Enhanced Efficiency of the Basal and Induced Apoptosis Process in Mucopolysaccharidosis IVA and IVB Human Fibroblasts
Abstract
:1. Introduction
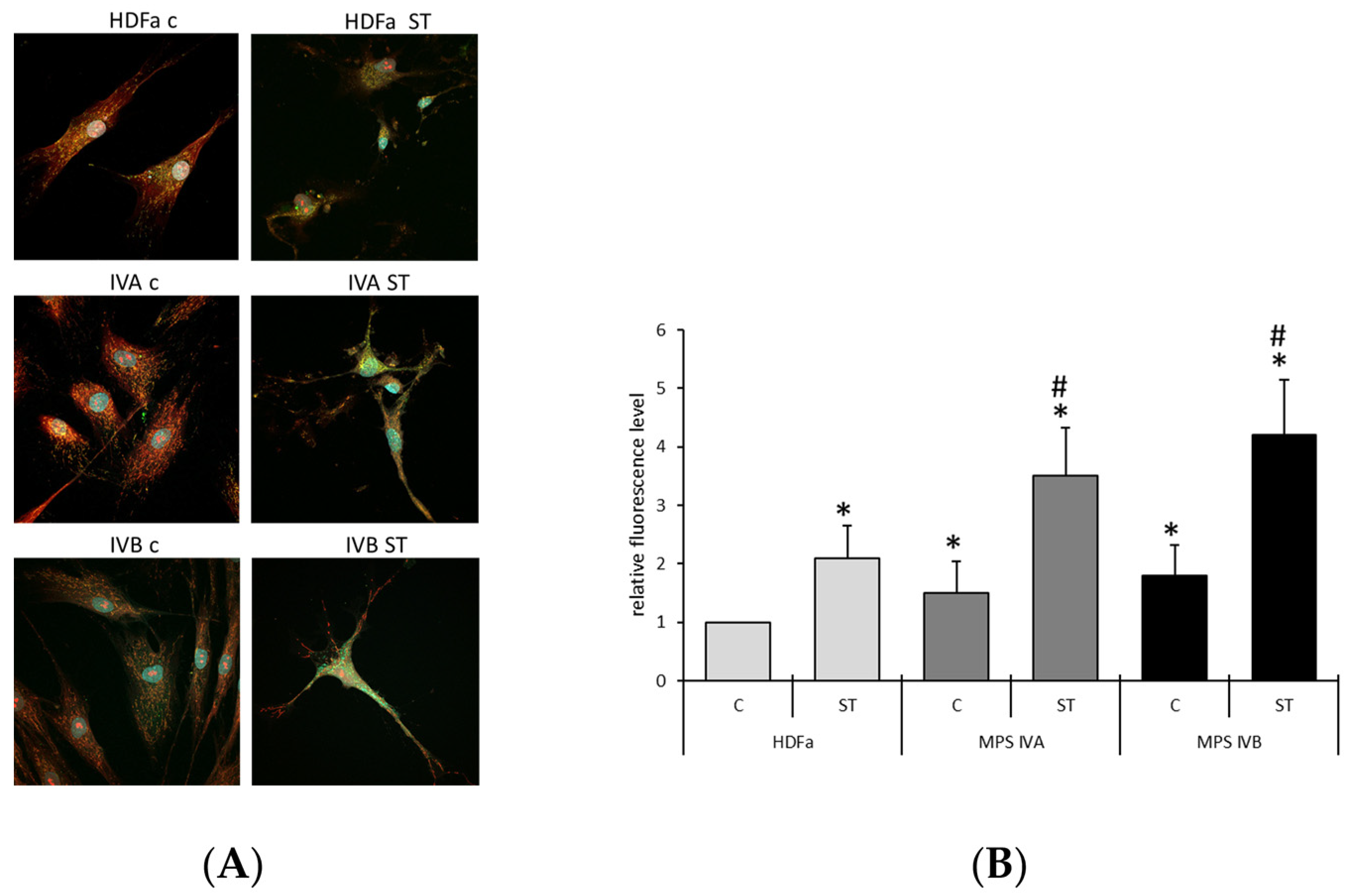
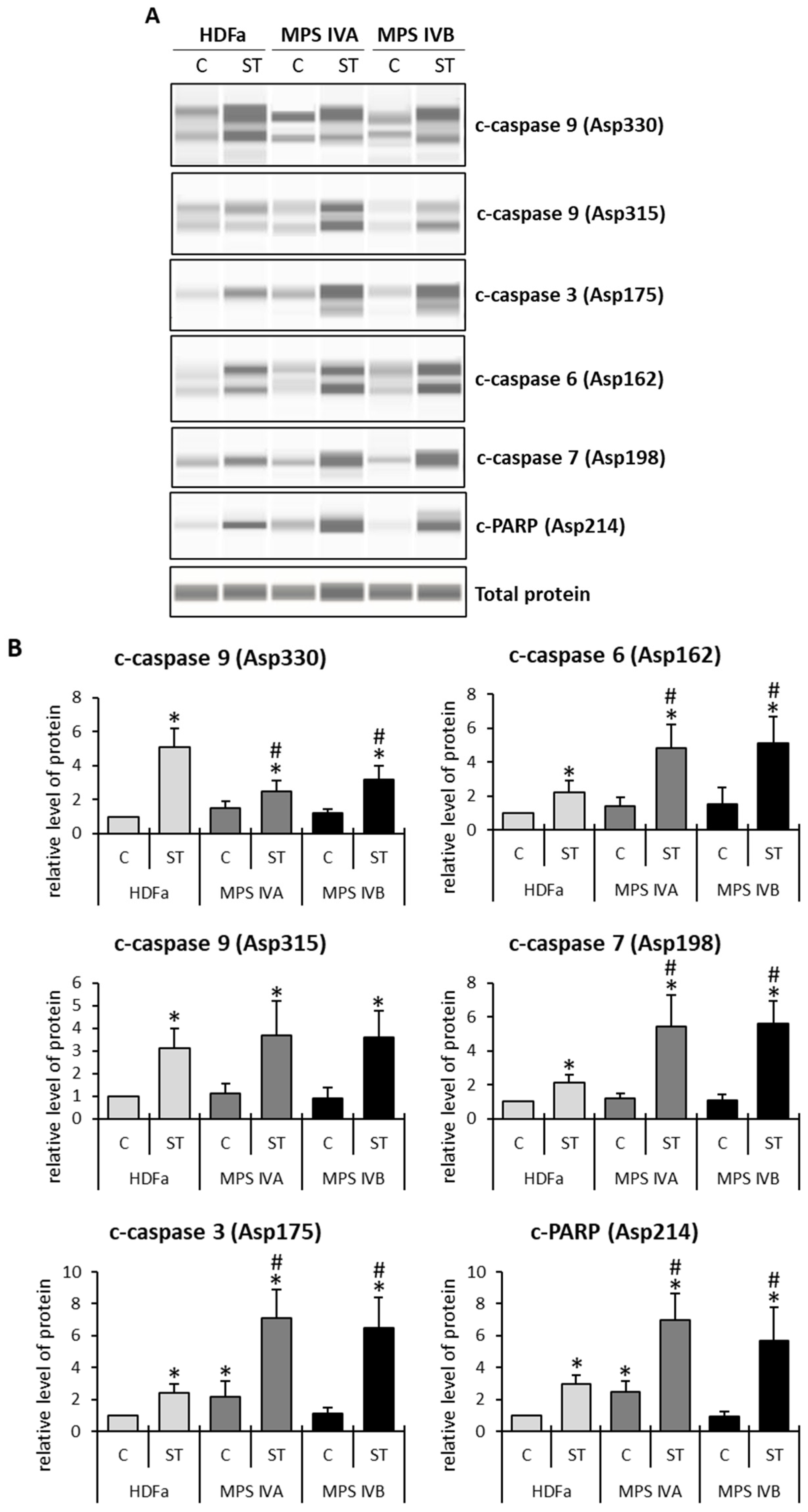
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Transcriptomic Studies
4.3. Immunofluorescence Microscopy
4.4. Western Blotting
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Scerra, G.; De Pasquale, V.; Scarcella, M.; Caporaso, M.G.; Pavone, L.M.; D’Agostino, M. Lysosomal positioning diseases: Beyond substrate storage. Open Biol. 2022, 12, 220155. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Lin, J.; Leung, W.T.; Wang, L. A basic understanding of mucopolysaccharidosis: Incidence, clinical features, diagnosis, and management. Intractable Rare Dis. Res. 2020, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Celik, B.; Tomatsu, S.C.; Tomatsu, S.; Khan, S.A. Epidemiology of Mucopolysaccharidoses Update. Diagnostics 2021, 11, 273. [Google Scholar] [CrossRef]
- Wiśniewska, K.; Wolski, J.; Gaffke, L.; Cyske, Z.; Pierzynowska, K.; Węgrzyn, G. Misdiagnosis in mucopolysaccharidoses. J. Appl. Genet. 2022, 63, 475–495. [Google Scholar] [CrossRef] [PubMed]
- Trabszo, C.; Ramms, B.; Chopra, P.; Lüllmann-Rauch, R.; Stroobants, S.; Spross, J.; Jeschke, A.; Schinke, T.; Boons, G.J.; Esko, J.D.; et al. Arylsulfatase K inactivation causes mucopolysaccharidosis due to deficient glucuronate desulfation of heparan and chondroitin sulfate. Biochem. J. 2020, 477, 3433–3451. [Google Scholar] [CrossRef] [PubMed]
- Velde, H.M.; Reurink, J.; Held, S.; Li, C.H.Z.; Yzer, S.; Oostrik, J.; Weeda, J.; Haer-Wigman, L.; Yntema, H.G.; Roosing, S.; et al. Usher syndrome type IV: Clinically and molecularly confirmed by novel ARSG variants. Hum. Genet. 2022, 141, 1723–1738. [Google Scholar] [CrossRef] [PubMed]
- Verheyen, S.; Blatterer, J.; Speicher, M.R.; Bhavani, G.S.; Boons, G.J.; Ilse, M.B.; Andrae, D.; Spross, J.; Vaz, F.M.; Kircher, S.G.; et al. Novel subtype of mucopolysaccharidosis caused by arylsulfatase K (ARSK) deficiency. J. Med. Genet. 2022, 59, 957–964. [Google Scholar] [CrossRef]
- Vasilev, F.; Sukhomyasova, A.; Otomo, T. Mucopolysaccharidosis-Plus Syndrome. Int. J. Mol. Sci. 2020, 21, 421. [Google Scholar] [CrossRef]
- Lipiński, P.; Szczałuba, K.; Buda, P.; Zakharova, E.Y.; Baydakova, G.; Ługowska, A.; Różdzyńska-Świątkowska, A.; Cyske, Z.; Węgrzyn, G.; Pollak, A.; et al. Mucopolysaccharidosis-Plus Syndrome: Report on a Polish Patient with a Novel VPS33A Variant with Comparison with Other Described Patients. Int. J. Mol. Sci. 2022, 23, 11424. [Google Scholar] [CrossRef]
