The Effect of Antimicrobial Photodynamic Inactivation on the Protein Profile of Dormant Mycolicibacterium smegmatis Containing Endogenous Porphyrins
Abstract
1. Introduction
2. Results
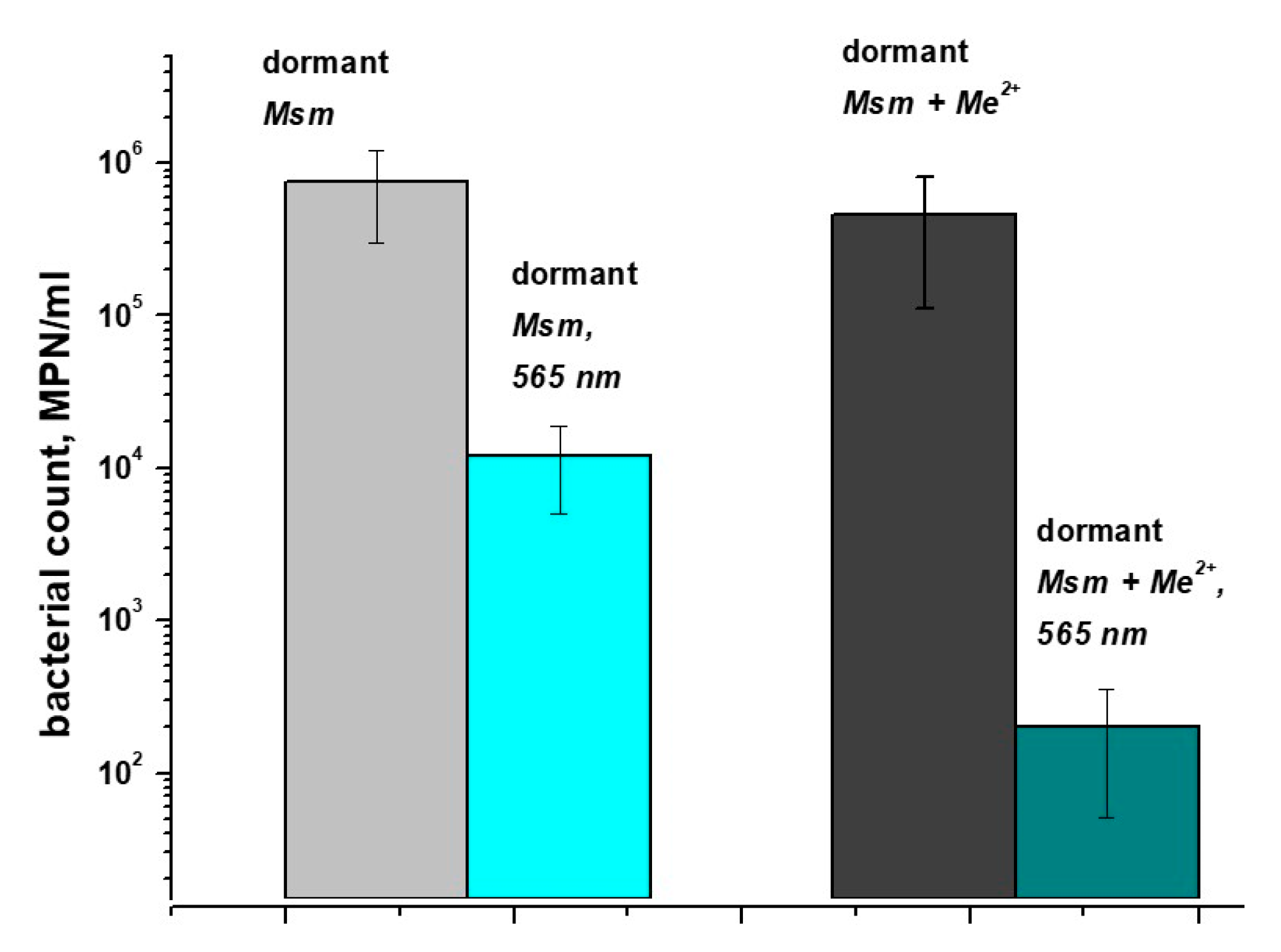
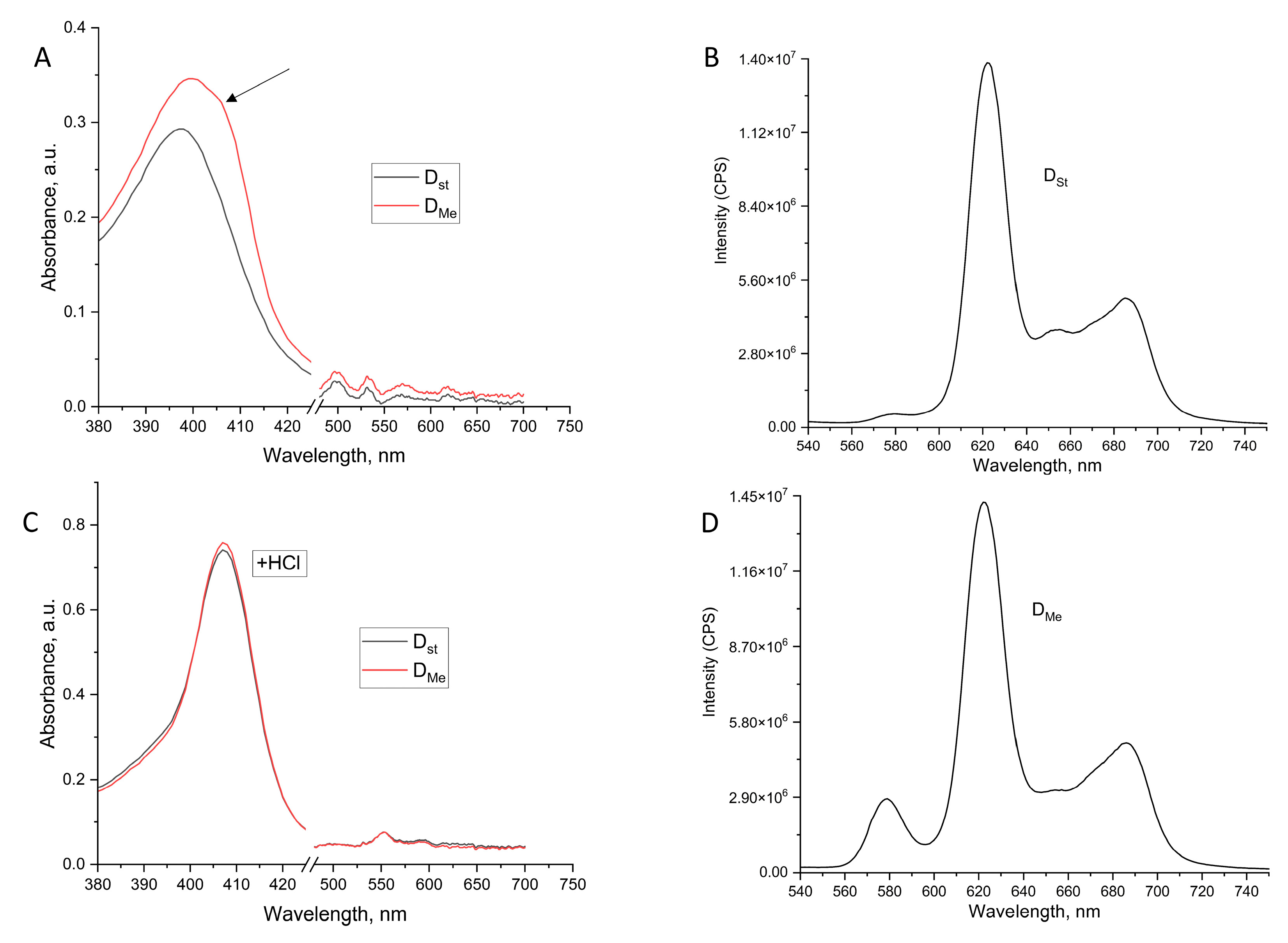
2.1. Influence of Metal Ions on the Accumulation of Porphyrins in Dormant Forms of M. smegmatis and Their Photosensitivity
2.2. Influence of Metals on the Representation of Proteins of Porphyrin Metabolism in Dormant M. smegmatis Cells
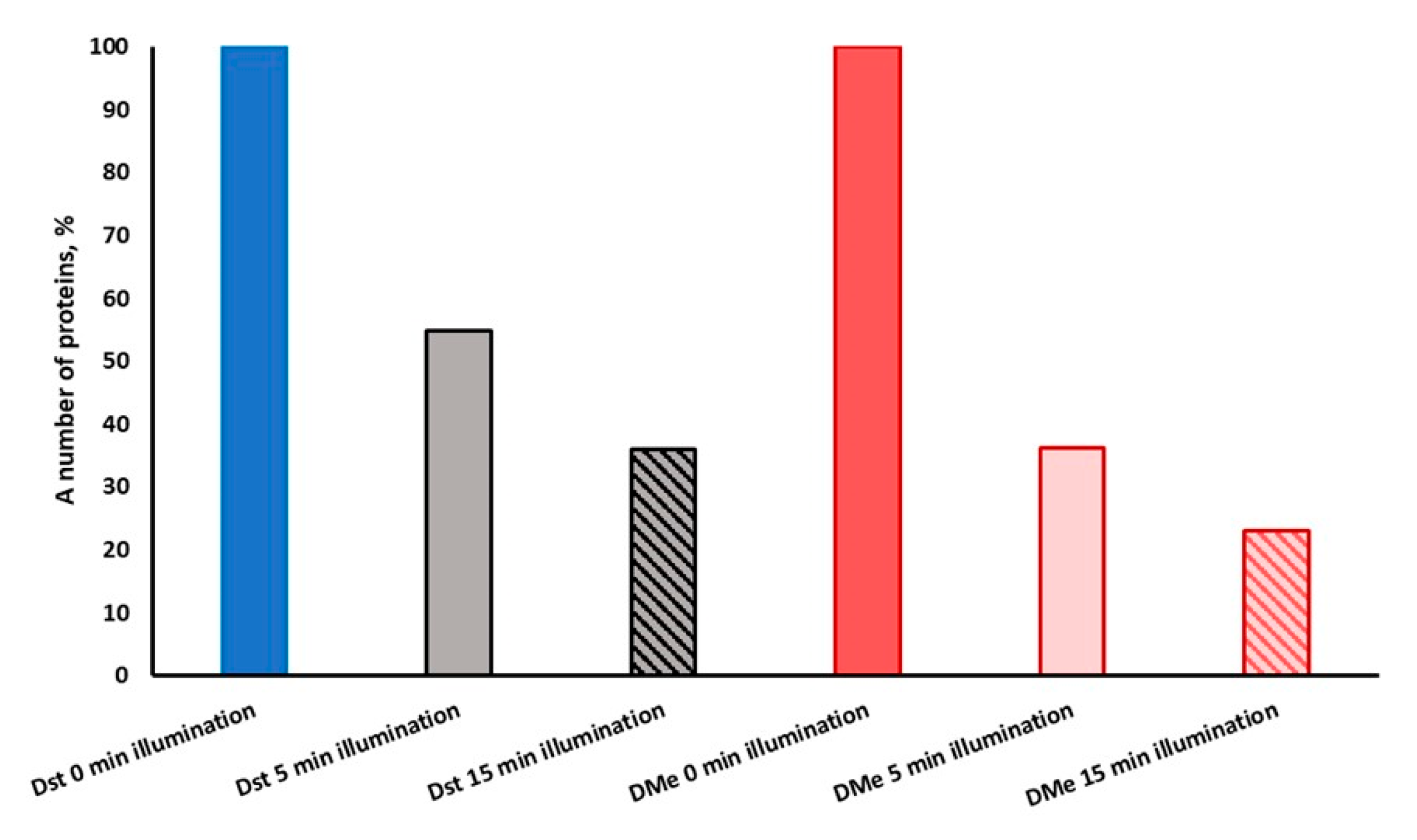
