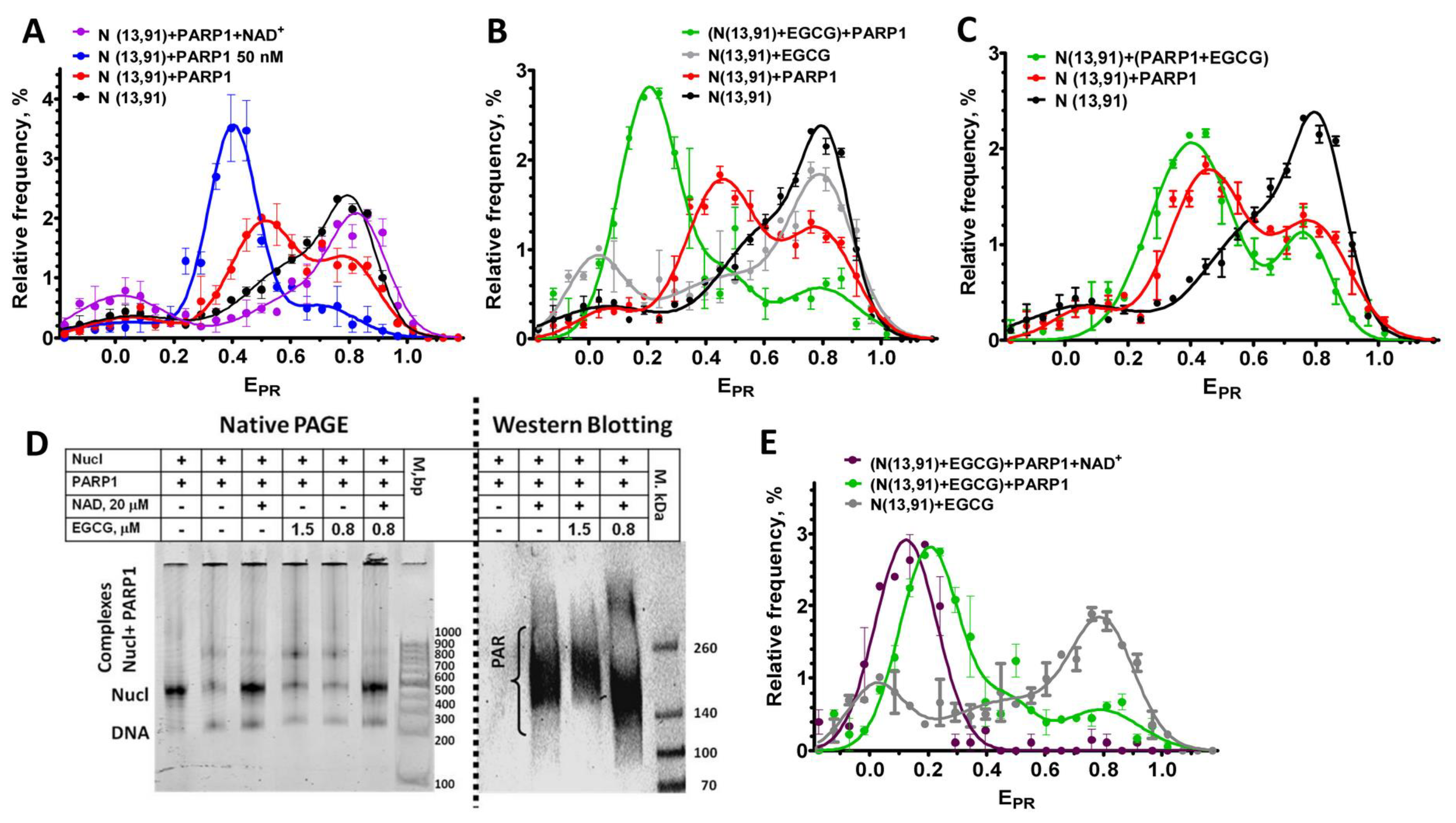
2.5. Effect of EGCG on the PARP1–Nucleosome Complexes
3. Discussion
4. Materials and Methods
4.1. Cellular Experiments
4.2. DNA
4.3. Proteins
4.4. Nucleosomes
4.5. Preparation of Complexes for spFRET Analysis
4.6. spFRET Microscopy
4.7. Electrophoresis
4.8. Western Blotting
4.9. Statistical Analysis
5. Conclusions
- EGCG very effectively penetrated into the cell nuclei and bound to DNA there.