- Fecarotta, S.; Tarallo, A.; Damiano, C.; Minopoli, N.; Parenti, G. Pathogenesis of Mucopolysaccharidoses, an Update. Int. J. Mol. Sci. 2020, 21, 2515. [Google Scholar] [CrossRef]
- Gaffke, L.; Pierzynowska, K.; Podlacha, M.; Brokowska, J.; Węgrzyn, G. Changes in cellular processes occurring in mucopolysaccharidoses as underestimated pathomechanisms of these diseases. Cell Biol. Int. 2021, 45, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Leal, A.F.; Benincore-Flórez, E.; Rintz, E.; Herreño-Pachón, A.M.; Celik, B.; Ago, Y.; Alméciga-Díaz, C.J.; Tomatsu, S. Mucopolysaccharidoses: Cellular Consequences of Glycosaminoglycans Accumulation and Potential Targets. Int. J. Mol. Sci. 2022, 24, 477. [Google Scholar] [CrossRef] [PubMed]
- Gaffke, L.; Pierzynowska, K.; Podlacha, M.; Hoinkis, D.; Rintz, E.; Brokowska, J.; Cyske, Z.; Wegrzyn, G. Underestimated Aspect of Mucopolysaccharidosis Pathogenesis: Global Changes in Cellular Processes Revealed by Transcriptomic Studies. Int. J. Mol. Sci. 2020, 21, 1204. [Google Scholar] [CrossRef] [PubMed]
- Gaffke, L.; Pierzynowska, K.; Rintz, E.; Cyske, Z.; Giecewicz, I.; Węgrzyn, G. Gene Expression-Related Changes in Morphologies of Organelles and Cellular Component Organization in Mucopolysaccharidoses. Int. J. Mol. Sci. 2021, 22, 2766. [Google Scholar] [CrossRef] [PubMed]
- Gaffke, L.; Pierzynowska, K.; Krzelowska, K.; Piotrowska, E.; Węgrzyn, G. Changes in expressions of genes involved in the regulation of cellular processes in mucopolysaccharidoses as assessed by fibroblast culture-based transcriptomic analyses. Metab. Brain Dis. 2021, 35, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Pierzynowska, K.; Gaffke, L.; Jankowska, E.; Rintz, E.; Witkowska, J.; Wieczerzak, E.; Podlacha, M.; Węgrzyn, G. Proteasome Composition and Activity Changes in Cultured Fibroblasts Derived From Mucopolysaccharidoses Patients and Their Modulation by Genistein. Front. Cell Dev. Biol. 2020, 8, 540726. [Google Scholar] [CrossRef]
- Gaffke, L.; Szczudło, Z.; Podlacha, M.; Cyske, Z.; Rintz, E.; Mantej, J.; Krzelowska, K.; Węgrzyn, G.; Pierzynowska, K. Impaired ion homeostasis as a possible associate factor in mucopolysaccharidosis pathogenesis: Transcriptomic, cellular and animal studies. Metab. Brain Dis. 2022, 37, 299–310. [Google Scholar] [CrossRef]
