Influence of Clinical Factors on miR-3613-3p Expression in Colorectal Cancer
Abstract
1. Introduction
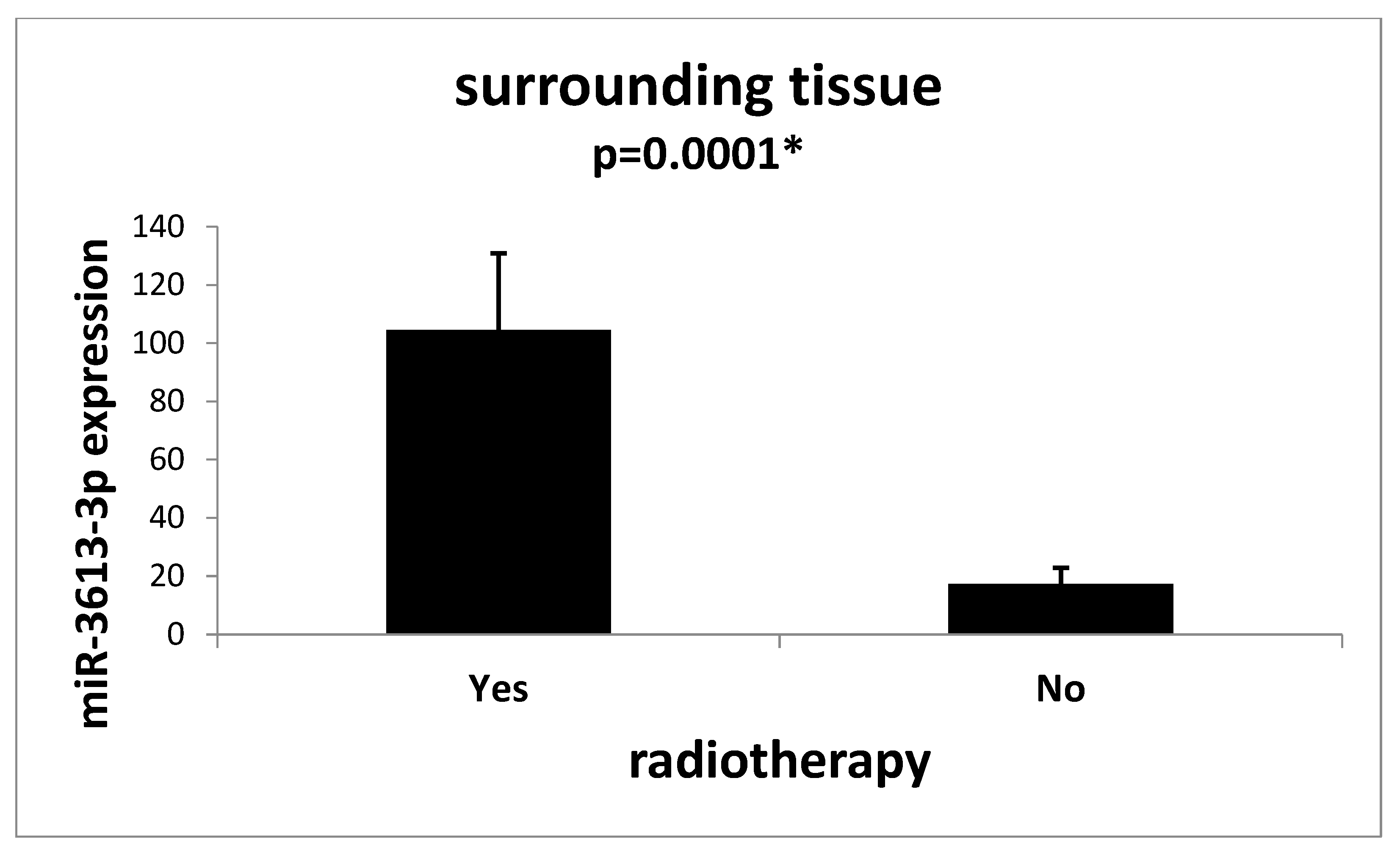
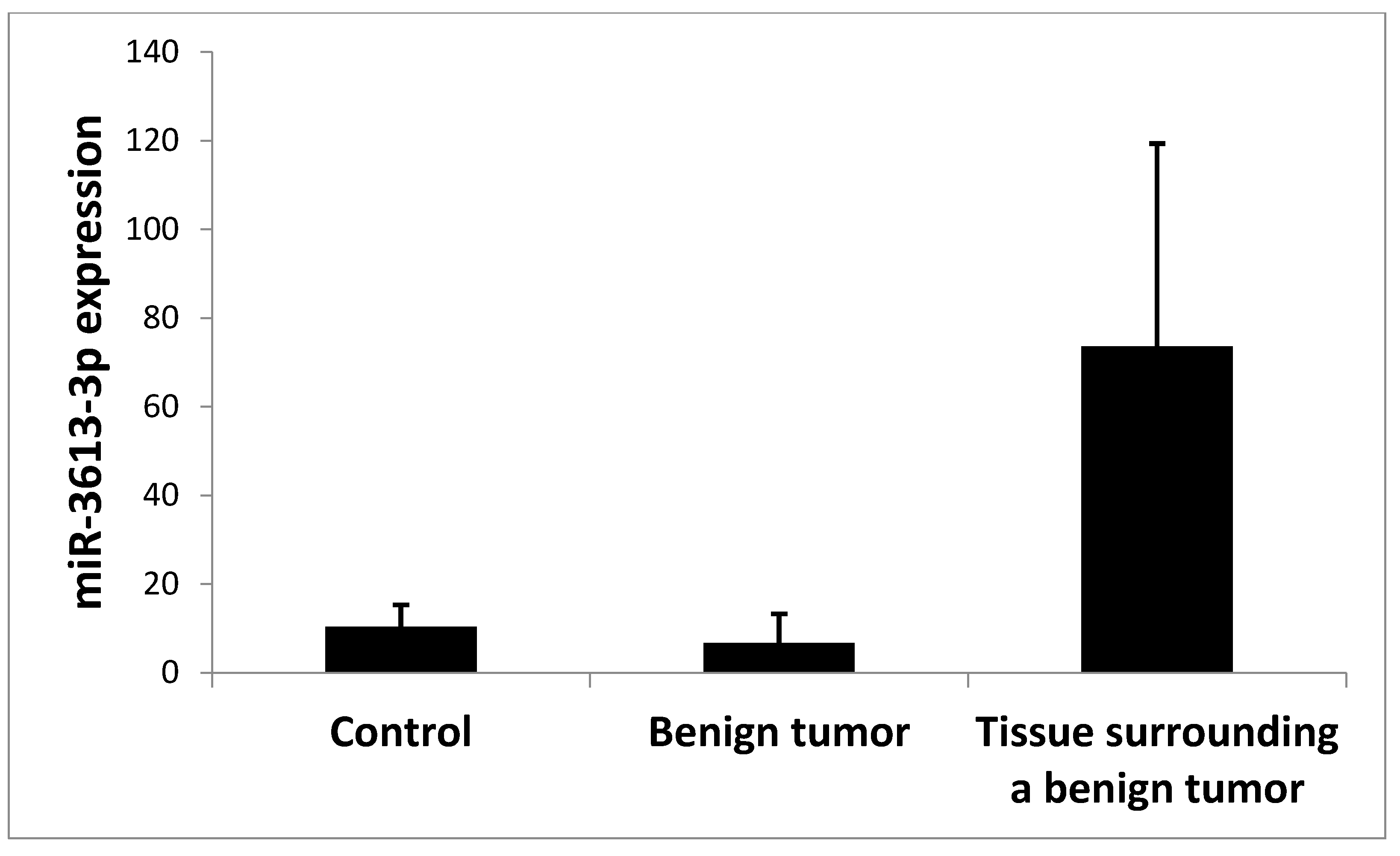
2. Results
3. Discussion
4. Materials and Methods
4.1. Sample Collecting
4.2. Total RNA Isolation
4.3. Reverse Transcription
4.4. qPCR Reaction
4.5. The miRNA Profiling
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. The global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2022, 72, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Dekker, E.; Chiu, H.M.; Lansdorp-Vogelaar, I.; Caro, L.E.; Dominitz, J.A.; Halloran, S.; Hassan, C.; Ismael, J.; Jover, R.; Kaminski, M.F.; et al. Colorectal Cancer Screening in the Novel Coronavirus Disease-2019 Era. Gastroenterology 2020, 159, 1998–2003. [Google Scholar] [CrossRef] [PubMed]
- Qaseem, A.; Crandall, C.J.; Mustafa, R.A.; Hicks, L.A.; Wilt, T.J. Screening for Colorectal Cancer in Asymptomatic Average-Risk Adults: A Guidance Statement From the American College of Physicians. Ann. Intern. Med. 2019, 171, 643–654. [Google Scholar] [CrossRef]
- Argilés, G.; Tabernero, J.; Labianca, R.; Hochhauser, D.; Salazar, R.; Iveson, T.; Laurent-Puig, P.; Quirke, P.; Yoshino, T.; Taieb, J.; et al. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis. treatment and follow-up. Ann. Oncol. 2020, 31, 1291–1305. [Google Scholar] [CrossRef]
- Li, F.Y.; Lai, M.D. Colorectal cancer. one entity or three. J. Zhejiang Univ. Sci. B 2009, 10, 219–229. [Google Scholar] [CrossRef]
- Redston, M. Carcinogenesis in the GI tract: From morphology to genetics and back again. Mod. Pathol. 2001, 14, 236–245. [Google Scholar] [CrossRef][Green Version]
- Ogino, S.; Goel, A. Molecular classification and correlates in colorectal cancer. J. Mol. Diagn. 2008, 10, 13–27. [Google Scholar] [CrossRef]
- Dienstmann, R.; Villacampa, G.; Sveen, A.; Mason, M.J.; Niedzwiecki, D.; Nesbakken, A.; Moreno, V.; Warren, R.S.; Lothe, R.A.; Guinney, J. Relative contribution of clinicopathological variables. genomic markers, transcriptomic subtyping and microenvironment features for outcome prediction in stage II/III colorectal cancer. Ann. Oncol. 2019, 30, 1622–1629. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Alwers, E.; Jia, M.; Kloor, M.; Bläker, H.; Brenner, H.; Hoffmeister, M. Associations Between Molecular Classifications of Colorectal Cancer and Patient Survival: A Systematic Review. Clin. Gastroenterol. Hepatol. 2019, 17, 402–410.e2. [Google Scholar] [CrossRef] [PubMed]