2.3. Primary Protein Targets for Photodynamic Inactivation of M. smegmatis
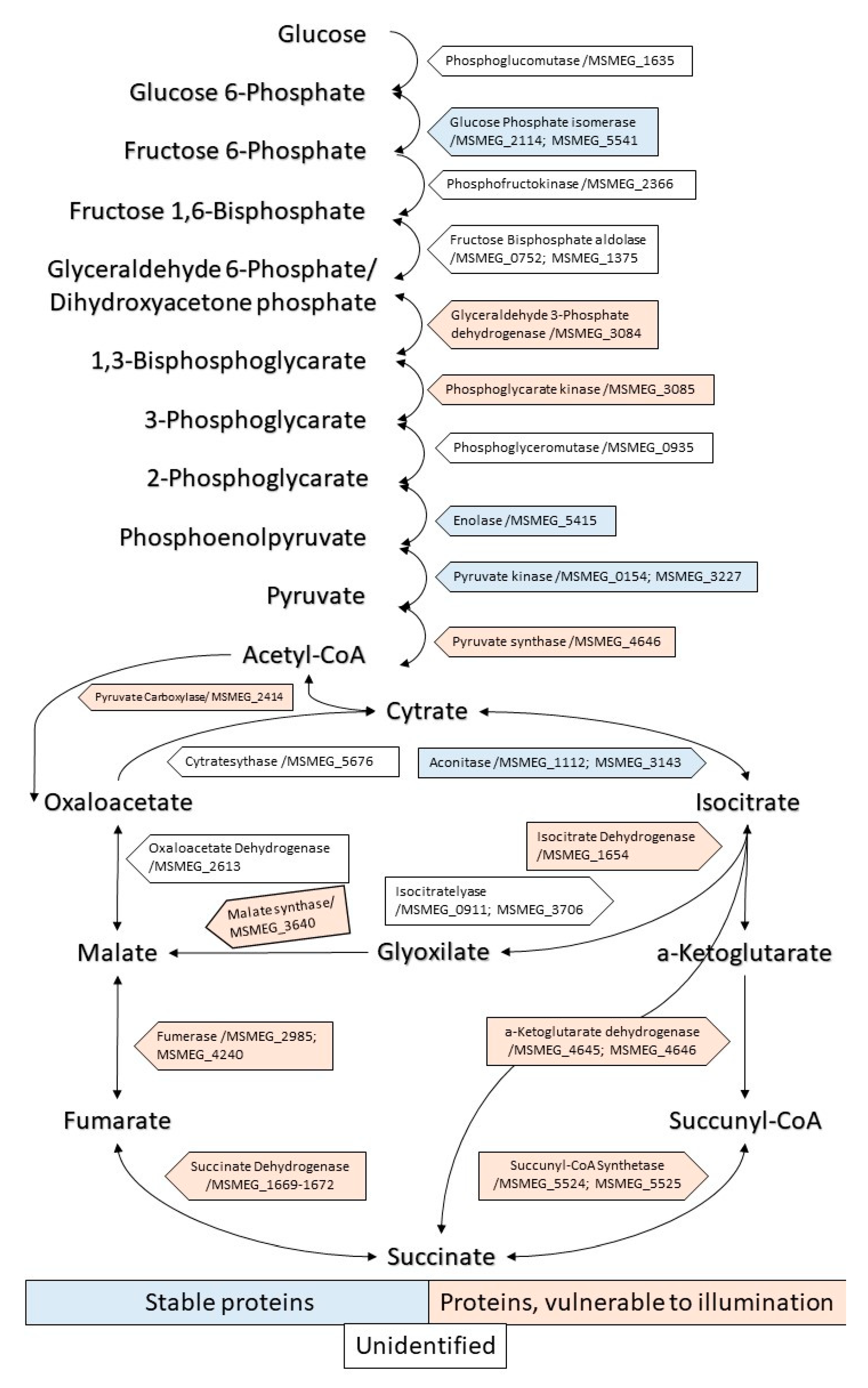
2.3.1. Enzymes of the Central Metabolism
2.3.2. Oxidoreductases
2.3.3. Various Metabolic Enzymes (Table 2)
2.3.4. Biopolymer Synthesis Enzymes
2.3.5. Regulatory Proteins (Table 3)
2.3.6. Enzymes of Defense Systems
- -
- Trigger factor/MSMEG_4674 (tig), which is involved in protein export. It acts as a chaperone by maintaining the newly synthesized protein in an open conformation.
- -
- MSMEG_3932/alpha-crystallin stress protein induced by anoxia. It has a proposed role in the maintenance of long-term viability during dormancy and latent infections.
- -
- Thioredoxin/MSMEG_4917 participates in various redox reactions through the reversible oxidation of its active center dithiol to a disulfide, and dithiol–disulfide exchange reactions. Copper/zinc superoxide dismutase/MSMEG_0835 (sodC), which destroys radicals which are normally produced within the cells and are toxic to biological systems. AhpC/TSA family protein/MSMEG_4753, which is a putative antioxidant protein.
- -
- Glyoxalase family protein/MSMEG_5680 which is important in the detoxification of methylglyoxal.