- Pierzynowska, K.; Żabińska, M.; Gaffke, L.; Cyske, Z.; Węgrzyn, G. Changes in expression of signal transduction-related genes, and formation of aggregates of GPER1 and OXTR receptors in mucopolysaccharidosis cells. Eur. J. Cell Biol. 2022, 101, 151232. [Google Scholar] [CrossRef]
- Brokowska, J.; Gaffke, L.; Pierzynowska, K.; Cyske, Z.; Węgrzyn, G. Cell cycle disturbances in mucopolysaccharidoses: Transcriptomic and experimental studies on cellular models. Exp. Biol. Med. 2022, 247, 1639–1649. [Google Scholar] [CrossRef]
- Gaffke, L.; Pierzynowska, K.; Cyske, Z.; Podlacha, M.; Węgrzyn, G. Contribution of vesicle trafficking dysregulation to the pathomechanism of mucopolysaccharidosis. Biochem. Biophys. Res. Commun. 2023, 665, 107–117. [Google Scholar] [CrossRef]
- Gaffke, L.; Rintz, E.; Pierzynowska, K.; Węgrzyn, G. Actin Cytoskeleton Polymerization and Focal Adhesion as Important Factors in the Pathomechanism and Potential Targets of Mucopolysaccharidosis Treatment. Cells 2023, 12, 1782. [Google Scholar] [CrossRef] [PubMed]
- Brokowska, J.; Pierzynowska, K.; Gaffke, L.; Rintz, E.; Węgrzyn, G. Expression of genes involved in apoptosis is dysregulated in mucopolysaccharidoses as revealed by pilot transcriptomic analyses. Cell Biol. Int. 2021, 45, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Padash, S.; Obaid, H.; Henderson, R.D.E.; Padash, Y.; Adams, S.J.; Miller, S.F.; Babyn, P. A pictorial review of the radiographic skeletal findings in Morquio syndrome (mucopolysaccharidosis type IV). Pediatr. Radiol. 2023, 53, 971–983. [Google Scholar] [CrossRef]
- Zanetti, A.; D’Avanzo, F.; AlSayed, M.; Brusius-Facchin, A.C.; Chien, Y.H.; Giugliani, R.; Izzo, E.; Kasper, D.C.; Lin, H.Y.; Lin, S.P.; et al. Molecular basis of mucopolysaccharidosis IVA (Morquio A syndrome): A review and classification of GALNS gene variants and reporting of 68 novel variants. Hum. Mutat. 2021, 42, 1384–1398. [Google Scholar] [CrossRef] [PubMed]
- Stütz, A.E.; Thonhofer, M.; Weber, P.; Wolfsgruber, A.; Wrodnigg, T.M. Pharmacological Chaperones for β-Galactosidase Related to GM1-Gangliosidosis and Morquio B: Recent Advances. Chem. Record. 2021, 21, 2980–2989. [Google Scholar] [CrossRef]
- Tam, Z.Y.; Cai, Y.H.; Gunawan, R. Elucidating cytochrome C release from mitochondria: Insights from an in silico three-dimensional model. Biophys. J. 2010, 99, 3155–3163. [Google Scholar] [CrossRef]
