Identification of SH2 Domain-Containing Protein 3C as a Novel, Putative Interactor of Dipeptidyl Peptidase 3
Abstract
1. Introduction
2. Results
2.1. Overexpressed DPP3 Protein Interacts with Isoforms 2 and 3 of SH2D3C
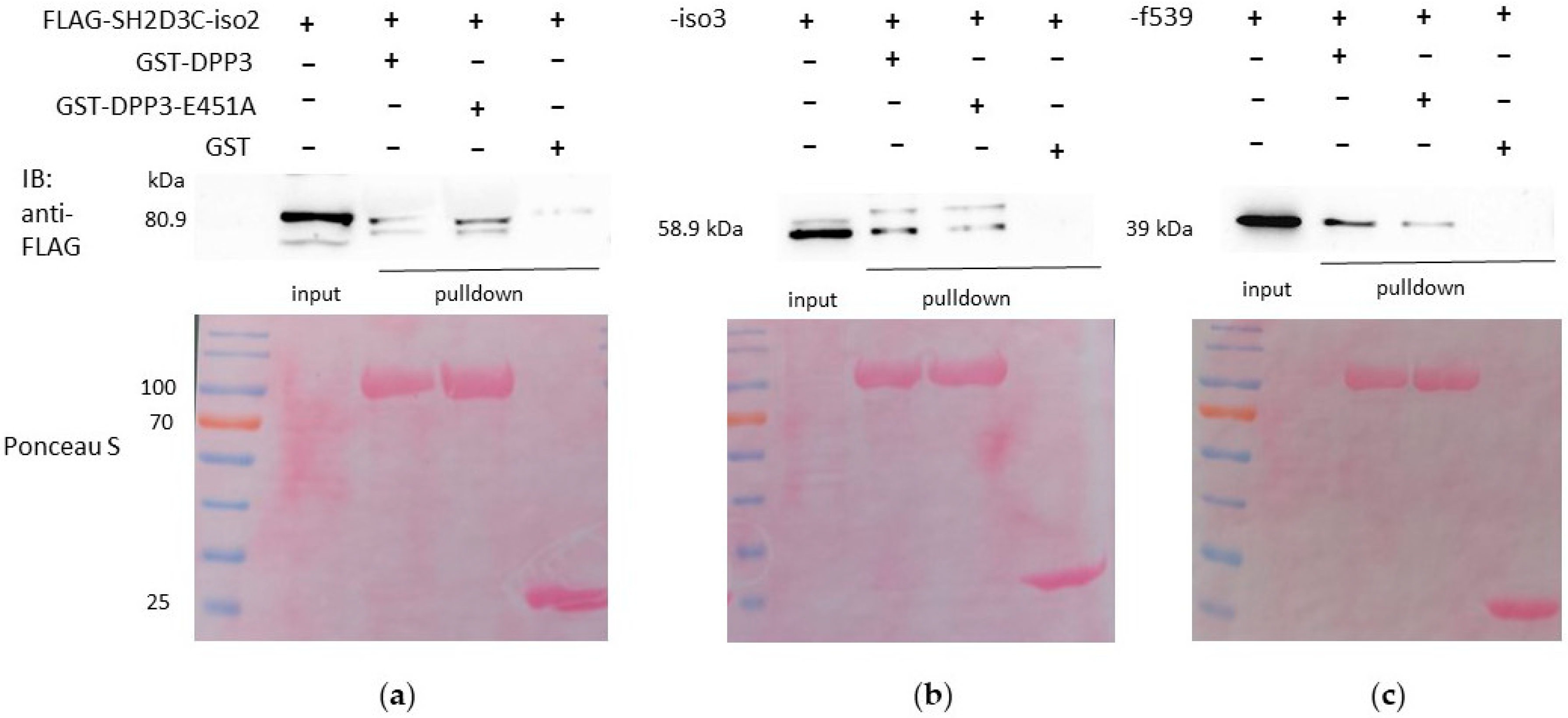
2.2. Both Isoforms 2 and 3 of SH2D3C, as Well as the C-Terminal Ras GEF-like Domain Alone, Interact with WT DPP3 and the Catalytically Inactive Variant E451A
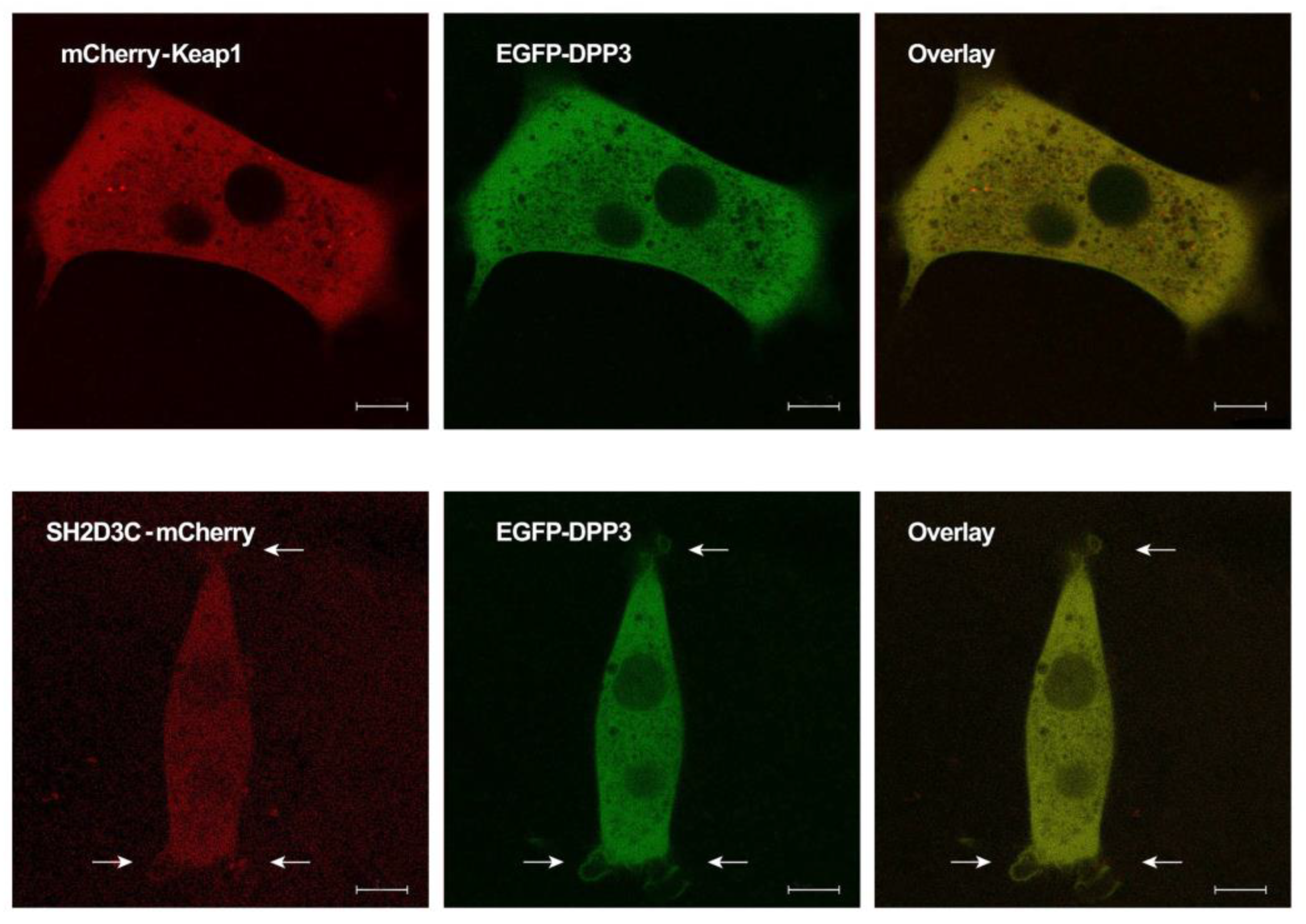
2.3. EGFP-DPP3 and SH2D3C-mCherry Colocalize in Cytosol, Nucleus, and Membrane Ruffles
2.4. DPP3 and SH2D3C Interact Specifically
2.5. SH2D3C Protein Does Not Show GEF Activity towards Small GTPase RRAS
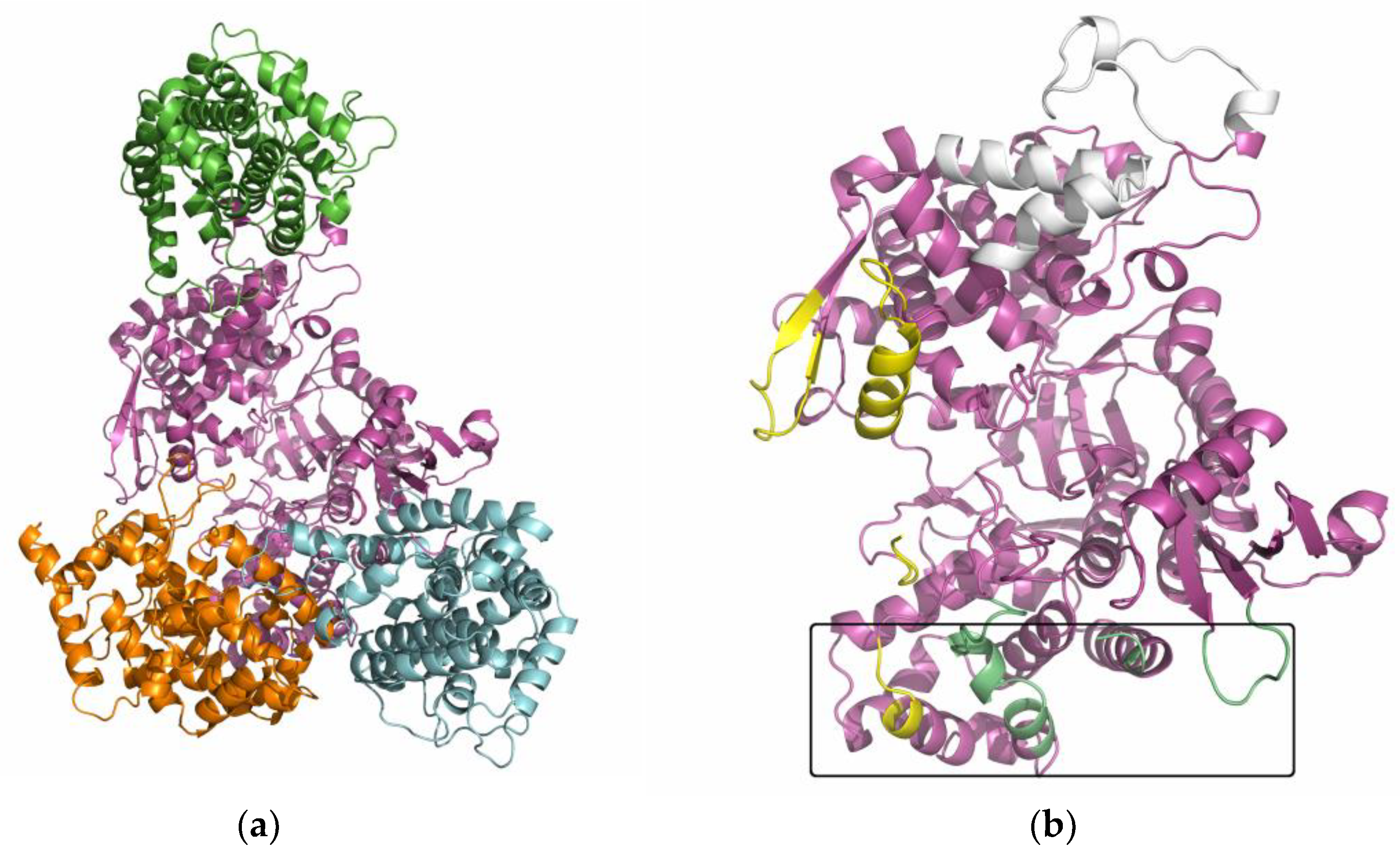
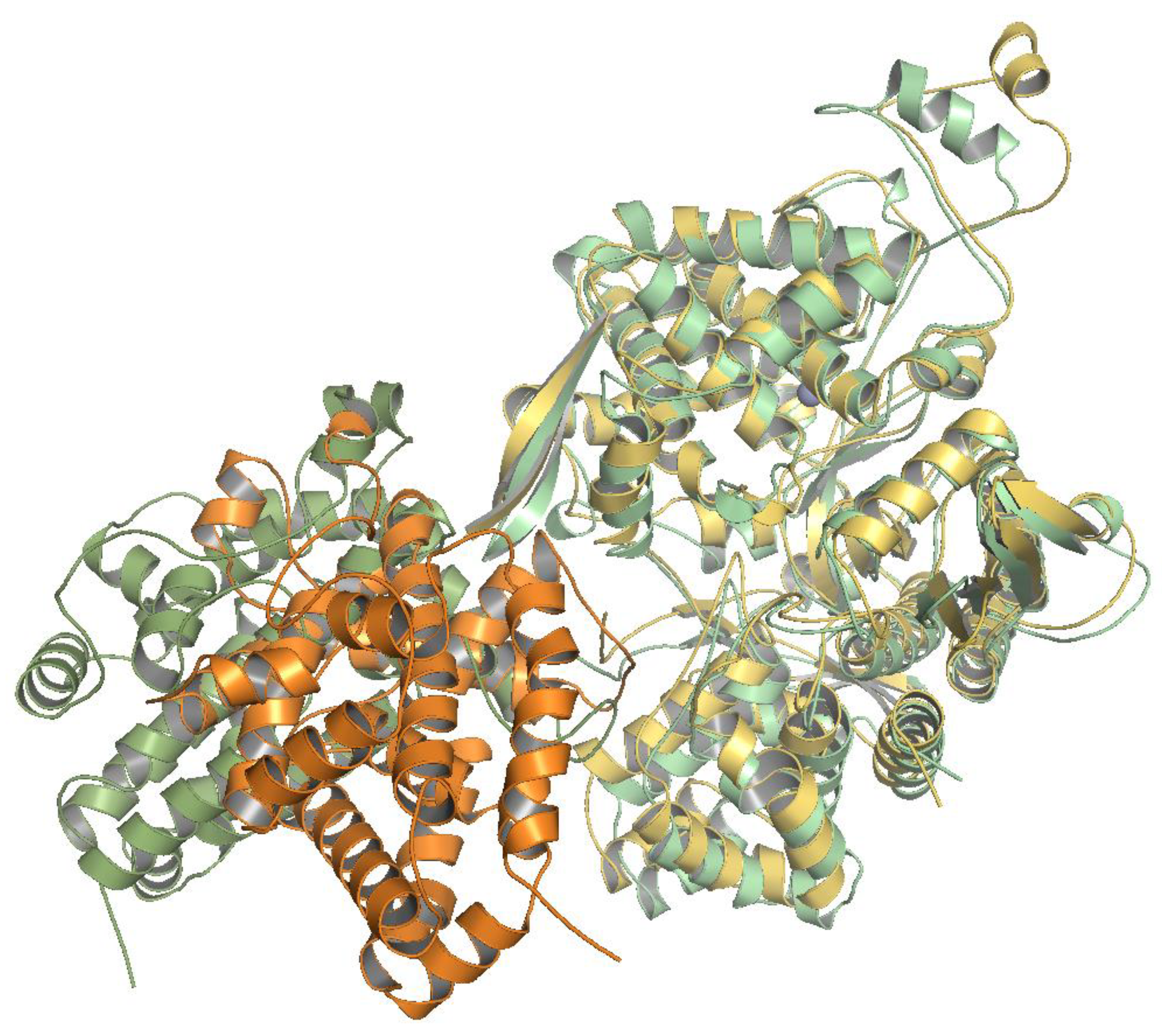
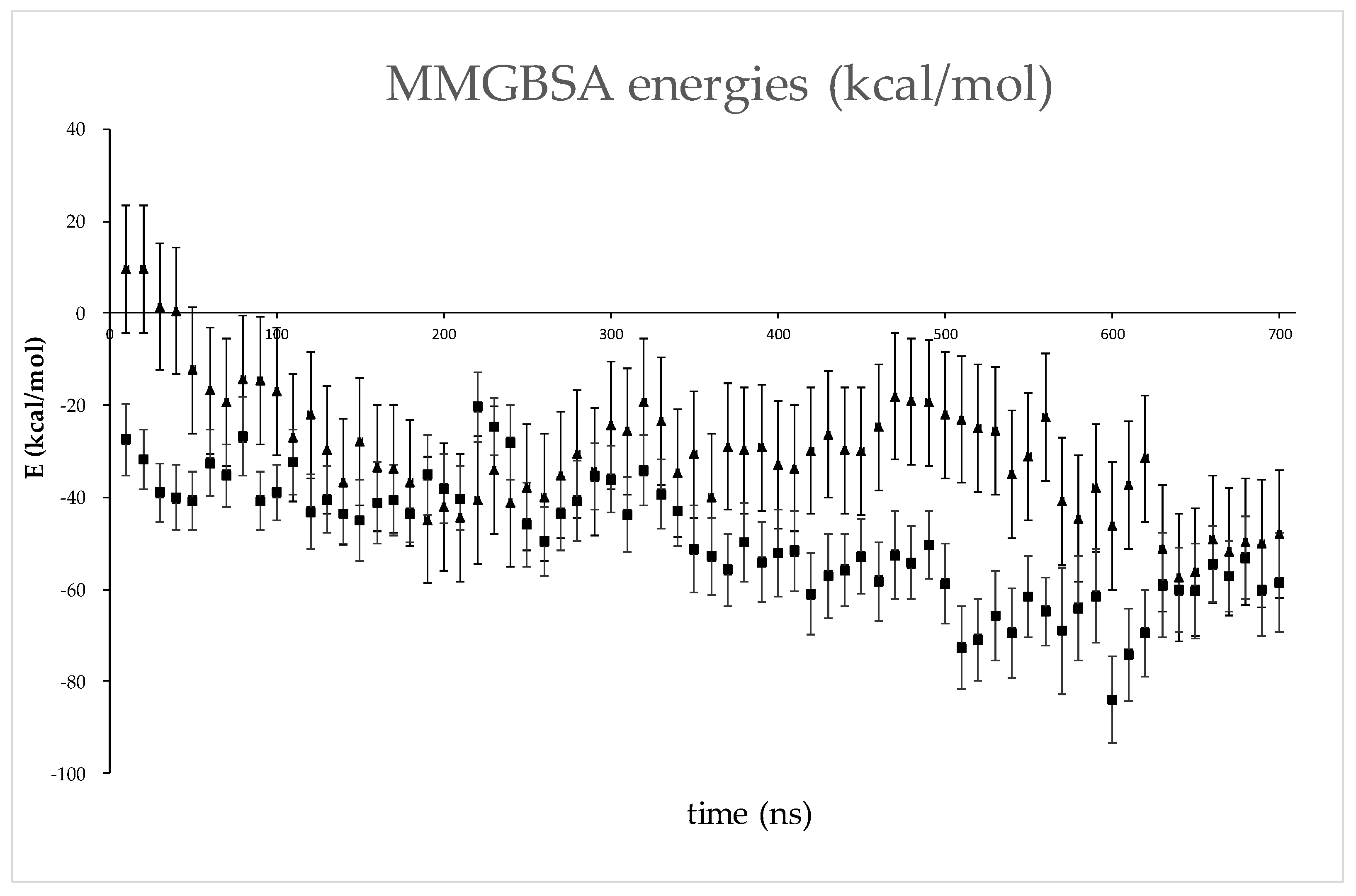
2.6. Several Models of Binding of DPP3 and C-Terminal Domain of SH2D3C Are Revealed by Protein Docking Analysis
3. Discussion
4. Materials and Methods
4.1. Experimental Study
4.1.1. Cell Culture and Transfection
4.1.2. Cloning
4.1.3. CoImmunoprecipitation
4.1.4. GST-Pulldown
4.1.5. Confocal Microscopy
4.1.6. Protein Purification
4.1.7. Guanine Nucleotide Exchange (GEF) Activity Test
4.2. Computational Study
4.2.1. Protein Docking
4.2.2. System Preparations
4.2.3. Classical MD Simulations
4.2.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karlsson, M.; Zhang, C.; Méar, L.; Zhong, W.; Digre, A.; Katona, B.; Sjöstedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A single–cell type transcriptomics map of human tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef]
- Prajapati, S.C.; Chauhan, S.S. Dipeptidyl peptidase III: A multifaceted oligopeptide N-end cutter. FEBS J. 2011, 278, 3256–3276. [Google Scholar] [CrossRef]