- EGCG induced a large-scale uncoiling of nucleosomal DNA from the histone octamer; this process was reversible and was not accompanied by the dissociation of core histones from DNA.
- EGCG reorganized the structure of chromatosomes, very likely without the dissociation of the linker histone.
- EGCG did not reduce the affinity of PARP1 to nucleosomes.
- EGCG affected the structure of the PARP1–nucleosome complex.
- The auto-PARylation activity of PARP1 did not change in the presence of sub- and low-micromolar concentrations of EGCG.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tang, G.; Xu, Y.; Zhang, C.; Wang, N.; Li, H.; Feng, Y. Green Tea and Epigallocatechin Gallate (EGCG) for the Management of Nonalcoholic Fatty Liver Diseases (NAFLD): Insights into the Role of Oxidative Stress and Antioxidant Mechanism. Antioxidants 2021, 10, 1076. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.; Nahashon, S.; Taka, E.; Adinew, G.M.; Soliman, K.F.A. Epigallocatechin-3-Gallate (EGCG): New Therapeutic Perspectives for Neuroprotection, Aging, and Neuroinflammation for the Modern Age. Biomolecules 2022, 12, 371. [Google Scholar] [CrossRef] [PubMed]
- Steinmann, J.; Buer, J.; Pietschmann, T.; Steinmann, E. Anti-Infective Properties of Epigallocatechin-3-Gallate (EGCG), a Component of Green Tea. Br. J. Pharmacol. 2013, 168, 1059–1073. [Google Scholar] [CrossRef] [PubMed]
- Gan, R.Y.; Li, H.B.; Sui, Z.Q.; Corke, H. Absorption, Metabolism, Anti-Cancer Effect and Molecular Targets of Epigallocatechin Gallate (EGCG): An Updated Review. Crit. Rev. Food Sci. Nutr. 2018, 58, 924–941. [Google Scholar] [CrossRef] [PubMed]
- Isbrucker, R.A.; Edwards, J.A.; Wolz, E.; Davidovich, A.; Bausch, J. Safety Studies on Epigallocatechin Gallate (EGCG) Preparations. Part 2: Dermal, Acute and Short-Term Toxicity Studies. Food Chem. Toxicol. 2006, 44, 636–650. [Google Scholar] [CrossRef]
- Hu, J.; Webster, D.; Cao, J.; Shao, A. The Safety of Green Tea and Green Tea Extract Consumption in Adults—Results of a Systematic Review. Regul. Toxicol. Pharmacol. 2018, 95, 412–433. [Google Scholar] [CrossRef]
- Isbrucker, R.A.; Edwards, J.A.; Wolz, E.; Davidovich, A.; Bausch, J. Safety Studies on Epigallocatechin Gallate (EGCG) Preparations. Part 3: Teratogenicity and Reproductive Toxicity Studies in Rats. Food Chem. Toxicol. 2006, 44, 651–661. [Google Scholar] [CrossRef]
- Mah, E.; Chen, O.; Liska, D.J.; Blumberg, J.B. Dietary Supplements for Weight Management: A Narrative Review of Safety and Metabolic Health Benefits. Nutrients 2022, 14, 1787. [Google Scholar] [CrossRef]
- Kim, H.S.; Quon, M.J.; Kim, J.A. New Insights into the Mechanisms of Polyphenols beyond Antioxidant Properties; Lessons from the Green Tea Polyphenol, Epigallocatechin 3-Gallate. Redox Biol. 2014, 2, 187–195. [Google Scholar] [CrossRef]
- Isbrucker, R.A.; Bausch, J.; Edwards, J.A.; Wolz, E. Safety Studies on Epigallocatechin Gallate (EGCG) Preparations. Part 1: Genotoxicity. Food Chem. Toxicol. 2006, 44, 626–635. [Google Scholar] [CrossRef]
- Ciesielski, O.; Biesiekierska, M.; Balcerczyk, A. Epigallocatechin-3-Gallate (EGCG) Alters Histone Acetylation and Methylation and Impacts Chromatin Architecture Profile in Human Endothelial Cells. Molecules 2020, 25, 2326. [Google Scholar] [CrossRef] [PubMed]