- Chae, H.J.; Kang, J.S.; Byun, J.O.; Han, K.S.; Kim, D.U.; Oh, S.M.; Kim, H.M.; Chae, S.W.; Kim, H.R. Molecular mechanism of staurosporine-induced apoptosis in osteoblasts. Pharmacol. Res. 2000, 42, 373–381. [Google Scholar] [CrossRef]
- Sahoo, G.; Samal, D.; Khandayataray, P.; Murthy, M.K. A Review on Caspases: Key Regulators of Biological Activities and Apoptosis. Mol. Neurobiol. 2023, 60, 5805–5837. [Google Scholar] [CrossRef]
- Park, M.Y.; Ha, S.E.; Vetrivel, P.; Kim, H.H.; Bhosale, P.B.; Abusaliya, A.; Kim, G.S. Differences of Key Proteins between Apoptosis and Necroptosis. BioMed Res. Int. 2021, 2021, 3420168. [Google Scholar] [CrossRef]
- Svandova, E.; Lesot, H.; Sharpe, P.; Matalova, E. Making the head: Caspases in life and death. Front. Cell Dev. Biol. 2023, 10, 1075751. [Google Scholar] [CrossRef]
- Boatright, K.M.; Deis, C.; Denault, J.B.; Sutherlin, D.P.; Salvesen, G.S. Activation of caspases-8 and -10 by FLIP(L). Biochem. J. 2004, 382, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Scott, F.L.; Stec, B.; Pop, C.; Dobaczewska, M.K.; Lee, J.J.; Monosov, E.; Robinson, H.; Salvesen, G.S.; Schwarzenbacher, R.; Riedl, S.J. The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature 2009, 457, 1019–1022. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.W.; Jeffrey, P.D.; Shi, Y. Mechanism of procaspase-8 activation by c-FLIPL. Proc. Natl. Acad. Sci. USA 2009, 106, 8169–8174. [Google Scholar] [CrossRef] [PubMed]
- Gonzalvez, F.; Lawrence, D.; Yang, B.; Yee, S.; Pitti, R.; Marsters, S.; Pham, V.C.; Stephan, J.P.; Lill, J.; Ashkenazi, A. TRAF2 Sets a threshold for extrinsic apoptosis by tagging caspase-8 with a ubiquitin shutoff timer. Mol. Cell 2012, 48, 888–899. [Google Scholar] [CrossRef]
- Contadini, C.; Ferri, A.; Cirotti, C.; Stupack, D.; Barilà, D. Caspase-8 and Tyrosine Kinases: A Dangerous Liaison in Cancer. Cancers 2023, 15, 3271. [Google Scholar] [CrossRef]
- Renatus, M.; Stennicke, H.R.; Scott, F.L.; Liddington, R.C.; Salvesen, G.S. Dimer formation drives the activation of the cell death protease caspase 9. Proc. Natl. Acad. Sci. USA 2001, 98, 14250–14255. [Google Scholar] [CrossRef]
- Kim, H.E.; Du, F.; Fang, M.; Wang, X. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc. Natl. Acad. Sci. USA 2005, 102, 17545–17550. [Google Scholar] [CrossRef]
- Malladi, S.; Challa-Malladi, M.; Fearnhead, H.O.; Bratton, S.B. The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J. 2009, 28, 1916–1925. [Google Scholar] [CrossRef]