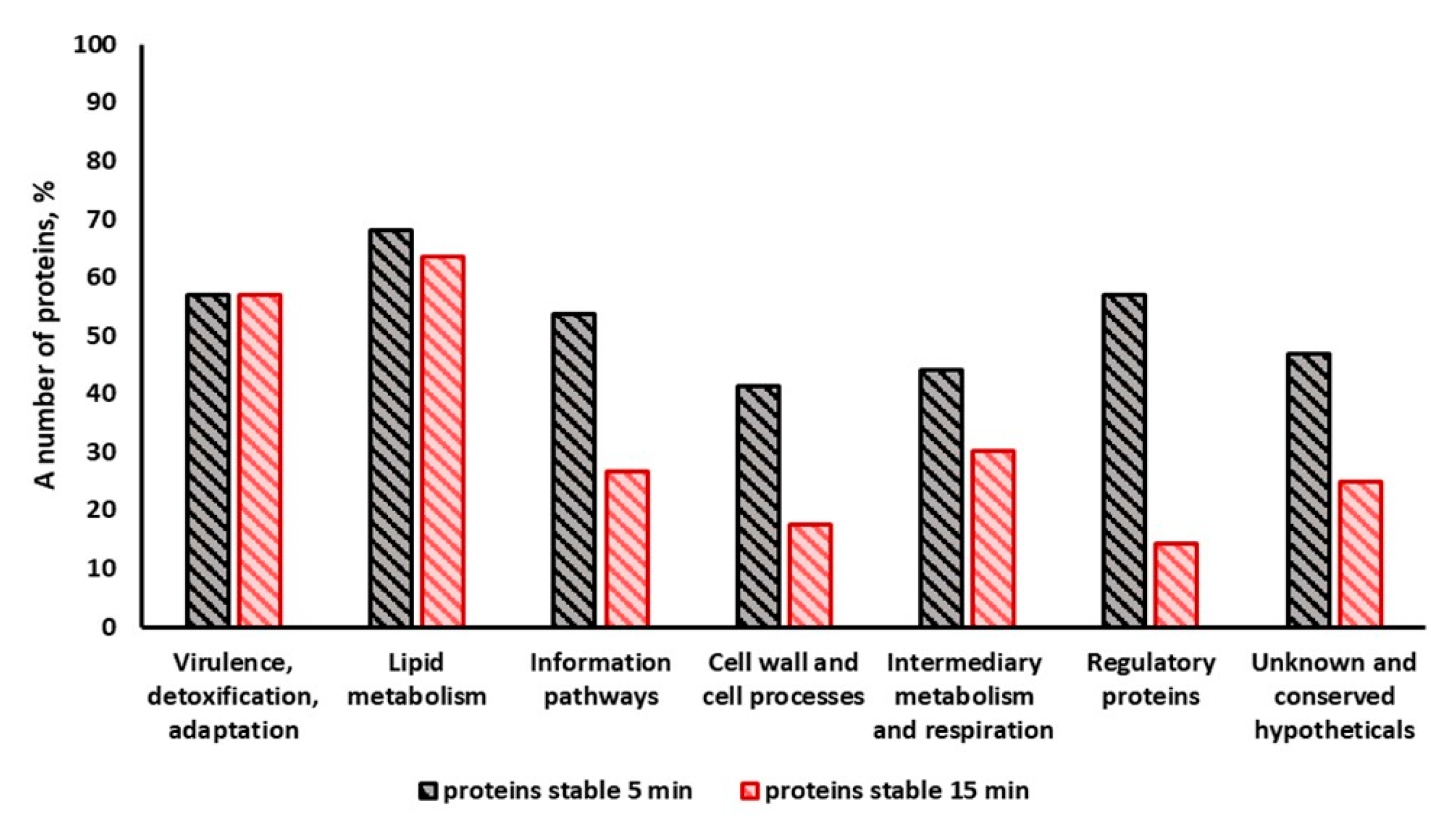
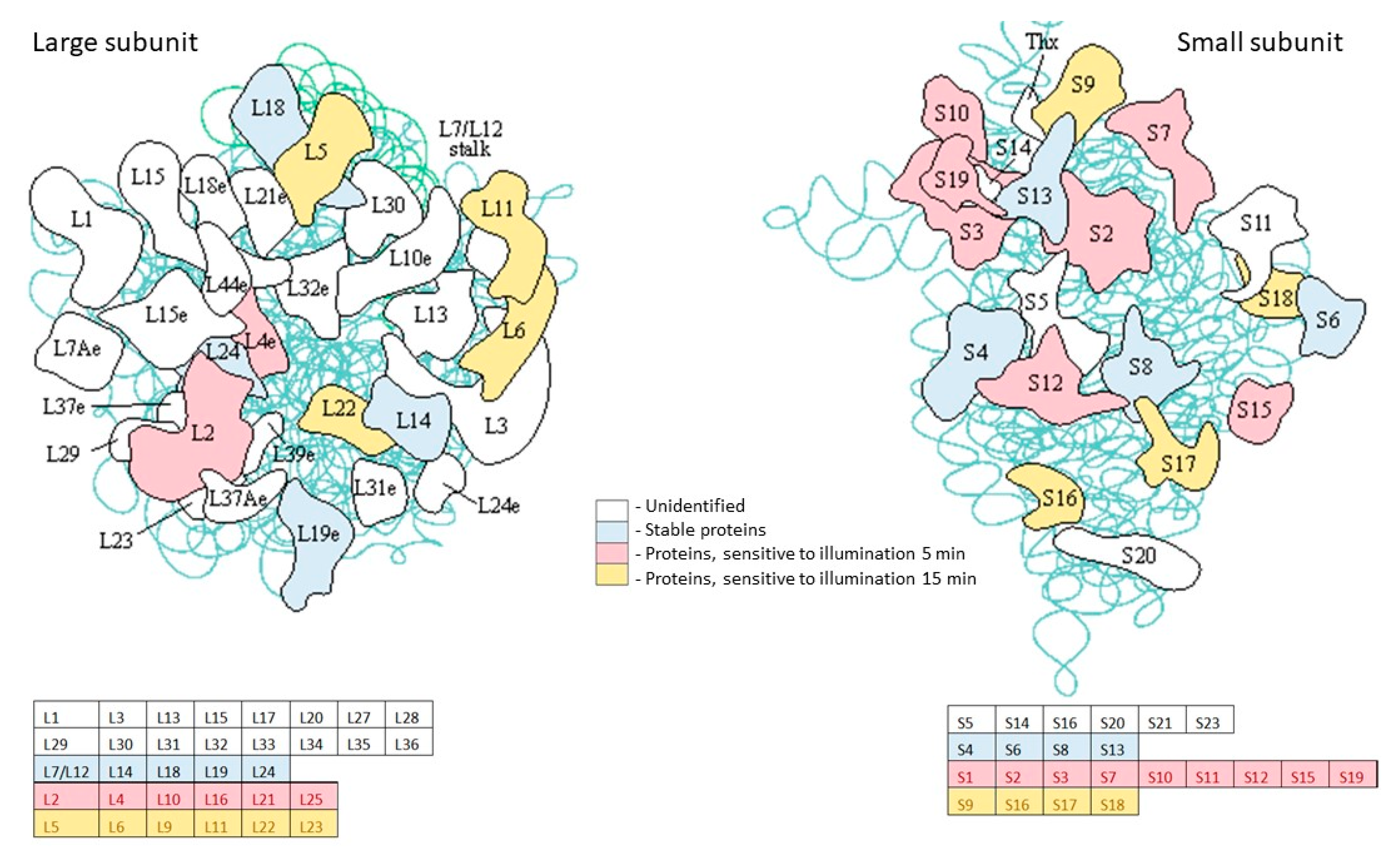
2.4. Enzymes Stable under Light Exposure
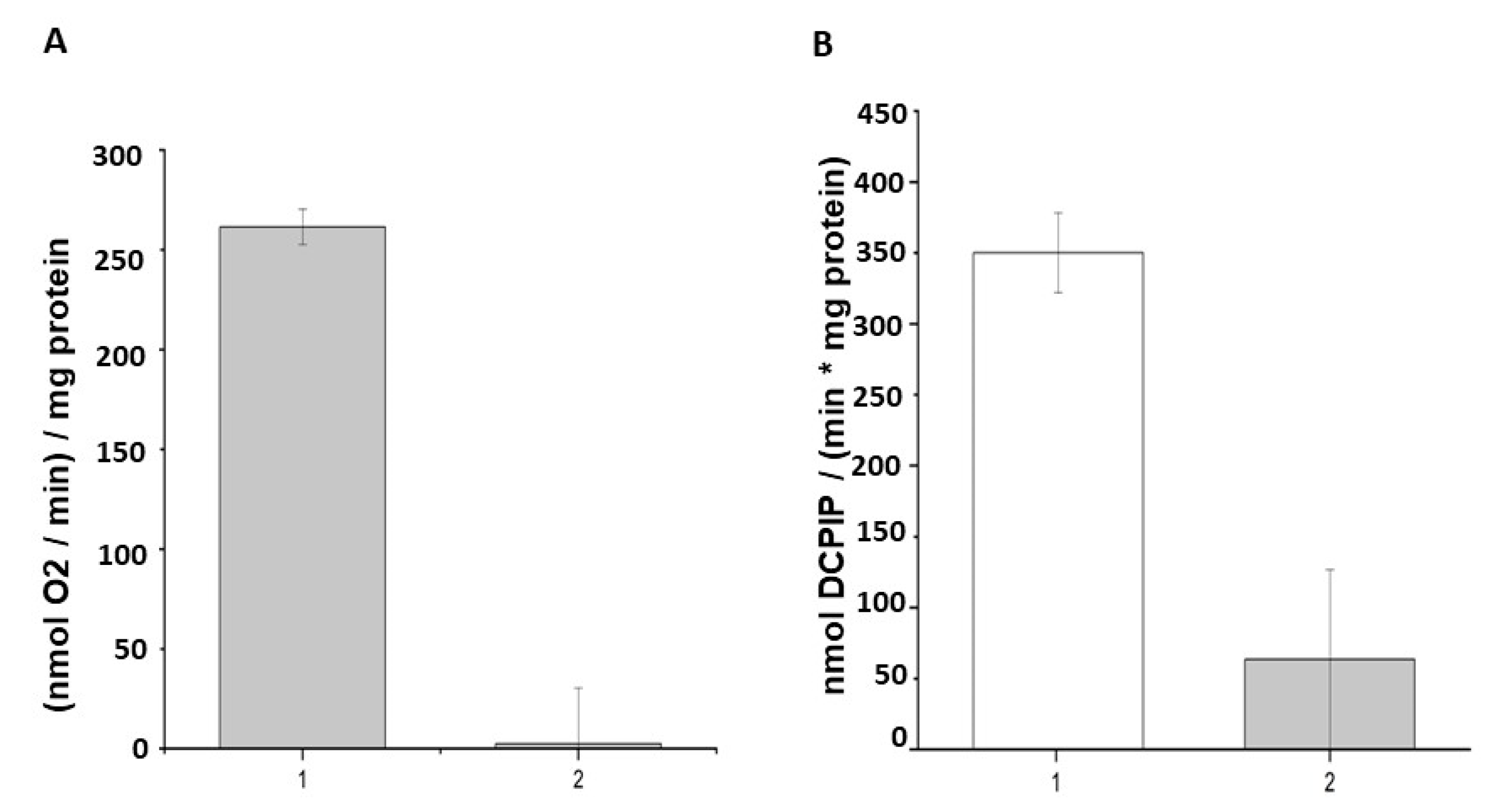
2.5. Influence of Photodynamic Inactivation on DCPIP Reduction by M. smegmatis Cells
3. Discussion
4. Materials and Methods
4.1. Formation of the Dormant Forms of M. smegmatis upon the Medium Self-Acidification
4.2. Viability Estimation by MPN
4.3. Infection of Macrophages with Dormant M. smegmatis Forms
4.4. Pigment Extraction from the Cells
4.5. Spectrophotometric and Fluorescence Analysis of Porphyrin in Extracts
4.6. Porphyrin Estimation by HPLC
4.7. Photodynamic Inactivation
4.8. Sample Preparation for SDS Electrophoresis
4.9. Protein Amount Determination
4.10. SDS Electrophoresis
4.11. Protein Identification by MALDI-TOF
4.12. Respiration, DCPIP Reduction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shleeva, M.O.; Kaprelyants, A.S. Hypobiosis of Mycobacteria: Biochemical Aspects. Biochemistry 2023, 88, S52–S74. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Kaur, M.; Singh, L.V.; Aggarwal, D.; Verma, I.; Radotra, B.D.; Sharma, S. Reactivation of Latent Tuberculosis through Modulation of Resuscitation Promoting Factors by Diabetes. Sci. Rep. 2021, 11, 19700. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.W.; Ruan, Q.L.; Liu, Q.H.; Zhang, W.H. Updates on the Risk Factors for Latent Tuberculosis Reactivation and Their Managements. Emerg. Microbes Infect. 2016, 5, e10. [Google Scholar] [CrossRef] [PubMed]
- Patra, K.; Batabyal, S.; Mandal, K.; Ghose, D.; Sarkar, J. Tuberculosis and COVID-19: A Combined Global Threat to Human Civilization. Clin. Epidemiol. Glob. Health 2022, 15, 101031. [Google Scholar] [CrossRef]
- Nikitushkin, V.D.; Shleeva, M.O.; Zinin, A.I.; Trutneva, K.A.; Ostrovsky, D.N.; Kaprelyants, A.S. The Main Pigment of the Dormant Mycobacterium Smegmatis Is Porphyrin. FEMS Microbiol. Lett. 2016, 363, fnw206. [Google Scholar] [CrossRef]
- Shleeva, M.O.; Savitsky, A.P.; Nikitushkin, V.D.; Soloviev, I.D.; Trutneva, K.A. Effect of Photodynamic Inactivation against Dormant Forms and Active Growing Cells of Mycobacterium Smegmatis. Appl. Biochem. Microbiol. 2020, 56, 242–249. [Google Scholar] [CrossRef]
- Meyer, M.; Bown, S.G.; Speight, P. A Study of the Effects of Photodynamic Therapy on the Normal Tissues of the Rabbit Jaw. Br. J. Cancer 1991, 64, 1093–1097. [Google Scholar] [CrossRef][Green Version]
