Association between the Cytosine Hydroxymethylation and the Expression of microRNA in Multiple Sclerosis in Polish Population
Abstract
:1. Introduction
2. Results
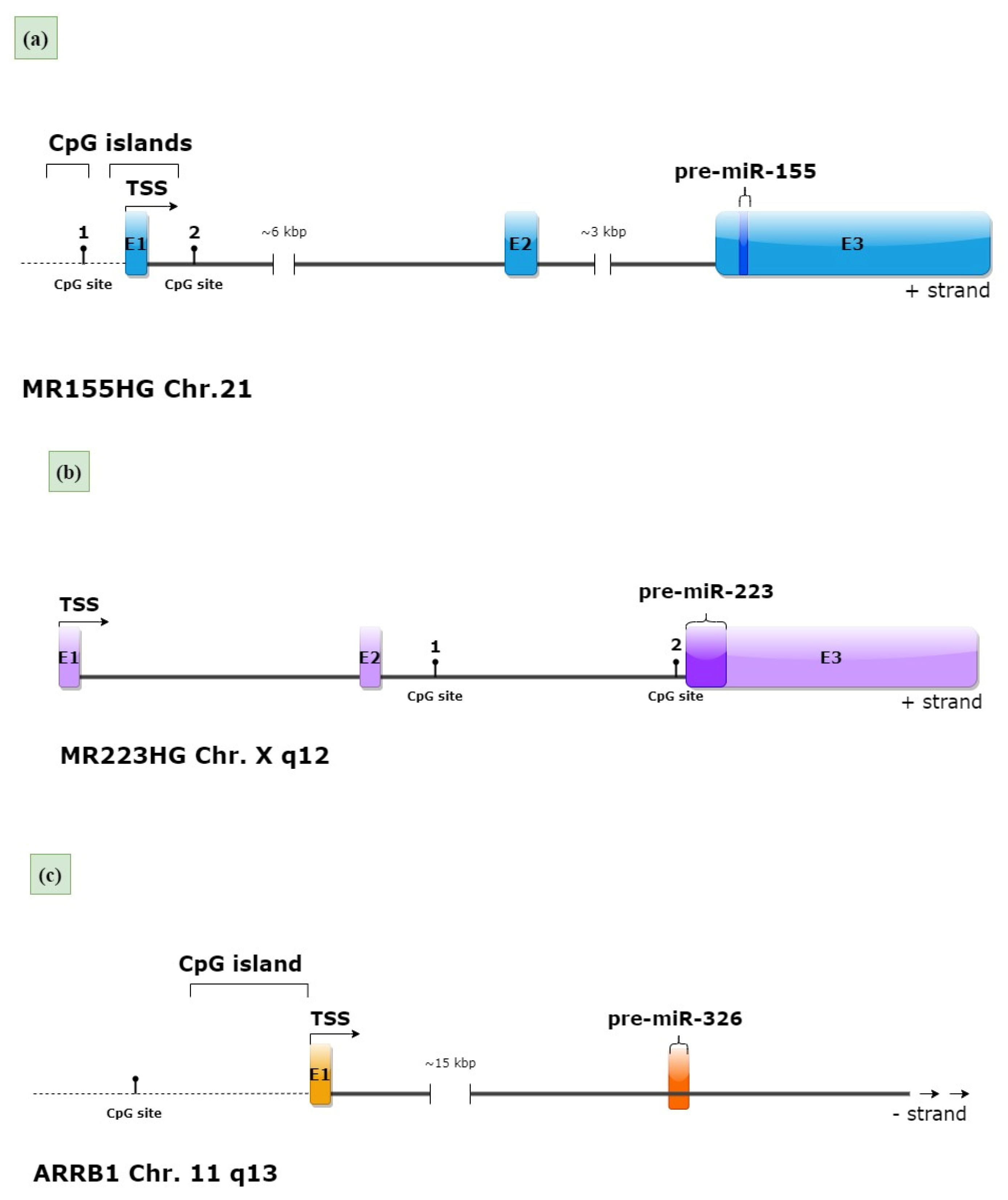
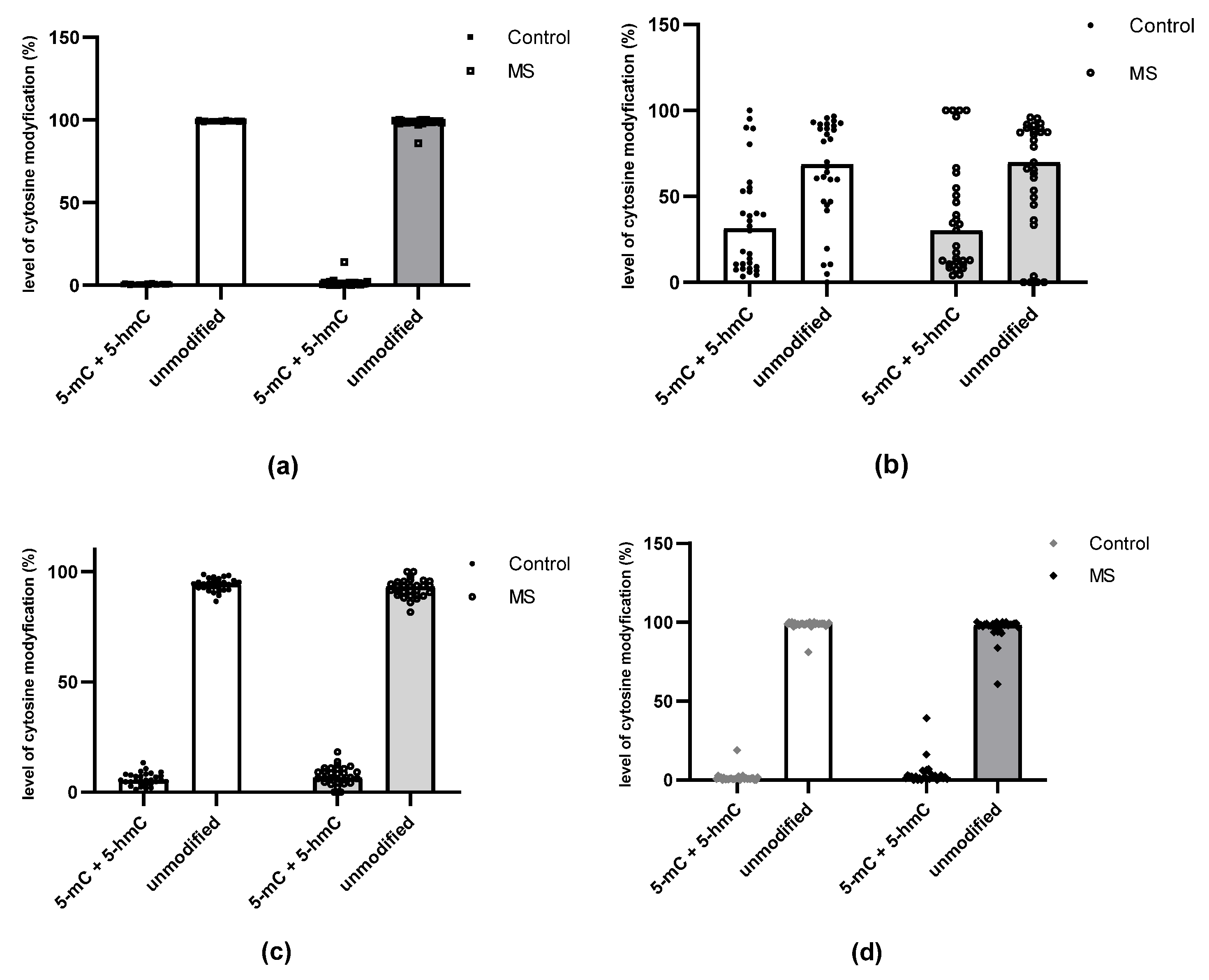
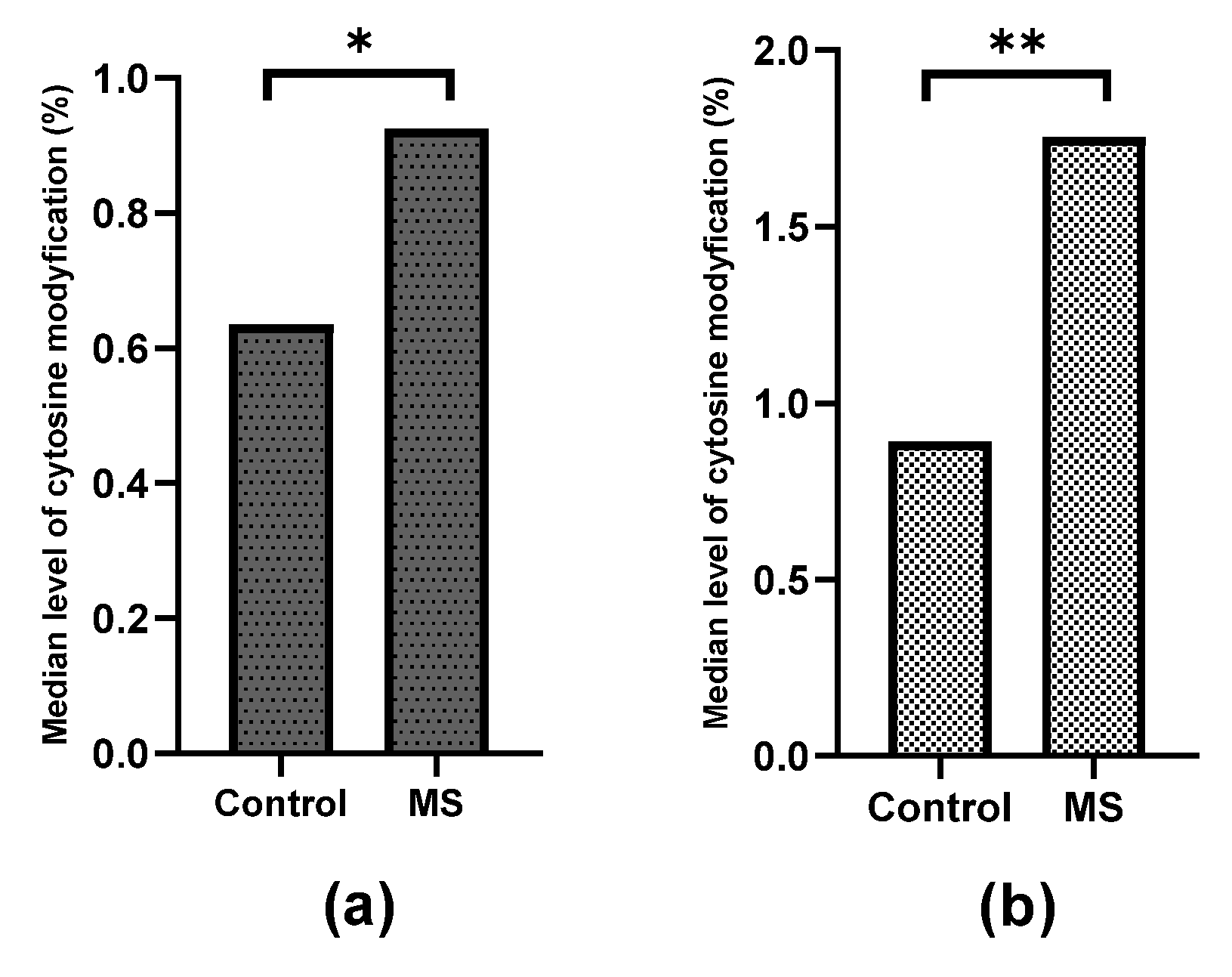
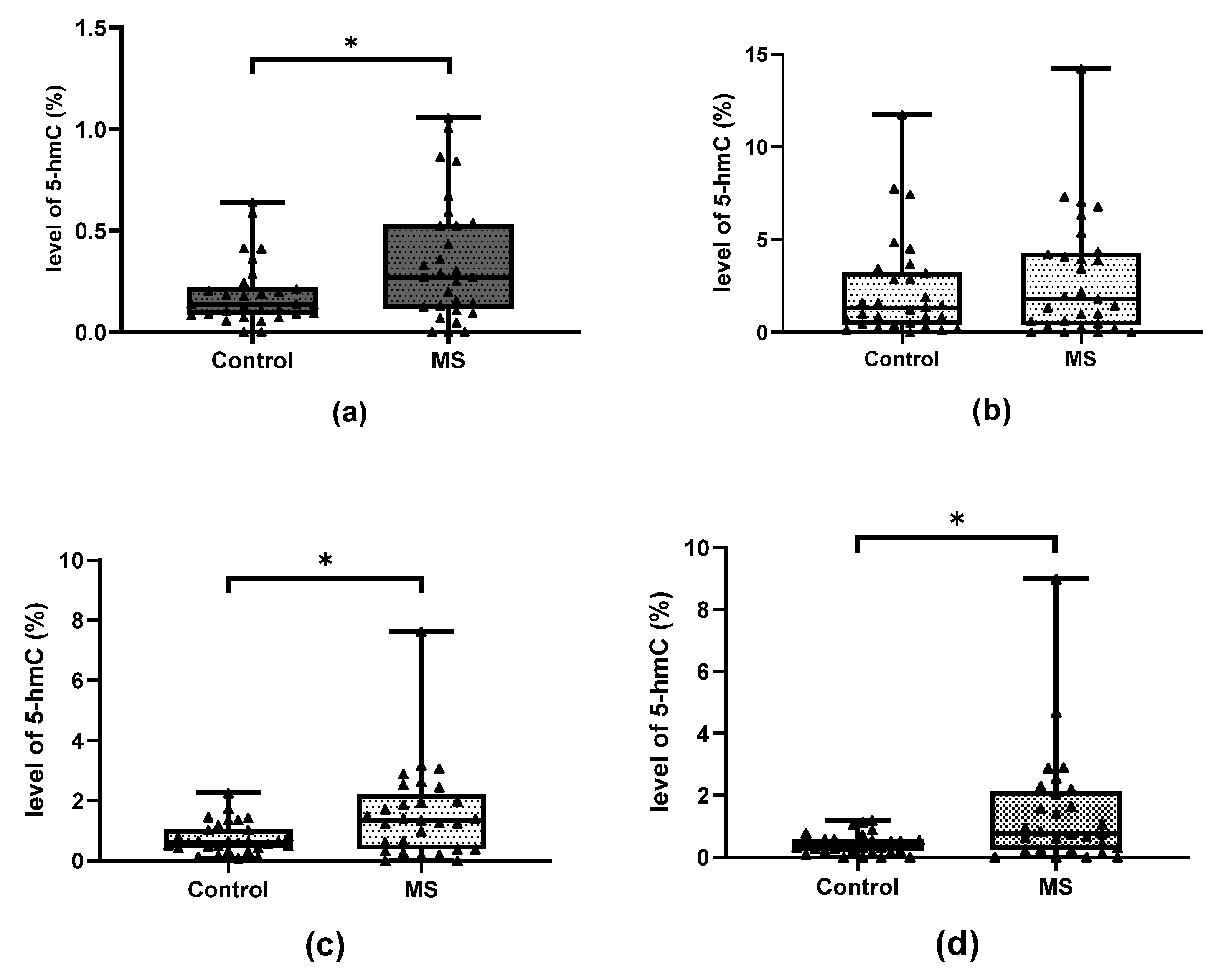
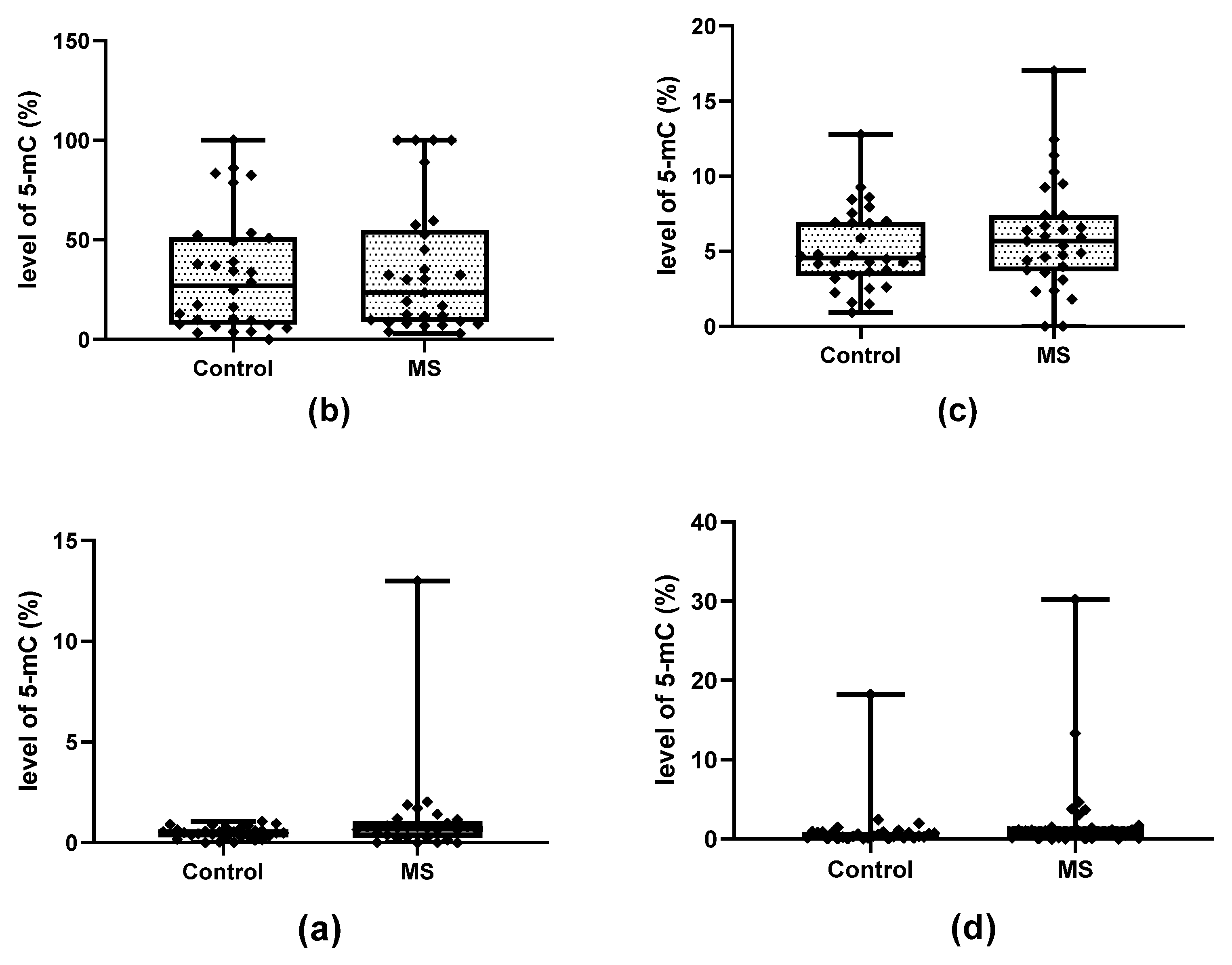
2.1. Methylation and Hydroxymethylation Levels of miR-155, miR-223 and miR-326 CpG Sites
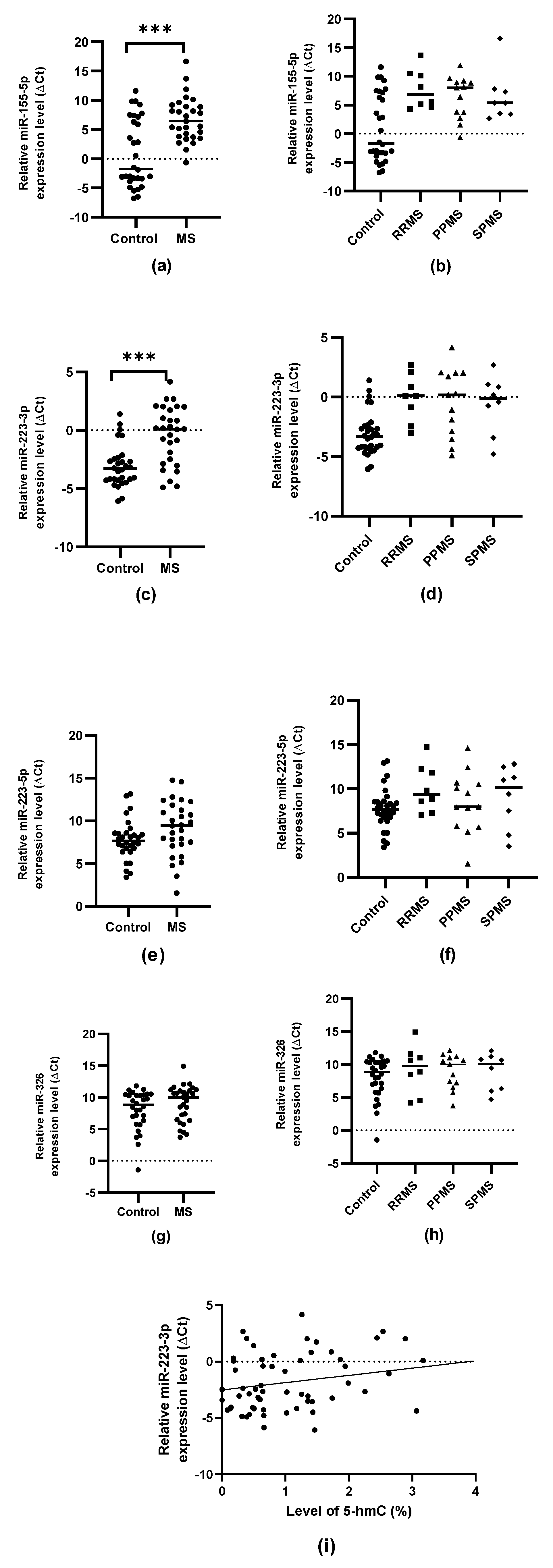
2.2. Expression Levels of miR-155-5p, miR-223-3p, miR-223-5p and miR-326
3. Discussion
4. Materials and Methods
4.1. PBMCs DNA and RNA Purification
4.2. Analysis of the Level of Methylation and Hydroxymethylation
4.3. Analysis miRNA Expression
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ståhl, D.; Bjereld, Y.; Dunér, A. Disabled in Society—A Scoping Review on Persons Living with Multiple Sclerosis and Disability. J. Multidiscip. Healthc. 2022, 15, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Myhr, K.M. Diagnosis and treatment of multiple sclerosis. Acta Neurol. Scand. 2008, 188, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Høglund, R.A.; Maghazachi, A.A. Multiple sclerosis and the role of immune cells. World J. Exp. Med. 2014, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Field, J.; Browning, S.R.; Johnson, L.J.; Danoy, P.; Varney, M.D.; Tait, B.D.; Gandhi, K.S.; Charlesworth, J.C.; Heard, R.N.; The Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene); et al. A Polymorphism in the HLA-DPB1 Gene Is Associated with Susceptibility to Multiple Sclerosis. PLoS ONE 2010, 5, e13454. [Google Scholar] [CrossRef] [PubMed]
- Handel, A.E.; Williamson, A.J.; Disanto, G.; Handunnetthi, L.; Giovannoni, G.; Ramagopalan, S.V. An updated meta-analysis of risk of multiple sclerosis following infectious mononucleosis. PLoS ONE 2010, 5, e12496. [Google Scholar] [CrossRef] [PubMed]