- Bratton, S.B.; Salvesen, G.S. Regulation of the Apaf-1-caspase-9 apoptosome. J. Cell Sci. 2010, 123, 3209–3214. [Google Scholar] [CrossRef]
- Wu, C.C.; Bratton, S.B. Regulation of the intrinsic apoptosis pathway by reactive oxygen species. Antioxid. Redox Signal. 2013, 19, 546–558. [Google Scholar] [CrossRef]
- Park, H.; Kam, T.I.; Dawson, T.M.; Dawson, V.L. Poly (ADP-ribose) (PAR)-dependent cell death in neurodegenerative diseases. Int. Rev. Cell Mol. Biol. 2020, 353, 1–29. [Google Scholar] [PubMed]
- Rose, M.; Burgess, J.T.; O’Byrne, K.; Richard, D.J.; Bolderson, E. PARP Inhibitors: Clinical Relevance, Mechanisms of Action and Tumor Resistance. Front. Cell Dev. Biol. 2020, 8, 564601. [Google Scholar] [CrossRef] [PubMed]
- Villani, G.R.; Chierchia, A.; Di Napoli, D.; Di Natale, P. Unfolded protein response is not activated in the mucopolysaccharidoses but protein disulfide isomerase 5 is deregulated. J. Inherit. Metab. Dis. 2012, 35, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Pereira, V.G.; Gazarini, M.L.; Rodrigues, L.C.; da Silva, F.H.; Han, S.W.; Martins, A.M.; Tersariol, I.L.; D’Almeida, V. Evidence of lysosomal membrane permeabilization in mucopolysaccharidosis type I: Rupture of calcium and proton homeostasis. J. Cell. Physiol. 2010, 223, 335–342. [Google Scholar] [CrossRef]
- Noguti, J.; Pereira, V.G.; Guilheiro, J.M.; Martins, A.M.; D’Almeida, V.; Ribeiro, D.A. Apoptosis status and proliferative activity in mucopolysaccharidosis type I mice tongue mucosa cells. Dent. Res. J. 2012, 9 (Suppl. 1), S69–S74. [Google Scholar]
- Viana, G.M.; Buri, M.V.; Paredes-Gamero, E.J.; Martins, A.M.; D’Almeida, V. Impaired Hematopoiesis and Disrupted Monocyte/Macrophage Homeostasis in Mucopolysaccharidosis Type I Mice. J. Cell. Physiol. 2016, 231, 698–707. [Google Scholar] [CrossRef]
- Viana, G.M.; do Nascimento, C.C.; Paredes-Gamero, E.J.; D’Almeida, V. Altered Cellular Homeostasis in Murine MPS I Fibroblasts: Evidence of Cell-Specific Physiopathology. JIMD Rep. 2017, 36, 109–116. [Google Scholar]
- Liu, D.; Jiang, Z.; Deng, L.; Li, H.; Jiang, H. Identification of an α-l-iduronidase (IDUA) M1T mutation in a Chinese family with autosomal recessive mucopolysaccharidosis I. Ann. N. Y. Acad. Sci. 2023, 1526, 114–125. [Google Scholar] [CrossRef]
- Fusar Poli, E.; Zalfa, C.; D’Avanzo, F.; Tomanin, R.; Carlessi, L.; Bossi, M.; Nodari, L.R.; Binda, E.; Marmiroli, P.; Scarpa, M.; et al. Murine neural stem cells model Hunter disease in vitro: Glial cell-mediated neurodegeneration as a possible mechanism involved. Cell Death Dis. 2013, 4, e906. [Google Scholar] [CrossRef]