- Hast, B.E.; Cloer, E.W.; Goldfarb, D.; Li, H.; Siesser, P.F.; Yan, F.; Walter, V.; Zheng, N.; Hayes, D.N.; Major, M.B. Cancer-derived mutations in KEAP1 impair NRF2 degradation but not ubiquitination. Cancer Res. 2014, 74, 808–817. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Alcivar, A.L.; Ma, J.; Foo, T.K.; Zywea, S.; Huo, Y.; Kensler, T.W.; Gatza, M.L.; Xia, B. DPP3 in NRF2 Signaling and Breast Cancer. Free Radic. Biol. Med. 2017, 100, S132. [Google Scholar] [CrossRef]
- Menale, C.; Robinson, L.J.; Palagano, E.; Rigoni, R.; Erreni, M.; Almarza, A.J.; Strina, D.; Mantero, S.; Lizier, M.; Forlino, A.; et al. Absence of Dipeptidyl Peptidase 3 Increases Oxidative Stress and Causes Bone Loss. J. Bone Miner. Res. 2019, 34, 2133–2148. [Google Scholar] [CrossRef]
- Jha, S.; Taschler, U.; Domenig, O.; Poglitsch, M.; Bourgeois, B.; Pollheimer, M.; Pusch, L.M.; Malovan, G.; Frank, S.; Madl, T.; et al. Dipeptidyl peptidase 3 modulates the renin–Angiotensin system in mice. J. Biol. Chem. 2020, 295, 13711–13723. [Google Scholar] [CrossRef]
- Rehfeld, L.; Funk, E.; Jha, S.; Macheroux, P.; Melander, O.; Bergmann, A. Novel Methods for the Quantification of Dipeptidyl Peptidase 3 (DPP3) Concentration and Activity in Human Blood Samples. J. Appl. Lab. Med. 2019, 3, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Takagi, K.; Blet, A.; Levy, B.; Deniau, B.; Azibani, F.; Feliot, E.; Bergmann, A.; Santos, K.; Hartmann, O.; Gayat, E.; et al. Circulating dipeptidyl peptidase 3 and alteration in haemodynamics in cardiogenic shock: Results from the OptimaCC trial. Eur. J. Heart Fail. 2019, 22, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Dodelet, V.C.; Pazzagli, C.; Zisch, A.H.; Hauser, C.A.; Pasquale, E.B. A novel signaling intermediate, SHEP1, directly couples Eph receptors to R-Ras and Rap1A. J. Biol. Chem. 1999, 274, 31941–31946. [Google Scholar] [CrossRef]