- Gacias, M.; Casaccia, P. Epigenetic Mechanisms in Multiple Sclerosis. Rev. Esp. Escler. Mult. 2014, 6, 25. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6107087/ (accessed on 14 December 2022). [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Lee, E.; Li, T.; Rampersaud, M. Role of miRNAs in Neurodegeneration: From Disease Cause to Tools of Biomarker Discovery and Therapeutics. Genes 2022, 13, 425. [Google Scholar] [CrossRef]
- Gaudet, A.D.; Fonken, L.K.; Watkins, L.R.; Nelson, R.J.; Popovich, P.G. microRNAs: Roles in regulating neuroinflammation. Neuroscientist 2018, 24, 221. [Google Scholar] [CrossRef]
- Baghi, M.; Yadegari, E.; Delavar, M.R.; Peymani, M.; Ganjalikhani-Hakemi, M.; Salari, M.; Nasr-Esfahani, M.H.; Megraw, T.L.; Ghaedi, K. MiR-193b deregulation is associated with Parkinson’s disease. J. Cell. Mol. Med. 2021, 25, 6348. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, Y.; Wang, J.; Yan, Y.; Peng, L.; Qiu, W. Dysregulated microRNA involvement in multiple sclerosis by induction of T helper 17 cell differentiation. Front. Immunol. 2018, 9, 1256. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Chaudhuri, A.A.; Rao, D.S.; Baltimore, D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. USA 2009, 106, 7113–7118. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Huffaker, T.B.; Kagele, D.A.; Runtsch, M.C.; Bake, E.; Chaudhuri, A.A.; Round, J.L.; O’connell, R.M. MicroRNA-155 Confers Encephalogenic Potential to Th17 Cells by Promoting Effector Gene Expression. J. Immunol. 2013, 190, 5972–5980. [Google Scholar] [CrossRef] [PubMed]