- Jao, Y.; Ding, S.; Chen, C. Antimicrobial Photodynamic Therapy for the Treatment of Oral Infections: A Systematic Review. J. Dent. Sci. 2023; in press. [Google Scholar] [CrossRef]
- Mitchell, L.W.; Jaffe, E.K. Porphobilinogen Synthase from Escherichia Coli Is a Zn(II) Metalloenzyme Stimulated by Mg(II). Arch. Biochem. Biophys. 1993, 300, 169–177. [Google Scholar] [CrossRef]
- Petrovieh, R.M.; Litwin, S.; Jaffe, E.K. Bradyrhizobium Japonicum Porphobilinogen Synthase Uses Two Mg(II) and Monovalent Cations. J. Biol. Chem. 1996, 271, 8692–8699. [Google Scholar] [CrossRef]
- Connell, N.D. Chapter 6: Mycobacterium: Isolation, Maintenance, Transformation, and Mutant Selection. Methods Cell Biol. 1994, 45, 107–125. [Google Scholar] [CrossRef] [PubMed]
- Falk, J.E. Porphyrins and Metalloporphyrins; Elsevier: Amsterdam, The Netherlands, 1964. [Google Scholar]
- Dailey, H.A.; Dailey, T.A.; Gerdes, S.; Jahn, D.; Jahn, M.; O’Brian, M.R.; Warren, M.J. Prokaryotic Heme Biosynthesis: Multiple Pathways to a Common Essential Product Harry. Microbiol. Mol. Biol. Rev. 2017, 81, e00048-16. [Google Scholar] [CrossRef] [PubMed]
- Vatansever, F.; de Melo, W.C.M.A.; Avci, P.; Vecchio, D.; Sadasivam, M.; Gupta, A.; Chandran, R.; Karimi, M.; Parizotto, N.A.; Yin, R.; et al. Antimicrobial Strategies Centered around Reactive Oxygen Species—Bactericidal Antibiotics, Photodynamic Therapy, and Beyond. FEMS Microbiol. Rev. 2013, 37, 955–989. [Google Scholar] [CrossRef] [PubMed]
- Portevin, D.; De Sousa-D’Auria, C.; Houssin, C.; Grimaldi, C.; Chami, M.; Daffé, M.; Guilhot, C. A Polyketide Synthase Catalyzes the Last Condensation Step of Mycolic Acid Biosynthesis in Mycobacteria and Related Organisms. Proc. Natl. Acad. Sci. USA 2004, 101, 314–319. [Google Scholar] [CrossRef]
- Seshadri, A.; Singh, N.S.; Varshney, U. Recycling of the Posttermination Complexes of Mycobacterium Smegmatis and Escherichia Coli Ribosomes Using Heterologous Factors. J. Mol. Biol. 2010, 401, 854–865. [Google Scholar] [CrossRef]
- Trutneva, K.; Shleeva, M.; Nikitushkin, V.; Demina, G.; Kaprelyants, A. Protein Composition of Mycobacterium Smegmatis Differs Significantly between Active Cells and Dormant Cells with Ovoid Morphology. Front. Microbiol. 2018, 9, 2083. [Google Scholar] [CrossRef]
- Shleeva, M.; Savitsky, A.; Kaprelyants, A. Photoinactivation of Mycobacteria to Combat Infection Diseases: Current State and Perspectives. Appl. Microbiol. Biotechnol. 2021, 105, 4099–4109. [Google Scholar] [CrossRef]