- Phipps, A.I.; Limburg, P.J.; Baron, J.A.; Burnett-Hartman, A.N.; Weisenberger, D.J.; Laird, P.W.; Sinicrope, F.A.; Rosty, C.; Buchanan, D.D.; Potter, J.D.; et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 2015, 148, 77–87.e2. [Google Scholar] [CrossRef]
- Di Martino, M.T.; Arbitrio, M.; Caracciolo, D.; Cordua, A.; Cuomo, O.; Grillone, K.; Riillo, C.; Caridà, G.; Scionti, F.; Labanca, C.; et al. miR-221/222 as biomarkers and targets for therapeutic intervention on cancer and other diseases: A systematic review. Mol. Ther. Nucleic Acids 2022, 27, 1191–1224. [Google Scholar] [CrossRef] [PubMed]
- Pichler, M.; Ress, A.L.; Winter, E.; Stiegelbauer, V.; Karbiener, M.; Schwarzenbacher, D.; Scheideler, M.; Ivan, C.; Jahn, S.W.; Kiesslich, T.; et al. MiR-200a regulates epithelial to mesenchymal transition-related gene expression and determines prognosis in colorectal cancer patients. Br. J. Cancer 2014, 110, 1614–1621. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis. Mechanisms of Actions and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Pong, S.K.; Gullerova, M. Noncanonical functions of microRNA pathway enzymes—Drosha. DGCR8. Dicer and Ago proteins. FEBS Lett. 2018, 592, 2973–2986. [Google Scholar] [CrossRef]
- Weiland, M.; Gao, X.H.; Zhou, L.; Mi, Q.S. Small RNAs have a large impact: Circulating microRNAs as biomarkers for human diseases. RNA Biol. 2012, 9, 850–859. [Google Scholar] [CrossRef]
- Pathak, S.; Banerjee, A. Emerging Importance of microRNA in Early Detection of Colorectal Cancer. Endocr. Metab Immune Disord. Drug Targets 2021, 21, 2–3. [Google Scholar] [CrossRef]
- Bhome, R.; Emaduddin, M.; James, V.; House, L.M.; Thirdborough, S.M.; Mellone, M.; Tulkens, J.; Primrose, J.N.; Thomas, G.J.; De Wever, O.; et al. Epithelial to mesenchymal transition influences fibroblast phenotype in colorectal cancer by altering miR-200 levels in extracellular vesicles. J. Extracell. Vesicles 2022, 11, e12226. [Google Scholar] [CrossRef]
- Lai, P.S.; Chang, W.M.; Chen, Y.Y.; Lin, Y.F.; Liao, H.F.; Chen, C.Y. Circulating microRNA-762 upregulation in colorectal cancer may be accompanied by Wnt-1/β-catenin signaling. Cancer Biomark. 2021, 32, 111–122. [Google Scholar] [CrossRef]