- Mycko, M.P.; Cichalewska, M.; Cwiklinska, H.; Selmaj, K.W. miR-155-3p drives the development of autoimmune demyelination by regulation of heat shock protein 40. J. Neurosci. 2015, 35, 16504–16515. [Google Scholar] [CrossRef] [PubMed]
- Orefice, N.S.; Guillemot-Legris, O.; Capasso, R.; Bottemanne, P.; Hantraye, P.; Caraglia, M.; Orefice, G.; Alhouayek, M.; Muccioli, G.G. Mirna profile is altered in a modified eae mouse model of multiple sclerosis featuring cortical lesions. eLife 2020, 9, e56916. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Ramirez, M.A.; Wu, D.; Pryce, G.; Simpson, J.E.; Reijerkerk, A.; Kay, O.; Vries, H.E.; Hirst, M.C.; Sharrack, B.; Baker, D.; et al. MicroRNA-155 negatively affects blood-brain barrier function during neuroinflammation. FASEB J. 2014, 28, 2551–2565. [Google Scholar] [CrossRef] [PubMed]
- McCoy, C.E. miR-155 dysregulation and therapeutic intervention in multiple sclerosis. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2017; Volume 1024, pp. 111–131. [Google Scholar]
- Zhang, J.; Cheng, Y.; Cui, W.; Li, M.; Li, B.; Guo, L. MicroRNA-155 modulates Th1 and Th17 cell differentiation and is associated with multiple sclerosis and experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2014, 266, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Mazloumfard, F.; Mirian, M.; Eftekhari, S.M.; Aliomrani, M. Hydroxychloroquine effects on miR-155-3p and miR-219 expression changes in animal model of multiple sclerosis. Metab. Brain Dis. 2020, 35, 1299–1307. [Google Scholar] [CrossRef]
- Cantoni, C.; Cignarella, F.; Ghezzi, L.; Mikesell, B.; Bollman, B.; Berrien-Elliott, M.M.; Ireland, A.R.; Fehniger, T.A.; Wu, G.F.; Piccio, L. Mir-223 regulates the number and function of myeloid-derived suppressor cells in multiple sclerosis and experimental autoimmune encephalomyelitis. Acta Neuropathol. 2017, 133, 61–77. [Google Scholar] [CrossRef]