- Dailey, H.A.; Gerdes, S.; Dailey, T.A.; Burch, J.S.; Phillips, J.D. Noncanonical Coproporphyrin-Dependent Bacterial Heme Biosynthesis Pathway That Does Not Use Protoporphyrin. Proc. Natl. Acad. Sci. USA 2015, 112, 2210–2215. [Google Scholar] [CrossRef]
- Alves, E.; Faustino, M.A.F.; Neves, M.G.P.M.S.; Cunha, A.; Tome, J.; Almeida, A. An Insight on Bacterial Cellular Targets of Photodynamic Inactivation. Future Med. Chem. 2014, 6, 141–164. [Google Scholar] [CrossRef]
- Sabino, C.P.; Ribeiro, M.S.; Wainwright, M.; dos Anjos, C.; Sellera, F.P.; Dropa, M.; Nunes, N.B.; Brancini, G.T.P.; Braga, G.U.L.; Arana-Chavez, V.E.; et al. The Biochemical Mechanisms of Antimicrobial Photodynamic Therapy. Photochem. Photobiol. 2023, 99, 742–750. [Google Scholar] [CrossRef]
- Almeida, A.; Faustino, M.A.; Tome, J.P. Photodynamic Inactivation of Bacteria: Finding the Effective Targets. Future Med. Chem. 2015, 7, 1221–1224. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wan, M.; Zhang, L.; Dai, Y.; Hai, Y.; Yue, C.; Xu, J.; Ding, Y.; Wang, M.; Xie, J.; et al. ALA_PDT Promotes Ferroptosis-Like Death of Mycobacterium abscessus and Antibiotic Sterilization via Oxidative Stress. Antioxidants 2022, 11, 546. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chu, Z.; Ruan, Z.; Wang, X.; Dai, T.; Hu, X. Changes of Intracellular Porphyrin, Reactive Oxygen Species, and Fatty Acids Profiles During Inactivation of Methicillin-Resistant Staphylococcus aureus by Antimicrobial Blue Light. Front. Physiol. 2018, 9, 1658. [Google Scholar] [CrossRef] [PubMed]
- Awad, M.M.; Tovmasyan, A.; Craik, J.D.; Batinic-Haberle, I.; Benov, L.T. Important Cellular Targets for Antimicrobial Photodynamic Therapy. Appl. Microbiol. Biotechnol. 2016, 100, 7679–7688. [Google Scholar] [CrossRef]
- dos Anjos, C.; Leanse, L.G.; Ribeiro, M.S.; Sellera, F.P.; Dropa, M.; Arana-Chavez, V.E.; Lincopan, N.; Baptista, M.S.; Pogliani, F.C.; Dai, T.; et al. New Insights into the Bacterial Targets of Antimicrobial Blue Light. Microbiol. Spectr. 2023, 11, e02833-22. [Google Scholar] [CrossRef] [PubMed]
- Muehler, D.; Brandl, E.; Hiller, K.A.; Cieplik, F.; Maisch, T. Membrane Damage as Mechanism of Photodynamic Inactivation Using Methylene Blue and TMPyP in Escherichia coli and Staphylococcus aureus. Photochem. Photobiol. Sci. 2022, 21, 209–220. [Google Scholar] [CrossRef]
- Dosselli, R.; Millioni, R.; Puricelli, L.; Tessari, P.; Arrigoni, G.; Franchin, C.; Segalla, A.; Teardo, E.; Reddi, E. Molecular Targets of Antimicrobial Photodynamic Therapy Identified by a Proteomic Approach. J. Proteom. 2012, 77, 329–343. [Google Scholar] [CrossRef]