- Sakakibara, A.; Hattori, S. Chat, a Cas / HEF1-associated Adaptor Protein That Integrates Multiple Signaling Pathways *. J. Biol. Chem. 2000, 275, 6404–6410. [Google Scholar] [CrossRef]
- Dail, M.; Kalo, M.S.; Seddon, J.A.; Côté, J.F.; Vuori, K.; Pasquale, E.B. SHEP1 function in cell migration is impaired by a single amino acid mutation that disrupts association with the scaffolding protein cas but not with ras GTPases. J. Biol. Chem. 2004, 279, 41892–41902. [Google Scholar] [CrossRef] [PubMed]
- Regelmann, A.G.; Danzl, N.M.; Wanjalla, C.; Alexandropoulos, K. The Hematopoietic Isoform of Cas-Hef1-Associated Signal Transducer Regulates Chemokine-Induced Inside-Out Signaling and T Cell Trafficking. Immunity 2006, 25, 907–918. [Google Scholar] [CrossRef] [PubMed]
- Vervoort, V.S.; Roselli, S.; Oshima, R.G.; Pasquale, E.B. Splice variants and expression patterns of SHEP1, BCAR3 and NSP1, a gene family involved in integrin and receptor tyrosine kinase signaling. Gene 2007, 391, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Mace, P.D.; Wallez, Y.; Dobaczewska, M.K.; Lee, J.J.; Robinson, H.; Pasquale, E.B.; Riedl, S.J. NSP-Cas protein structures reveal a promiscuous interaction module in cell signaling. Nat. Struct. Mol. Biol. 2011, 18, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Vervoort, V.; Wallez, Y.; Coré, N.; Cremer, H.; Pasquale, E.B. The Src homology 2 domain protein shep1 plays an important role in the penetration of olfactory sensory axons into the forebrain. J. Neurosci. 2010, 30, 13201–13210. [Google Scholar] [CrossRef]