- Zaki, A.; Fawzy, A.; Akel, S.Y.; Gamal, H.; Elshimy, R.A.A. Evaluation of microRNA 92a Expression and Its Target Protein Bim in Colorectal Cancer. Asian Pac. J. Cancer Prev. 2022, 23, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, X.; Li, Y.; Ren, P.; Zhang, C.; Wang, L.; Du, X.; Xing, B. miR-6716-5p promotes metastasis of colorectal cancer through downregulating NAT10 expression. Cancer Manag. Res. 2019, 11, 5317–5332. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Chen, J.; Li, Z.; Li, L.; Chen, J.; Guo, Y. microRNA-96 promotes occurrence and progression of colorectal cancer via regulation of the AMPKα2-FTO-m6A/MYC axis. J. Exp. Clin. Cancer Res. 2020, 39, 240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhao, L.; Shen, Q.; Lv, Q.; Jin, M.; Ma, H.; Nie, X.; Zheng, X.; Huang, S.; Zhou, P.; et al. Down-regulation of KIAA1199/CEMIP by miR-216a suppresses tumor invasion and metastasis in colorectal cancer. Int. J. Cancer 2017, 140, 2298–2309. [Google Scholar] [CrossRef]
- Cao, R.; Yang, F.; Ma, S.C.; Liu, L.; Zhao, Y.; Li, Y.; Wu, D.H.; Wang, T.; Lu, W.J.; Cai, W.J.; et al. Development and interpretation of a pathomics-based model for the prediction of microsatellite instability in Colorectal Cancer. Theranostics 2020, 10, 11080–11091. [Google Scholar] [CrossRef]
- Chen, C.; Yuan, W.; Zhou, Q.; Shao, B.; Guo, Y.; Wang, W.; Yang, S.; Guo, Y.; Zhao, L.; Dang, Q.; et al. N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics 2021, 11, 4298–4315. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, T.; Ruszkowska, M.; Danielewicz, A.; Niedźwiedzka, E.; Arłukowicz, T.; Przybyłowicz, K.E. A Review of Colorectal Cancer in Terms of Epidemiology. Risk Factors. Development. Symptoms and Diagnosis. Cancers 2021, 13, 2025. [Google Scholar] [CrossRef]
- Ding, L.; Lan, Z.; Xiong, X.; Ao, H.; Feng, Y.; Gu, H.; Yu, M.; Cui, Q. The Dual Role of MicroRNAs in Colorectal Cancer Progression. Int. J. Mol. Sci. 2018, 19, 2791. [Google Scholar] [CrossRef]
- Yan, W.; Yang, W.; Liu, Z.; Wu, G. Characterization of microRNA expression in primary human colon adenocarcinoma cells (SW480) and their lymph node metastatic derivatives (SW620). OncoTargets Ther. 2018, 11, 4701–4709. [Google Scholar] [CrossRef]
- Bibi, F.; Naseer, M.I.; Alvi, S.A.; Yasir, M.; Jiman-Fatani, A.A.; Sawan, A.; Abuzenadah, A.M.; Al-Qahtani, M.H.; Azhar, E.I. microRNA analysis of gastric cancer patients from Saudi Arabian population. BMC Genom. 2016, 17, 751. [Google Scholar] [CrossRef]