- Kobolák, J.; Molnár, K.; Varga, E.; Bock, I.; Jezsó, B.; Téglási, A.; Zhou, S.; Lo Giudice, M.; Hoogeveen-Westerveld, M.; Pijnappel, W.P.; et al. Modelling the neuropathology of lysosomal storage disorders through disease-specific human induced pluripotent stem cells. Exp. Cell Res. 2019, 380, 216–233. [Google Scholar] [CrossRef]
- Arfi, A.; Richard, M.; Gandolphe, C.; Bonnefont-Rousselot, D.; Thérond, P.; Scherman, D. Neuroinflammatory and oxidative stress phenomena in MPS IIIA mouse model: The positive effect of long-term aspirin treatment. Mol. Genet. Metabol. 2011, 103, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Villani, G.R.; Gargiulo, N.; Faraonio, R.; Castaldo, S.; Gonzalez, Y.; Reyero, E.; Di Natale, P. Cytokines, neurotrophins, and oxidative stress in brain disease from mucopolysaccharidosis IIIB. J. Neurosci. Res. 2007, 85, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Simonaro, C.M.; Haskins, M.E.; Schuchman, E.H. Articular chondrocytes from animals with a dermatan sulfate storage disease undergo a high rate of apoptosis and release nitric oxide and inflammatory cytokines: A possible mechanism underlying degenerative joint disease in the mucopolysaccharidoses. Lab. Investig. 2001, 81, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Tessitore, A.; Faella, A.; O’Malley, T.; Cotugno, G.; Doria, M.; Kunieda, T.; Matarese, G.; Haskins, M.; Auricchio, A. Biochemical, pathological, and skeletal improvement of mucopolysaccharidosis VI after gene transfer to liver but not to muscle. Mol. Ther. 2008, 16, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Tessitore, A.; Pirozzi, M.; Auricchio, A. Abnormal autophagy, ubiquitination, inflammation and apoptosis are dependent upon lysosomal storage and are useful biomarkers of mucopolysaccharidosis VI. PathoGenetics 2009, 2, 4. [Google Scholar] [CrossRef] [PubMed]
- Broeders, M.; van Rooij, J.; Oussoren, E.; van Gestel, T.; Smith, C.; Kimber, S.; Verdijk, R.; Wagenmakers, M.; van den Hout, J.; van der Ploeg, A.; et al. Modeling cartilage pathology in mucopolysaccharidosis VI using iPSCs reveals early dysregulation of chondrogenic and metabolic gene expression. Front. Bioeng. Biotechnol. 2022, 10, 949063. [Google Scholar] [CrossRef]
- Simonaro, C.M.; D’Angelo, M.; Haskins, M.E.; Schuchman, E.H. Joint and bone disease in mucopolysaccharidoses VI and VII: Identification of new therapeutic targets and biomarkers using animal models. Pediatr. Res. 2005, 57, 701–707. [Google Scholar] [CrossRef]