- Al-Shami, A.; Wilkins, C.; Crisostomo, J.; Seshasayee, D.; Martin, F.; Xu, N.; Suwanichkul, A.; Anderson, S.J.; Oravecz, T. The Adaptor Protein Sh2d3c Is Critical for Marginal Zone B Cell Development and Function. J. Immunol. 2010, 185, 327–334. [Google Scholar] [CrossRef]
- Gomi, F.; Uchida, Y.; Endo, S. Up-regulation of NSP3 by Oligomeric Aβ Accelerates Neuronal Death Through Cas-independent Rap1A Activation. Neuroscience 2018, 386, 182–193. [Google Scholar] [CrossRef]
- Stumpf, M.P.H.; Thorne, T.; De Silva, E.; Stewart, R.; Hyeong, J.A.; Lappe, M.; Wiuf, C. Estimating the size of the human interactome. Proc. Natl. Acad. Sci. USA 2008, 105, 6959–6964. [Google Scholar] [CrossRef]
- Hast, B.E.; Goldfarb, D.; Mulvaney, K.M.; Hast, M.A.; Siesser, P.F.; Yan, F.; Hayes, D.N.; Major, M.B. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Res. 2013, 73, 2199–2210. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef]
- Tong, Y.; Huang, Y.; Zhang, Y.; Zeng, X.; Yan, M.; Xia, Z.; Lai, D. DPP3/CDK1 contributes to the progression of colorectal cancer through regulating cell proliferation, cell apoptosis, and cell migration. Cell Death Dis. 2021, 12, 529. [Google Scholar] [CrossRef] [PubMed]
- Malovan, G.; Hierzberger, B.; Suraci, S.; Schaefer, M.; Santos, K.; Jha, S.; Macheroux, P. The emerging role of dipeptidyl peptidase 3 in pathophysiology. FEBS J. 2022, 290, 2246–2262. [Google Scholar] [CrossRef]
- Sobočanec, S.; Filić, V.; Matovina, M.; Majhen, D.; Šafranko, Ž.M.; Hadžija, M.P.; Krsnik, Ž.; Kurilj, A.G.; Šarić, A.; Abramić, M.; et al. Prominent role of exopeptidase DPP III in estrogen-mediated protection against hyperoxia in vivo. Redox Biol. 2016, 8, 149–159. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sakakibara, A.; Hattori, S.; Nakamura, S.; Katagiri, T. A novel hematopoietic adaptor protein, chat-H, positively regulates T cell receptor-mediated interleukin-2 production by jurkat cells. J. Biol. Chem. 2003, 278, 6012–6017. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Yamashita, T. Inactivation of Ras by p120GAP via focal adhesion kinase dephosphorylation mediates RGMa-induced growth cone collapse. J. Neurosci. 2009, 29, 6649–6662. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- Liu, J.K.; Abudula, A.; Yang, H.T.; Xu, L.X.; Nuerrula, Y.; Bai, G.; Tulahong, A.; Eli, M. DPP3 expression promotes cell proliferation and migration in vitro and tumour growth in vivo, which is associated with poor prognosis of oesophageal carcinoma. Oncol. Rep. 2023, 49, 9. [Google Scholar] [CrossRef]
- Jimi, E.; Honda, H.; Nakamura, I. The unique function of p130Cas in regulating the bone metabolism. Pharmacol. Ther. 2022, 230, 107965. [Google Scholar] [CrossRef]
- Lepur, A.; Kovačević, L.; Belužić, R.; Vugrek, O. Combining Unique Multiplex Gateway Cloning and Bimolecular Fluorescence Complementation (BiFC) for High-Throughput Screening of Protein-Protein Interactions. J. Biomol. Screen. 2016, 21, 1100–1111. [Google Scholar] [CrossRef]
- Špoljarić, J.; Tomić, A.; Vukelić, B.; Salopek-Sondi, B.; Agić, D.; Tomić, S.; Abramić, M. Human Dipeptidyl Peptidase III: The Role of Asn406 in Ligand Binding and Hydrolysis. Croat. Chem. Acta 2011, 84, 259–268. [Google Scholar] [CrossRef]
- Tomić, A.; Berynskyy, M.; Wade, R.C.; Tomić, S.; Tomic, A.; Berynskyy, M.; Wade, R.C.; Tomić, S. Molecular simulations reveal that the long range fluctuations of human DPP III change upon ligand binding. Mol. Biosyst. 2015, 11, 3068–3080. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef] [PubMed]