- Chen, C.; Pan, Y.; Bai, L.; Chen, H.; Duan, Z.; Si, Q.; Zhu, R.; Chuang, T.H.; Luo, Y. MicroRNA-3613-3p functions as a tumor suppressor and represents a novel therapeutic target in breast cancer. Breast Cancer Res. 2021, 23, 12. [Google Scholar] [CrossRef] [PubMed]
- Pu, Q.; Huang, Y.; Lu, Y.; Peng, Y.; Zhang, J.; Feng, G.; Wang, C.; Liu, L.; Dai, Y. Tissue-specific and plasma microRNA profiles could be promising biomarkers of histological classification and TNM stage in non-small cell lung cancer. Thorac. Cancer 2016, 7, 348–354. [Google Scholar] [CrossRef]
- Chong, G.O.; Jeon, H.S.; Han, H.S.; Son, J.W.; Lee, Y.H.; Hong, D.G.; Lee, Y.S.; Cho, Y.L. Differential microRNA expression profiles in primary and recurrent epithelial ovarian cancer. Anticancer Res. 2015, 35, 2611–2617. [Google Scholar] [PubMed]
- Ji, H.; Chen, M.; Greening, D.W.; He, W.; Rai, A.; Zhang, W.; Simpson, R.J. Deep sequencing of RNA from three different extracellular vesicle (EV) subtypes released from the human LIM1863 colon cancer cell line uncovers distinct miRNA-enrichment signatures. PLoS ONE 2014, 9, e110314. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, E.; Kang, J.; Yang, X.; Liu, H. MiR-3613-3p affects cell proliferation and cell cycle in hepatocellular carcinoma. Oncotarget 2017, 8, 93014–93028. [Google Scholar] [CrossRef]
- Xiang, F.; Xu, X. CirRNA F-circEA-2a Suppresses the Role of miR-3613-3p in Colorectal Cancer by Direct Sponging and Predicts Poor Survival. Cancer Manag. Res. 2022, 14, 1825–1833. [Google Scholar] [CrossRef]
- Avsar, R.; Gurer, T.; Aytekin, A. Bioinformatics and Expression Analyses of miR-639, miR-641, miR-1915-3p and miR-3613-3p in Colorectal Cancer Pathogenesis. J. Cancer 2023, 14, 2399–2409. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Y.; Du, J.; Lin, D.; Li, F. MiR-3613-3p from carcinoma-associated fibroblasts exosomes promoted breast cancer cell proliferation and metastasis by regulating SOCS2 expression. IUBMB Life 2020, 72, 1705–1714. [Google Scholar] [CrossRef]
- Fricke, A.; Cimniak, A.F.V.; Ullrich, P.V.; Becherer, C.; Bickert, C.; Pfeifer, D.; Heinz, J.; Stark, G.B.; Bannasch, H.; Braig, D.; et al. Whole blood miRNA expression analysis reveals miR-3613-3p as a potential biomarker for dedifferentiated liposarcoma. Cancer Biomark. Sect. A Dis. Markers 2018, 22, 199–207. [Google Scholar] [CrossRef]