- Thakur, V.S.; Gupta, K.; Gupta, S. Green Tea Polyphenols Causes Cell Cycle Arrest and Apoptosis in Prostate Cancer Cells by Suppressing Class I Histone Deacetylases. Carcinogenesis 2012, 33, 377–384. [Google Scholar] [CrossRef] [PubMed]
- López-Lázaro, M.; Calderón-Montaño, J.M.; Burgos-Morón, E.; Austin, C.A. Green Tea Constituents (-)-Epigallocatechin-3-Gallate (EGCG) and Gallic Acid Induce Topoisomerase I- and Topoisomerase II-DNA Complexes in Cells Mediated by Pyrogallol-Induced Hydrogen Peroxide. Mutagenesis 2011, 26, 489–498. [Google Scholar] [CrossRef]
- Suzuki, K.; Yahara, S.; Hashimoto, F.; Uyeda, M. Inhibitory Activities of (-)-Epigallocatechin-3-O-Gallate against Topoisomerases I and II. Biol. Pharm. Bull. 2001, 24, 1088–1090. [Google Scholar] [CrossRef][Green Version]
- Bandele, O.J.; Clawson, S.J.; Osheroff, N. Dietary Polyphenols as Topoisomerase II Poisons: B Ring and C Ring Substituents Determine the Mechanism of Enzyme-Mediated DNA Cleavage Enhancement. Chem. Res. Toxicol. 2008, 21, 1253–1260. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, S.; Welton, K.; Gius, J.P.; Elmegerhi, S.; Kato, T.A. The Effect of Green and Black Tea Polyphenols on BRCA2 Deficient Chinese Hamster Cells by Synthetic Lethality through PARP Inhibition. Int. J. Mol. Sci. 2019, 20, 1274. [Google Scholar] [CrossRef]
- Ray Chaudhuri, A.; Nussenzweig, A. The Multifaceted Roles of PARP1 in DNA Repair and Chromatin Remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610. [Google Scholar] [CrossRef]
- Spiegel, J.O.; Van Houten, B.; Durrant, J.D. PARP1: Structural Insights and Pharmacological Targets for Inhibition. DNA Repair. 2021, 103, 103125. [Google Scholar] [CrossRef]
- Cohen-Armon, M. A Long-Lasting PARP1-Activation Mediates Signal-Induced Gene Expression. Cells 2022, 11, 1576. [Google Scholar] [CrossRef]
- Kuzuhara, T.; Sei, Y.; Yamaguchi, K.; Suganuma, M.; Fujiki, H. DNA and RNA as New Binding Targets of Green Tea Catechins. J. Biol. Chem. 2006, 281, 17446–17456. [Google Scholar] [CrossRef] [PubMed]
- Chanphai, P.; Tajmir-Riahi, H.A. Structural Dynamics of DNA Binding to Tea Catechins. Int. J. Biol. Macromol. 2019, 125, 238–243. [Google Scholar] [CrossRef]
- Galindo-Murillo, R.; Cheatham, T.E. Computational DNA Binding Studies of (–)-Epigallocatechin-3-Gallate. J. Biomol. Struct. Dyn. 2018, 36, 3311–3323. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chen, A.; Hoshi, T.; Anzai, J.I.; Li, G. Electrochemical Studies of (-)-Epigallocatechin Gallate and Its Interaction with DNA. Anal. Bioanal. Chem. 2006, 386, 1913–1919. [Google Scholar] [CrossRef]
- Ghosh, K.S.; Sahoo, B.K.; Jana, D.; Dasgupta, S. Studies on the Interaction of Copper Complexes of (-)-Epicatechin Gallate and (-)-Epigallocatechin Gallate with Calf Thymus DNA. J. Inorg. Biochem. 2008, 102, 1711–1718. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Yin, X.; Duan, H.; Liang, L. Molecular Complexes of Calf Thymus DNA with Various Bioactive Compounds: Formation and Characterization. Int. J. Biol. Macromol. 2021, 168, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Sriram, M.; Van der Marel, G.A.; Roelen, H.L.P.F.; Van Boom, J.H.; Wang, A.H.J. Conformation of B-DNA Containing O6-Ethyl-G-C Base Pairs Stabilized by Minor Groove Binding Drugs: Molecular Structure of d(CGC[E6G]AATTCGCG Complexed with Hoechst 33258 or Hoechst 33342. EMBO J. 1992, 11, 225–232. [Google Scholar] [CrossRef]
- Arndt Jovin, D.J.; Jovin, T.M. Analysis and Sorting of Living Cells According to Deoxyribonucleic Acid Content. J. Histochem. Cytochem. 1977, 25, 585–589. [Google Scholar] [CrossRef]