- Du, C.; Liu, C.; Kang, J.; Zhao, G.; Ye, Z.; Huang, S.; Li, Z.; Wu, Z.; Pei, G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 2009, 10, 1252–1259. [Google Scholar] [CrossRef]
- Keller, A.; Leidinger, P.; Lange, J.; Borries, A.; Schroers, H.; Scheffler, M.; Lenhof, H.-P.; Ruprecht, K.; Meese, E. Multiple Sclerosis: MicroRNA Expression Profiles Accurately Differentiate Patients with Relapsing-Remitting Disease from Healthy Controls. PLoS ONE 2009, 4, e7440. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, A.; Ghaedi, K.; Tanhaei, S.; Ganjalikhani-Hakemi, M.; Teimuri, S.; Etemadifar, M.; Esfahani, M.H.N. Upregulation of CD4+T-Cell Derived MiR-223 in The Relapsing Phase of Multiple Sclerosis Patients. Cell J. 2016, 18, 371–380. [Google Scholar] [CrossRef]
- De Santis, G.; Ferracin, M.; Biondani, A.; Caniatti, L.; Tola, M.R.; Castellazzi, M.; Zagatti, B.; Battistini, L.; Borsellino, G.; Fainardi, E.; et al. Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J. Neuroimmunol. 2010, 226, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Wu, Q.; Wang, Z.; Che, Y.; Zheng, S.; Chen, Y.; Zhong, X.; Shi, F. miR-223: An Immune Regulator in Infectious Disorders. Front. Immunol. 2021, 12, 5346. [Google Scholar] [CrossRef] [PubMed]
- Moles, R.; Bellon, M.; Nicot, C. STAT1: A Novel Target of miR-150 and miR-223 Is Involved in the Proliferation of HTLV-I–Transformed and ATL Cells. Neoplasia 2015, 17, 449. [Google Scholar] [CrossRef] [PubMed]
- Honardoost, M.A.; Kiani-Esfahani, A.; Ghaedi, K.; Etemadifar, M.; Salehi, M. MiR-326 and miR-26a, two potential markers for diagnosis of relapse and remission phases in patient with relapsing-remitting multiple sclerosis. Gene 2014, 544, 128–133. [Google Scholar] [CrossRef]