- Golda, A.; Jurecka, A.; Gajda, K.; Tylki-Szymańska, A.; Lalik, A. Human pulmonary artery endothelial cells in the model of mucopolysaccharidosis VI present a prohypertensive phenotype. Mol. Genet. Metab. Rep. 2015, 3, 11–17. [Google Scholar] [CrossRef]
- Richard, M.; Arfi, A.; Rhinn, H.; Gandolphe, C.; Scherman, D. Identification of new markers for neurodegeneration process in the mouse model of Sly disease as revealed by expression profiling of selected genes. J. Neurosci. Res. 2008, 86, 3285–3294. [Google Scholar] [CrossRef]
- Yamashita, T.; Fujii, T.; Yamauchi, I.; Ueda, Y.; Hirota, K.; Kanai, Y.; Yasoda, A.; Inagaki, N. C-Type Natriuretic Peptide Restores Growth Impairment Under Enzyme Replacement in Mice With Mucopolysaccharidosis VII. Endocrinology 2020, 161, bqaa008. [Google Scholar] [CrossRef]
- Bar, S.; Prasad, M.; Datta, R. Neuromuscular degeneration and locomotor deficit in a Drosophila model of mucopolysaccharidosis VII is attenuated by treatment with resveratrol. Dis. Models Mech. 2018, 11, dmm036954. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, V.J.; Bravo, S.B.; Chantada-Vazquez, M.P.; Colón, C.; De Castro, M.J.; Morales, M.; Vitoria, I.; Tomatsu, S.; Otero-Espinar, F.J.; Couce, M.L. Characterization of New Proteomic Biomarker Candidates in Mucopolysaccharidosis Type IVA. Int. J. Mol. Sci. 2021, 22, 226. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cui, J.; Li, Z.; Fu, X.; Li, J.; Li, H.; Wang, S.; Zhang, M. ORP8 induces apoptosis by releasing cytochrome c from mitochondria in non small cell lung cancer. Oncol. Rep. 2020, 43, 1516–1524. [Google Scholar] [CrossRef] [PubMed]
| Log2FC of Levels of Selected Transcripts in MPS IV Subtypes vs. HDFa Line | |||||
|---|---|---|---|---|---|
| Transcript * | MPS IVA | MPS IVB | Transcript * | MPS IVA | MPS IVB |
| Up-Regulated Transcripts | Down-Regulated Transcripts | ||||
| CLU (tr. 1) | 2.26 | 3.84 | BCAP29 | −1.02 | −0.90 |
| CLU (tr. 2) | 1.73 | 3.46 | BNIP3 | −0.71 | −0.74 |
| CLU (tr. 3) | 1.11 | 2.89 | C1D | −1.38 | −1.38 |
| CLU (tr. 4) | 1.90 | 3.43 | CDKN1A | −2.42 | −2.40 |
| CLU (tr. 5) | 1.86 | 3.46 | CLPTM1L | −1.33 | −1.04 |
| CRYAB | 0.38 | −0.31 | GAPDH | −0.67 | −0.78 |
| COMP | 2.58 | 1.67 | GPER1 | −0.96 | −1.51 |
| CRIP1 | 3.18 | 2.01 | IGFBP3 | −5.17 | −1.49 |
| ERCC2 | 0.47 | 0.39 | PRKCD | −0.65 | −0.90 |
| ERCC6 | 0.72 | 0.97 | RYBP | −0.77 | −0.76 |