- Lensink, M.F.; Nadzirin, N.; Velankar, S.; Wodak, S.J. Modeling protein-protein, protein-peptide, and protein-oligosaccharide complexes: CAPRI 7th edition. Proteins Struct. Funct. Bioinform. 2020, 88, 916–938. [Google Scholar] [CrossRef]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071–1081.e3. [Google Scholar] [CrossRef]
- Vajda, S.; Yueh, C.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Kozakov, D. New additions to the ClusPro server motivated by CAPRI. Proteins Struct. Funct. Bioinform. 2017, 85, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hallc, D.R.; Xiab, B.; Porterb, K.A.; Padhornya, D.; Yuehb, C.; Beglovb, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; Van Dijk, M.; De Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Honorato, R.V.; Koukos, P.I.; Jiménez-García, B.; Tsaregorodtsev, A.; Verlato, M.; Giachetti, A.; Rosato, A.; Bonvin, A.M.J.J. Structural Biology in the Clouds: The WeNMR-EOSC Ecosystem. Front. Mol. Biosci. 2021, 8, 729513. [Google Scholar] [CrossRef]
- Katchalskikatzir, E.; Shariv, I.; Eisenstein, M.; Friesem, A.A.; Aflalo, C.; Vakser, I.A. Molecular surface recognition: Determination of geometric fit between proteins and their ligands by correlation techniques. Proc. Natl. Acad. Sci. USA 1992, 89, 2195–2199. [Google Scholar] [CrossRef]
- Tovchigrechko, A.; Vakser, I.A. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006, 34, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Anandakrishnan, R.; Aguilar, B.; Onufriev, A. V H++3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012, 40, W537–W541. [Google Scholar] [CrossRef] [PubMed]
- Tomić, A.; Kovačević, B.; Tomić, S. Concerted nitrogen inversion and hydrogen bonding to Glu451 are responsible for protein-controlled suppression of the reverse reaction in human DPP III. Phys. Chem. Chem. Phys. 2016, 18, 27245–27256. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef]