- Okabe, S.; Suganuma, M.; Hayashi, M.; Sueoka, E.; Komori, A.; Fujiki, H. Mechanisms of Growth Inhibition of Human Lung Cancer Cell Line, PC-9, by Tea Polyphenols. Jpn. J. Cancer Res. 1997, 88, 639–643. [Google Scholar] [CrossRef]
- Kudryashova, K.S.; Chertkov, O.V.; Nikitin, D.V.; Pestov, N.A.; Kulaeva, O.I.; Efremenko, A.V.; Solonin, A.S.; Kirpichnikov, M.P.; Studitsky, V.M.; Feofanov, A.V. Preparation of Mononucleosomal Templates for Analysis of Transcription with RNA Polymerase Using SpFRET. Methods Mol. Biol. 2015, 1288, 395–412. [Google Scholar] [CrossRef]
- Lyubitelev, A.V.; Kudryashova, K.S.; Mikhaylova, M.S.; Malyuchenko, N.V.; Chertkov, O.V.; Studitsky, V.M.; Feofanov, A.V.; Kirpichnikov, M.P. Change in Linker DNA Conformation upon Histone H1.5 Binding to Nucleosome: Fluorescent Microscopy of Single Complexes. Mosc. Univ. Biol. Sci. Bull. 2016, 71, 108–113. [Google Scholar] [CrossRef]
- Buning, R.; Van Noort, J. Single-Pair FRET Experiments on Nucleosome Conformational Dynamics. Biochimie 2010, 92, 1729–1740. [Google Scholar] [CrossRef]
- Valieva, M.E.; Armeev, G.A.; Kudryashova, K.S.; Gerasimova, N.S.; Shaytan, A.K.; Kulaeva, O.I.; McCullough, L.L.; Formosa, T.; Georgiev, P.G.; Kirpichnikov, M.P.; et al. Large-Scale ATP-Independent Nucleosome Unfolding by a Histone Chaperone. Nat. Struct. Mol. Biol. 2016, 23, 1111–1116. [Google Scholar] [CrossRef]
- Almajano, M.P.; Delgado, M.E.; Gordon, M.H. Changes in the Antioxidant Properties of Protein Solutions in the Presence of Epigallocatechin Gallate. Food Chem. 2007, 101, 126–130. [Google Scholar] [CrossRef]
- Zhou, B.R.; Feng, H.; Kale, S.; Fox, T.; Khant, H.; de Val, N.; Ghirlando, R.; Panchenko, A.R.; Bai, Y. Distinct Structures and Dynamics of Chromatosomes with Different Human Linker Histone Isoforms. Mol. Cell 2021, 81, 166–182.e6. [Google Scholar] [CrossRef]
- Maluchenko, N.V.; Nilov, D.K.; Pushkarev, S.V.; Kotova, E.Y.; Gerasimova, N.S.; Kirpichnikov, M.P.; Langelier, M.-F.; Pascal, J.M.; Akhtar, M.S.; Feofanov, A.V.; et al. Mechanisms of Nucleosome Reorganization by PARP1. Int. J. Mol. Sci. 2021, 22, 12127. [Google Scholar] [CrossRef]
- Lambert, J.D.; Lee, M.J.; Diamond, L.; Ju, J.; Hong, J.; Bose, M.; Newmark, H.L.; Yang, C.S. Dose-Dependent Levels of Epigallocatechin-3-Gallate in Human Colon Cancer Cells and Mouse Plasma and Tissues. Drug Metab. Dispos. 2006, 34, 8–11. [Google Scholar] [CrossRef]
- Kiser, J.R.; Monk, R.W.; Smalls, R.L.; Petty, J.T. Hydration Changes in the Association of Hoechst 33258 with DNA. Biochemistry 2005, 44, 16988–16997. [Google Scholar] [CrossRef] [PubMed]
- Hayat, K.; Iqbal, H.; Malik, U.; Bilal, U.; Mushtaq, S. Tea and Its Consumption: Benefits and Risks. Crit. Rev. Food Sci. Nutr. 2015, 55, 939–954. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhi, F.; Chen, P.; Zhao, K.; Xiang, H.; Mao, Q.; Wang, X.; Zhang, X. Green Tea and the Risk of Prostate Cancer: A Systematic Review and Meta-Analysis. Medicine 2017, 96, e6426. [Google Scholar] [CrossRef] [PubMed]
- Reygaert, W.C. Green Tea Catechins: Their Use in Treating and Preventing Infectious Diseases. Biomed. Res. Int. 2018, 2018, 9105261. [Google Scholar] [CrossRef]