| FNIP2 | 0.92 | 0.97 | SGK1 | −1.33 | 0.18 |
| HIC1 | 0.35 | 0.74 | TNFAIP8 | −0.52 | −0.26 |
| HIP1 | 0.84 | 0.99 | ACAA2 | −0.20 | −0.59 |
| MAX | 1.11 | 1.33 | ARL6IP5 | −0.38 | −0.72 |
| MLLT11 | 0.78 | 0.69 | CAV1 (tr. 1) | −0.31 | −0.89 |
| MUL1 | 0.10 | 0.64 | CAV1 (tr. 2) | −0.40 | −1.05 |
| NLRP1 | 0.84 | 1.85 | CAV1 (tr. 3) | −2.95 | −4.74 |
| PLK3 | 0.50 | 0.59 | CYCS | −0.81 | −0.76 |
| PPARD | 0.38 | 0.57 | FOXO3 | −0.57 | −0.51 |
| RHOB | 0.52 | 0.94 | GAS6 | −0.39 | −1.29 |
| RNF216 | 0.38 | 0.44 | HTATIP2 | 0.02 | −0.97 |
| RPS3 | 0.97 | 1.00 | KPNB1 | −1.99 | −1.58 |
| SMAD3 | 0.61 | 1.21 | KREMEN1 | −1.77 | −2.54 |
| STPG1 | −0.19 | 0.72 | MSH5 | −0.93 | −1.33 |
| TMEM109 | 0.29 | 0.33 | PDK1 | −0.90 | −1.39 |
| TOP2A | 1.64 | 1.68 | PDK2 | −0.73 | −0.80 |
| TRIM35 | 0.29 | 0.51 | PERP | −0.55 | −0.78 |
| UACA | 0.21 | 0.99 | PLSCR1 | −0.44 | −0.67 |
| XIAP | 0.48 | 1.15 | PSMD10 | −0.37 | −0.45 |
| ZC3H12A | 0.71 | 1.02 | RIPK2 | 0.11 | −0.56 |
| SGPP1 | −0.42 | −0.22 | |||
| SH3GLB1 (tr. 1) | −0.15 | −0.31 | |||
| SH3GLB1 (tr. 2) | −0.63 | −0.63 | |||
| SOD2 | −1.34 | −1.73 | |||
| TGFBR1 | −0.26 | −1.09 | |||
| TGFBR2 | −0.33 | −0.74 | |||
| TMBIM4 | −0.46 | −0.92 | |||
| TNFRSF11B | −0.74 | −1.23 | |||
| TRAF3IP2 | −0.33 | −1.04 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brokowska, J.; Gaffke, L.; Pierzynowska, K.; Węgrzyn, G. Enhanced Efficiency of the Basal and Induced Apoptosis Process in Mucopolysaccharidosis IVA and IVB Human Fibroblasts. Int. J. Mol. Sci. 2023, 24, 14119. https://doi.org/10.3390/ijms241814119
Brokowska J, Gaffke L, Pierzynowska K, Węgrzyn G. Enhanced Efficiency of the Basal and Induced Apoptosis Process in Mucopolysaccharidosis IVA and IVB Human Fibroblasts. International Journal of Molecular Sciences. 2023; 24(18):14119. https://doi.org/10.3390/ijms241814119
Chicago/Turabian StyleBrokowska, Joanna, Lidia Gaffke, Karolina Pierzynowska, and Grzegorz Węgrzyn. 2023. "Enhanced Efficiency of the Basal and Induced Apoptosis Process in Mucopolysaccharidosis IVA and IVB Human Fibroblasts" International Journal of Molecular Sciences 24, no. 18: 14119. https://doi.org/10.3390/ijms241814119
APA StyleBrokowska, J., Gaffke, L., Pierzynowska, K., & Węgrzyn, G. (2023). Enhanced Efficiency of the Basal and Induced Apoptosis Process in Mucopolysaccharidosis IVA and IVB Human Fibroblasts. International Journal of Molecular Sciences, 24(18), 14119. https://doi.org/10.3390/ijms241814119