- Tomić, A.; Horvat, G.; Ramek, M.; Agić, D.; Brkić, H.; Tomić, S. New Zinc Ion Parameters Suitable for Classical MD Simulations of Zinc Metallopeptidases. J. Chem. Inf. Model. 2019, 59, 3437–3453. [Google Scholar] [CrossRef]
- Izadi, S.; Anandakrishnan, R.; Onufriev, A. V Building Water Models: A Different Approach. J. Phys. Chem. Lett. 2014, 5, 3863–3871. [Google Scholar] [CrossRef]
- Sengupta, A.; Li, Z.; Song, L.F.; Li, P.; Merz, K.M., Jr. Parameterization of Monovalent Ions for the OPC3, OPC, TIP3P-FB, and TIP4P-FB Water Models. J. Chem. Inf. Model. 2021, 61, 869–880. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E.I.; Tom, D.; Gohlke, H.; Luo, R.; Nerz, K.M.J.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An overview of the Amber biomolecular simulation package. WILEY Interdiscip. Rev. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Loncharich, R.J.; Brooks, B.R.; Pastor, R.W. Langevin dynamics of peptides: The frictional dependence of isomerization rates of N-acetylalanyl-N′-methylamide. Biopolymers 1992, 32, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. Model. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Matić, S.; Tomašić Paić, A.; Sobočanec, S.; Pinterić, M.; Pipalović, G.; Martinčić, M.; Matovina, M.; Tomić, S. Interdisciplinary Study of the Effects of Dipeptidyl-Peptidase III Cancer Mutations on the KEAP1-NRF2 Signaling Pathway. Int. J. Mol. Sci. 2022, 23, 1994. [Google Scholar] [CrossRef]
- Tomic, A.; Tomic, S. Hunting the human DPP III active conformation: Combined thermodynamic and QM/MM calculations. Dalt. Trans. 2014, 43, 15503–15514. [Google Scholar] [CrossRef]