- Liu, Y.; Cui, X.; Wang, S.; Liu, J.; Zhao, N.; Huang, M.; Qin, J.; Li, Y.; Shan, Z.; Teng, W. Elevated MicroRNA-326 Levels Regulate the IL-23/IL-23R/Th17 Cell Axis in Hashimoto’s Thyroiditis by Targeting a Disintegrin and Metalloprotease 17. Thyroid 2020, 30, 1327–1337. [Google Scholar] [CrossRef]
- Arif, K.M.T.; Elliot, E.K.; Haupt, L.M.; Griffiths, L.R. Regulatory Mechanisms of Epigenetic miRNA Relationships in Human Cancer and Potential as Therapeutic Targets. Cancers 2020, 12, 2922. [Google Scholar] [CrossRef]
- Kriaucionis, S.; Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in purkinje neurons and the brain. Science 2009, 324, 929–930. [Google Scholar] [CrossRef]
- Hahn, M.A.; Qiu, R.; Wu, X.; Li, A.X.; Zhang, H.; Wang, J.; Jui, J.; Jin, S.-G.; Jiang, Y.; Pfeifer, G.P.; et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 2013, 3, 291. [Google Scholar] [CrossRef]
- Pan, Z.; Zhang, M.; Ma, T.; Xue, Z.-Y.; Li, G.-F.; Hao, L.-Y.; Zhu, L.-J.; Li, Y.-Q.; Ding, H.-L.; Cao, J.-L. Hydroxymethylation of microRNA-365-3p Regulates Nociceptive Behaviors via Kcnh2. J. Neurosci. 2016, 36, 2769–2781. [Google Scholar] [CrossRef] [PubMed]
- Kolarz, B.; Ciesla, M.; Dryglewska, M.; Rosenthal, A.K.; Majdan, M. Hypermethylation of the miR-155 gene in the whole blood and decreased plasma level of miR-155 in rheumatoid arthritis. PLoS ONE 2020, 15, e0233897. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, F.; Zhou, X.; Zeng, S.; Zhan, Q.; Yuan, M.; Yang, Q.; Liu, Y.; Xia, J. Promoter hypomethylation of microRNA223 gene is associated with atherosclerotic cerebral infarction. Atherosclerosis 2017, 263, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Po, A.; Begalli, F.; Abballe, L.; Alfano, V.; Besharat, Z.M.; Catanzaro, G.; Vacca, A.; Napolitano, M.; Tafani, M.; Giangaspero, F.; et al. β-Arrestin1/miR-326 Transcription Unit Is Epigenetically Regulated in Neural Stem Cells Where It Controls Stemness and Growth Arrest. Stem Cells Int. 2017, 2017, 5274171. [Google Scholar] [CrossRef] [PubMed]