- Kun, E.; Kirsten, E.; Mendeleyev, J.; Ordahl, C.P. Regulation of the Enzymatic Catalysis of Poly(ADP-Ribose) Polymerase by DsDNA, Polyamines, Mg2+, Ca2+, Histones H1 and H3, and ATP†. Biochemistry 2003, 43, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Gillooly, J.F.; Hein, A.; Damiani, R. Nuclear DNA Content Varies with Cell Size across Human Cell Types. Cold Spring Harb. Perspect. Biol. 2015, 7, a019091. [Google Scholar] [CrossRef]
- Malinina, D.K.; Sivkina, A.L.; Korovina, A.N.; McCullough, L.L.; Formosa, T.; Kirpichnikov, M.P.; Studitsky, V.M.; Feofanov, A.V. Hmo1 Protein Affects the Nucleosome Structure and Supports the Nucleosome Reorganization Activity of Yeast FACT. Cells 2022, 11, 2931. [Google Scholar] [CrossRef] [PubMed]
- Malyuchenko, N.V.; Koshkina, D.O.; Korovina, A.N.; Gerasimova, N.S.; Kirpichnikov, M.P.; Studitsky, V.M.; Feofanov, A.V. The Effect of Gossypol on the Structure of Nucleosomes. Mosc. Univ. Biol. Sci. Bull. 2020, 75, 142–146. [Google Scholar] [CrossRef]
- Klinker, H.; Haas, C.; Harrer, N.; Becker, P.B.; Mueller-Planitz, F. Rapid Purification of Recombinant Histones. PLoS ONE 2014, 9, e104029. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.R.; Feng, H.; Ghirlando, R.; Li, S.; Schwieters, C.D.; Bai, Y. A Small Number of Residues Can Determine If Linker Histones Are Bound On or Off Dyad in the Chromatosome. J. Mol. Biol. 2016, 428, 3948–3959. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Rechsteiner, T.J.; Richmond, T.J. Expression and Purification of Recombinant Histones and Nucleosome Reconstitution. Methods Mol. Biol. 1999, 119, 1–16. [Google Scholar] [CrossRef]
- Gaykalova, D.A.; Kulaeva, O.I.; Bondarenko, V.A.; Studitsky, V.M. Preparation and Analysis of Uniquely Positioned Mononucleosomes. Methods Mol. Biol. 2009, 523, 109–123. [Google Scholar]
- Kotova, E.; Pinnola, A.D.; Tulin, A.V. Small-Molecule Collection and High-Throughput Colorimetric Assay to Identify PARP1 Inhibitors. Methods Mol. Biol. 2011, 780, 491–516. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andreeva, T.V.; Maluchenko, N.V.; Efremenko, A.V.; Lyubitelev, A.V.; Korovina, A.N.; Afonin, D.A.; Kirpichnikov, M.P.; Studitsky, V.M.; Feofanov, A.V. Epigallocatechin Gallate Affects the Structure of Chromatosomes, Nucleosomes and Their Complexes with PARP1. Int. J. Mol. Sci. 2023, 24, 14187. https://doi.org/10.3390/ijms241814187
Andreeva TV, Maluchenko NV, Efremenko AV, Lyubitelev AV, Korovina AN, Afonin DA, Kirpichnikov MP, Studitsky VM, Feofanov AV. Epigallocatechin Gallate Affects the Structure of Chromatosomes, Nucleosomes and Their Complexes with PARP1. International Journal of Molecular Sciences. 2023; 24(18):14187. https://doi.org/10.3390/ijms241814187
Chicago/Turabian StyleAndreeva, Tatiana V., Natalya V. Maluchenko, Anastasiya V. Efremenko, Alexander V. Lyubitelev, Anna N. Korovina, Dmitry A. Afonin, Mikhail P. Kirpichnikov, Vasily M. Studitsky, and Alexey V. Feofanov. 2023. "Epigallocatechin Gallate Affects the Structure of Chromatosomes, Nucleosomes and Their Complexes with PARP1" International Journal of Molecular Sciences 24, no. 18: 14187. https://doi.org/10.3390/ijms241814187
APA StyleAndreeva, T. V., Maluchenko, N. V., Efremenko, A. V., Lyubitelev, A. V., Korovina, A. N., Afonin, D. A., Kirpichnikov, M. P., Studitsky, V. M., & Feofanov, A. V. (2023). Epigallocatechin Gallate Affects the Structure of Chromatosomes, Nucleosomes and Their Complexes with PARP1. International Journal of Molecular Sciences, 24(18), 14187. https://doi.org/10.3390/ijms241814187