- Wu, R.L.; Ali, S.; Bandyopadhyay, S.; Alosh, B.; Hayek, K.; Daaboul, M.F.; Winer, I.; Sarkar, F.H.; Ali-Fehmi, R. Comparative Analysis of Differentially Expressed miRNAs and their Downstream mRNAs in Ovarian Cancer and its Associated Endometriosis. J. Cancer Sci. Ther. 2015, 7, 258–265. [Google Scholar] [CrossRef]
- Song, J.; Wang, W.; Wang, Y.; Qin, Y.; Wang, Y.; Zhou, J.; Wang, X.; Zhang, Y.; Wang, Q. Epithelial-mesenchymal transition markers screened in a cell-based model and validated in lung adenocarcinoma. BMC Cancer 2019, 19, 680. [Google Scholar] [CrossRef]
- Janani, B.; Vijayakumar, M.; Priya, K.; Kim, J.H.; Prabakaran, D.S.; Shahid, M.; Al-Ghamdi, S.; Alsaidan, M.; Othman Bahakim, N.; Hassan Abdelzaher, M.; et al. EGFR-Based Targeted Therapy for Colorectal Cancer-Promises and Challenges. Vaccines 2022, 10, 499. [Google Scholar] [CrossRef] [PubMed]
- Unson, S.; Chang, T.-C.; Yang, Y.-N.; Wang, S.-H.; Huang, C.-H.; Crawford, D.R.; Huang, H.-M.; Li, Z.-L.; Lin, H.-Y.; Whang-Peng, J.; et al. Heteronemin and Tetrac Induce Anti-Proliferation by Blocking EGFR-Mediated Signaling in Colorectal Cancer Cells. Mar. Drugs 2022, 20, 482. [Google Scholar] [CrossRef]
- Machmouchi, A.; Chehade, L.; Temraz, S.; Shamseddine, A. Overcoming EGFR Resistance in Metastatic Colorectal Cancer Using Vitamin C: A Review. Biomedicines 2023, 11, 678. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Liao, H.; Xie, R.; Zhang, Y.; Zheng, R.; Chen, J.; Zhang, B. Overexpression of miRNA-3613-3p Enhances the Sensitivity of Triple Negative Breast Cancer to CDK4/6 Inhibitor Palbociclib. Front. Oncol. 2020, 10, 590813. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 4, 402–408. [Google Scholar] [CrossRef]




| Group | N | Mean | SE | p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| Control | Tumor | Surrounding Tissue | Benign Tumor | Tissue Surrounding a Benign Tumor | ||||
| Control | 13 | 10.420 | 4.870 | 0.0020 * | 0.302 | 0.622 | 0.131 | |
| Tumor | 60 | 0.559 | 0.144 | 0.002 * | 0.00003 * | 0.020 * | 0.0004 * | |
| Surrounding tissue | 60 | 17.278 | 3.273 | 0.302 | 0.00003 * | 0.101 | 0.037 * | |
| Benign tumor | 5 | 6.674 | 6.592 | 0.622 | 0.0200 * | 0.101 | 0.026 * | |
| Tissue surrounding a benign tumor | 5 | 73.591 | 45.746 | 0.131 | 0.0004 * | 0.037 * | 0.026 * | |