- Richa, R.; Sinha, R.P. Hydroxymethylation of DNA: An epigenetic marker. Leibniz Research Centre for Working Environment and Human Factors. EXCLI J. 2014, 13, 592–610. [Google Scholar] [CrossRef]
- Calabrese, R.; Valentini, E.; Ciccarone, F.; Guastafierro, T.; Bacalini, M.; Ricigliano, V.; Zampieri, M.; Annibali, V.; Mechelli, R.; Franceschi, C.; et al. TET2 gene expression and 5-hydroxymethylcytosine level in multiple sclerosis peripheral blood cells. Biochim. Biophys. Acta 2014, 1842, 1130–1136. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Luo, M.; Pan, K.; Ahmad, T.; Zhou, T.; Miao, Z.; Zhou, H.; Sun, H.; Xu, X.; Namaka, M.; et al. DNA hydroxymethylation changes in response to spinal cord damage in a multiple sclerosis mouse model. Epigenomics 2019, 11, 323–335. [Google Scholar] [CrossRef]
- Keller, S.M.; Doherty, T.S.; Roth, T.L. Pharmacological manipulation of DNA methylation normalizes maternal behavior, DNA methylation, and gene expression in dams with a history of maltreatment. Sci. Rep. 2019, 91, 10253. [Google Scholar] [CrossRef]
- Ridolfi, E.; Fenoglio, C.; Cantoni, C.; Calvi, A.; De Riz, M.; Pietroboni, A.; Villa, C.; Serpente, M.; Bonsi, R.; Vercellino, M.; et al. Expression and Genetic Analysis of MicroRNAs Involved in Multiple Sclerosis. Int. J. Mol. Sci. 2013, 14, 4375–4384. [Google Scholar] [CrossRef]
- Waschbisch, A.; Atiya, M.; Linker, R.A.; Potapov, S.; Schwab, S.; Derfuss, T. Glatiramer acetate treatment normalizes deregulated microRNA expression in relapsing remitting multiple sclerosis. PLoS ONE 2011, 6, e24604. [Google Scholar] [CrossRef]