- Matić, S.; Kekez, I.; Tomin, M.; Bogár, F.; Šupljika, F.; Kazazić, S.; Hanić, M.; Jha, S.; Brkić, H.; Bourgeois, B.; et al. Binding of dipeptidyl peptidase III to the oxidative stress cell sensor Kelch-like ECH-associated protein 1 is a two-step process. J. Biomol. Struct. Dyn. 2021, 39, 6870–6881. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matovina, M.; Tomašić Paić, A.; Tomić, S.; Brkić, H.; Horvat, L.; Barbarić, L.; Filić, V.; Pinterić, M.; Jurić, S.; Kussayeva, A. Identification of SH2 Domain-Containing Protein 3C as a Novel, Putative Interactor of Dipeptidyl Peptidase 3. Int. J. Mol. Sci. 2023, 24, 14178. https://doi.org/10.3390/ijms241814178
Matovina M, Tomašić Paić A, Tomić S, Brkić H, Horvat L, Barbarić L, Filić V, Pinterić M, Jurić S, Kussayeva A. Identification of SH2 Domain-Containing Protein 3C as a Novel, Putative Interactor of Dipeptidyl Peptidase 3. International Journal of Molecular Sciences. 2023; 24(18):14178. https://doi.org/10.3390/ijms241814178
Chicago/Turabian StyleMatovina, Mihaela, Ana Tomašić Paić, Sanja Tomić, Hrvoje Brkić, Lucija Horvat, Lea Barbarić, Vedrana Filić, Marija Pinterić, Snježana Jurić, and Akmaral Kussayeva. 2023. "Identification of SH2 Domain-Containing Protein 3C as a Novel, Putative Interactor of Dipeptidyl Peptidase 3" International Journal of Molecular Sciences 24, no. 18: 14178. https://doi.org/10.3390/ijms241814178
APA StyleMatovina, M., Tomašić Paić, A., Tomić, S., Brkić, H., Horvat, L., Barbarić, L., Filić, V., Pinterić, M., Jurić, S., & Kussayeva, A. (2023). Identification of SH2 Domain-Containing Protein 3C as a Novel, Putative Interactor of Dipeptidyl Peptidase 3. International Journal of Molecular Sciences, 24(18), 14178. https://doi.org/10.3390/ijms241814178