| Variable | N | % |
|---|---|---|
| cT stage | ||
| T2 | 7 | 11 |
| T2/T3 | 3 | 5 |
| T3 | 34 | 52 |
| T4 | 16 | 24 |
| T4a | 1 | 2 |
| unknown | 4 | 6 |
| cN stage | ||
| N0 | 8 | 13 |
| N1 | 7 | 11 |
| N2 | 5 | 7 |
| Nx | 43 | 66 |
| Nx/0 | 1 | 2 |
| unknown | 7 | 11 |
| cM stage | ||
| M0 | 52 | 80 |
| M1 | 6 | 9 |
| Mx/0 | 1 | 2 |
| unknown | 6 | 9 |
| Variable | N | % |
|---|---|---|
| pT stage | ||
| T1 | 2 | 2.5 |
| T2 | 14 | 21 |
| T3 | 42 | 65 |
| T4 | 6 | 9 |
| T4a | 2 | 2.5 |
| pN stage | ||
| N0 | 31 | 48 |
| N1 | 10 | 15 |
| N1a | 3 | 5 |
| N1b | 7 | 11 |
| N2 | 3 | 5 |
| N2a | 5 | 7 |
| N2b | 6 | 9 |
| pM stage | ||
| M0 | 56 | 86 |
| M1 | 5 | 7 |
| M1a | 3 | 5 |
| M1b | 1 | 2 |
| Variable | N | % |
|---|---|---|
| Organ | ||
| rectum | 27 | 42 |
| hepatic fold | 5 | 8 |
| caecum | 5 | 8 |
| splenic fold | 3 | 4 |
| sigmoid | 12 | 19 |
| descending colon | 5 | 8 |
| transverse colon | 7 | 11 |
| Variable | N | % |
|---|---|---|
| Advancement according to TNM | ||
| I | 14 | 21 |
| IIA | 14 | 21 |
| IIB | 2 | 3 |
| IIIA | 2 | 3 |
| IIIB | 23 | 36 |
| IIIC | 2 | 3 |
| IV | 5 | 7 |
| IVA | 2 | 3 |
| IVB | 2 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil-Kulik, P.; Petniak, A.; Kluz, N.; Wallner, G.; Skoczylas, T.; Ciechański, A.; Kocki, J. Influence of Clinical Factors on miR-3613-3p Expression in Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 14023. https://doi.org/10.3390/ijms241814023
Gil-Kulik P, Petniak A, Kluz N, Wallner G, Skoczylas T, Ciechański A, Kocki J. Influence of Clinical Factors on miR-3613-3p Expression in Colorectal Cancer. International Journal of Molecular Sciences. 2023; 24(18):14023. https://doi.org/10.3390/ijms241814023
Chicago/Turabian StyleGil-Kulik, Paulina, Alicja Petniak, Natalia Kluz, Grzegorz Wallner, Tomasz Skoczylas, Aleksander Ciechański, and Janusz Kocki. 2023. "Influence of Clinical Factors on miR-3613-3p Expression in Colorectal Cancer" International Journal of Molecular Sciences 24, no. 18: 14023. https://doi.org/10.3390/ijms241814023
APA StyleGil-Kulik, P., Petniak, A., Kluz, N., Wallner, G., Skoczylas, T., Ciechański, A., & Kocki, J. (2023). Influence of Clinical Factors on miR-3613-3p Expression in Colorectal Cancer. International Journal of Molecular Sciences, 24(18), 14023. https://doi.org/10.3390/ijms241814023