- Sharaf-Eldin, W.E.; Kishk, N.A.; Gad, Y.Z.; Hassan, H.; Ali, M.A.; Zaki, M.S.; Mohamed, M.R.; Essawi, M.L. Extracellular miR-145, miR-223 and miR-326 expression signature allow for differential diagnosis of immune-mediated neuroinflammatory diseases. J. Neurol. Sci. 2017, 383, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Yu, J.; Yu, H.; Zhao, Y.; Chen, S.; Xu, C.; Chen, F. Evolutionary and Expression Analysis of miR-#-5p and miR-#-3p at the miRNAs/isomiRs Levels. BioMed Res. Int. 2015, 2015, 168358. [Google Scholar] [CrossRef] [PubMed]
- Lin, I.H.; Chen, Y.F.; Hsu, M.T. Correlated 5-Hydroxymethylcytosine (5hmC) and Gene Expression Profiles Underpin Gene and Organ-Specific Epigenetic Regulation in Adult Mouse Brain and Liver. PLoS ONE 2017, 12, e0170779. [Google Scholar] [CrossRef] [PubMed]
- Stroud, H.; Feng, S.; Kinney, S.M.; Pradhan, S.; Jacobsen, S.E. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011, 12, R54. [Google Scholar] [CrossRef] [PubMed]
- Valinluck, V.; Tsai, H.H.; Rogstad, D.K.; Burdzy, A.; Bird, A.; Sowers, L.C. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res. 2004, 32, 4100. [Google Scholar] [CrossRef]
- Della Ragione, F.; Vacca, M.; Fioriniello, S.; Pepe, G.; D’Esposito, M. MECP2, a multi-talented modulator of chromatin architecture. Brief. Funct. Genom. 2016, 15, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Glaich, O.; Parikh, S.; Bell, R.E.; Mekahel, K.; Donyo, M.; Leader, Y.; Shayevitch, R.; Sheinboim, D.; Yannai, S.; Hollander, D.; et al. DNA methylation directs microRNA biogenesis in mammalian cells. Nat. Commun. 2019, 10, 5657. [Google Scholar] [CrossRef] [PubMed]
- Law, P.P.; Holland, M.L. DNA methylation at the crossroads of gene and environment interactions. Essays Biochem. 2019, 63, 717. [Google Scholar] [CrossRef]
- Salameh, Y.; Bejaoui, Y.; El Hajj, N. DNA Methylation Biomarkers in Aging and Age-Related Diseases. Front. Genet. 2020, 11, 480672. [Google Scholar] [CrossRef]