- Valduga, G.; Breda, B.; Giacometti, G.M.; Jori, G.; Reddi, E. Photosensitization of Wild and Mutant Strains of Escherichia coli by Meso-Tetra (N-Methyl-4-Pyridyl)Porphine. Biochem. Biophys. Res. Commun. 1999, 256, 84–88. [Google Scholar] [CrossRef]
- Kudykina, Y.K.; Shleeva, M.O.; Artsabanov, V.Y.; Suzina, N.E.; Kaprelyants, A.S. Generation of Dormant Forms by Mycobacterium Smegmatis in the Poststationary Phase during Gradual Acidification of the Medium. Microbiology 2011, 80, 638–649. [Google Scholar] [CrossRef]
- de Man, J.C. The Probability of Most Probable Numbers. Eur. J. Appl. Microbiol. 1974, 1, 67–78. [Google Scholar] [CrossRef]
- Majorov, K.B.; Lyadova, I.V.; Kondratieva, T.K.; Eruslanov, E.B.; Rubakova, E.I.; Orlova, M.O.; Mischenko, V.V.; Apt, A.S. Different Innate Ability of I/St and A/Sn Mice to Combat Virulent Mycobacterium Tuberculosis: Phenotypes Expressed in Lung and Extrapulmonary Macrophages. Infect. Immun. 2003, 71, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Rimington, C. Spectral-Absorption Coefficients of Some Porphyrins in the Soret-Band Region. Biochem. J. 1960, 75, 620–623. [Google Scholar] [CrossRef]
- Diz, A.P.; Truebano, M.; Skibinski, D.O.F. The Consequences of Sample Pooling in Proteomics: An Empirical Study. Electrophoresis 2009, 30, 2967–2975. [Google Scholar] [CrossRef] [PubMed]
- Betts, J.C.; Lukey, P.T.; Robb, L.C.; Mcadam, R.A.; Duncan, K. Evaluation of a Nutrient Starvation Model of Mycobacterium Tuberculosis Persistence by Gene and Protein Expression Profiling. Mol. Microbiol. 2002, 43, 717–731. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Product | Time of Illuminations | ||
|---|---|---|---|---|
| 0 min | 5 min | 15 min | ||
| MSMEG_0777 | F420-dependent glucose-6-phosphate dehydrogenase | + | − | − |
| MSMEG_1030 | (NAD(P)/FAD-dependent oxidoreductase | + | − | − |
| MSMEG_3106 | quinone oxidoreductase | + | + | − |
| MSMEG_3232 | cytochrome D ubiquinol oxidase subunit II | + | + | + |
| MSMEG_3233 | cytochrome D ubiquinol oxidase subunit I | + | − | − |
| MSMEG_4645 | alpha oxoglutarate ferredoxin oxidoreductase, beta subunit | + | − | − |
| MSMEG_4935 | epsilon subunit ATP synthase F1-F0 | + | + | + |
| MSMEG_4936 | beta subunit ATP synthase F1-F0 | + | − | − |
| MSMEG_4937 | gamma subunit ATP synthase F1-F0 | + | + | + |
 Proteins disappeared after 5 min of illumination.