| Characteristic of Study Group | Total |
|---|---|
| Number of MS patients | 29 |
| RRMS | 8 |
| PPMS | 13 |
| SPMS | 8 |
| Females/males | 22/7 |
| Age (years, ± s) | 47.9 ± 14.1 |
| Disease duration (years, ± s) | 5.7 ± 2.6 |
| Expanded disability status scale (EDSS) (scale 1–10) 1 | 5.4 ± 0.8 |
| 0–5.5 | 16 (55.2%) |
| 6–6.5 | 13 (44.8%) |
| Characteristics of Control Group | Total |
|---|---|
| Number | 30 |
| Age (years, ± s) | 55.6 ± 17.7 |
| Females/males | 13/17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basak, J.; Piotrzkowska, D.; Kucharska-Lusina, A.; Majsterek, I. Association between the Cytosine Hydroxymethylation and the Expression of microRNA in Multiple Sclerosis in Polish Population. Int. J. Mol. Sci. 2023, 24, 13923. https://doi.org/10.3390/ijms241813923
Basak J, Piotrzkowska D, Kucharska-Lusina A, Majsterek I. Association between the Cytosine Hydroxymethylation and the Expression of microRNA in Multiple Sclerosis in Polish Population. International Journal of Molecular Sciences. 2023; 24(18):13923. https://doi.org/10.3390/ijms241813923
Chicago/Turabian StyleBasak, Justyna, Danuta Piotrzkowska, Aleksandra Kucharska-Lusina, and Ireneusz Majsterek. 2023. "Association between the Cytosine Hydroxymethylation and the Expression of microRNA in Multiple Sclerosis in Polish Population" International Journal of Molecular Sciences 24, no. 18: 13923. https://doi.org/10.3390/ijms241813923
APA StyleBasak, J., Piotrzkowska, D., Kucharska-Lusina, A., & Majsterek, I. (2023). Association between the Cytosine Hydroxymethylation and the Expression of microRNA in Multiple Sclerosis in Polish Population. International Journal of Molecular Sciences, 24(18), 13923. https://doi.org/10.3390/ijms241813923