Proteins disappeared after 5 min of illumination.  Proteins stable after 15 min of illumination.
Proteins stable after 15 min of illumination.  Proteins disappeared after 15 min of illumination.
Proteins disappeared after 15 min of illumination.| Gene Name | Product | Time of Illuminations | ||
|---|---|---|---|---|
| 0 min | 5 min | 15 min | ||
| MSMEG_0029 | anthranilate synthase component 2 | + | − | − |
| MSMEG_1342 | (3R)-hydroxyacyl-ACP dehydratase subunit HadC | + | − | − |
| MSMEG_3564 | bacterioferritin | + | + | − |
| MSMEG_3767 | acyl-CoA synthetase | + | − | − |
| MSMEG_4340 | NAD/mycothiol-dependent formaldehyde dehydrogenase | + | − | − |
| MSMEG_5136 | pyridoxine 5’-phosphate (PNP) oxidase | + | − | − |
| MSMEG_5191 | siderophore binding protein | + | + | − |
| MSMEG_5243 | pyridoxine 5’-phosphate (PNP) oxidase | + | − | − |
| MSMEG_5471 | UTP--glucose-1-phosphate uridylyltransferase | + | − | − |
| MSMEG_5485 | molybdopterin biosynthesis protein | + | + | − |
| MSMEG_5702 | molybdenum cofactor synthesis domain protein | + | − | − |
| MSMEG_5824 | phosphoribosylformylglycinamidine synthase II | + | − | − |
| MSMEG_5892 | trehalose-6-phosphate synthase | + | − | − |
| MSMEG_6392 | polyketide synthase | + | − | − |
| MSMEG_6520 | orotate phosphoribosyltransferase | + | − | − |
 Proteins disappeared after 5 min of illumination.
Proteins disappeared after 5 min of illumination.  Proteins disappeared after 15 min of illumination.
Proteins disappeared after 15 min of illumination.| Gene Name | Product | Time of Illuminations | ||
|---|---|---|---|---|
| 0 min | 5 min | 15 min | ||
| MSMEG_0937 | DNA-binding response regulator RegX3 | + | − | − |
| MSMEG_0994 | regulators DNA-binding response regulator ResD | + | − | − |
| MSMEG_3246 | sensor part of a two-component regulatory system | + | − | − |
| MSMEG_3647 | glycogen accumulation regulator GarA | + | − | − |
| MSMEG_5488 | the DNA-binding response regulator | + | + | − |
| MSMEG_6091 | negative regulator of genetic competence ClpC/mecB | + | − | − |
| MSMEG_6896 | the single-stranded DNA-binding protein | + | + | − |
 Proteins disappeared after 5 min of illumination.
Proteins disappeared after 5 min of illumination.  Proteins disappeared after 15 min of illumination.
Proteins disappeared after 15 min of illumination.| Gene Name | Product | Time of Illuminations | ||
|---|---|---|---|---|
| 0 min | 5 min | 15 min | ||
| MSMEG_0835 | copper/zinc superoxide dismutase | + | − | − |
| MSMEG_0880 | chaperonin GroEL | + | − | − |
| MSMEG_3932 | alpha-crystallin stress protein | + | − | − |
| MSMEG_4674 | trigger factor | + | − | − |
| MSMEG_4753 | antioxidant protein | + | − | − |
| MSMEG_4917 | thioredoxin | + | − | − |
| MSMEG_5680 | glyoxalase family protein | + | − | − |
 Proteins disappeared after 5 min of illumination.
Proteins disappeared after 5 min of illumination.| LED | Power Density (mW/cm2) | Time of Illuminations (min) and Light Doses (J/cm2) | |||
|---|---|---|---|---|---|
| 565 nm | 180 | 5 min | 15 min | 30 min | 60 min |
| 54 J/cm2 | 162 J/cm2 | 324 J/cm2 | 648 J/cm2 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shashin, D.M.; Demina, G.R.; Linge, I.A.; Vostroknutova, G.N.; Kaprelyants, A.S.; Savitsky, A.P.; Shleeva, M.O. The Effect of Antimicrobial Photodynamic Inactivation on the Protein Profile of Dormant Mycolicibacterium smegmatis Containing Endogenous Porphyrins. Int. J. Mol. Sci. 2023, 24, 13968. https://doi.org/10.3390/ijms241813968
Shashin DM, Demina GR, Linge IA, Vostroknutova GN, Kaprelyants AS, Savitsky AP, Shleeva MO. The Effect of Antimicrobial Photodynamic Inactivation on the Protein Profile of Dormant Mycolicibacterium smegmatis Containing Endogenous Porphyrins. International Journal of Molecular Sciences. 2023; 24(18):13968. https://doi.org/10.3390/ijms241813968
Chicago/Turabian StyleShashin, Denis M., Galina R. Demina, Irina A. Linge, Galina N. Vostroknutova, Arseny S. Kaprelyants, Alexander P. Savitsky, and Margarita O. Shleeva. 2023. "The Effect of Antimicrobial Photodynamic Inactivation on the Protein Profile of Dormant Mycolicibacterium smegmatis Containing Endogenous Porphyrins" International Journal of Molecular Sciences 24, no. 18: 13968. https://doi.org/10.3390/ijms241813968
APA StyleShashin, D. M., Demina, G. R., Linge, I. A., Vostroknutova, G. N., Kaprelyants, A. S., Savitsky, A. P., & Shleeva, M. O. (2023). The Effect of Antimicrobial Photodynamic Inactivation on the Protein Profile of Dormant Mycolicibacterium smegmatis Containing Endogenous Porphyrins. International Journal of Molecular Sciences, 24(18), 13968. https://doi.org/10.3390/ijms241813968






