Signaling and Resistosome Formation in Plant Innate Immunity to Viruses: Is There a Common Mechanism of Antiviral Resistance Conserved across Kingdoms?
Abstract
1. Introduction
2. Virus-Specific Proteins Involved in the Hypersensitive Response
2.1. Virus RdRp Proteins
2.1.1. Tobacco Mosaic Virus (TMV) p50
2.1.2. Potyviral RdRp (NIb)
2.1.3. Plantago Asiatica Mosaic Virus (PlAMV) RdRp
2.1.4. Tomato Yellow Leaf Curl Virus (TYLCV) Rep/C1
2.1.5. Cucumber Mosaic Virus (CMV) 2a
2.2. Virus Coat Proteins (CP)
2.2.1. Potato Virus X (PVX) CP
2.2.2. Turnip Crinkle Virus (TCV) CP
2.2.3. Potato Virus Y (PVY) CP
2.2.4. Cucumber Mosaic Virus (CMV) CP
2.3. Virus Movement Proteins (MP)
2.3.1. Tomato Mosaic Virus (ToMV) MP
2.3.2. Tobacco Rattle Virus (TRV) MP
2.3.3. Tomato Spotted Wilt Virus (TSWV) MP
2.3.4. Cauliflower Mosaic Virus (CaMV) MP
2.4. Other Viral Proteins Involved in the HR
2.4.1. Tomato Spotted Wilt Virus (TSWV) Silencing Suppressor Protein (NSs)
2.4.2. Tomato Leaf Curl New Delhi Virus (ToLCNDV) Silencing Suppressor Protein AC4
2.4.3. Soybean Mosaic Virus (SMV) Proteins CI, P3 and HC-Pro
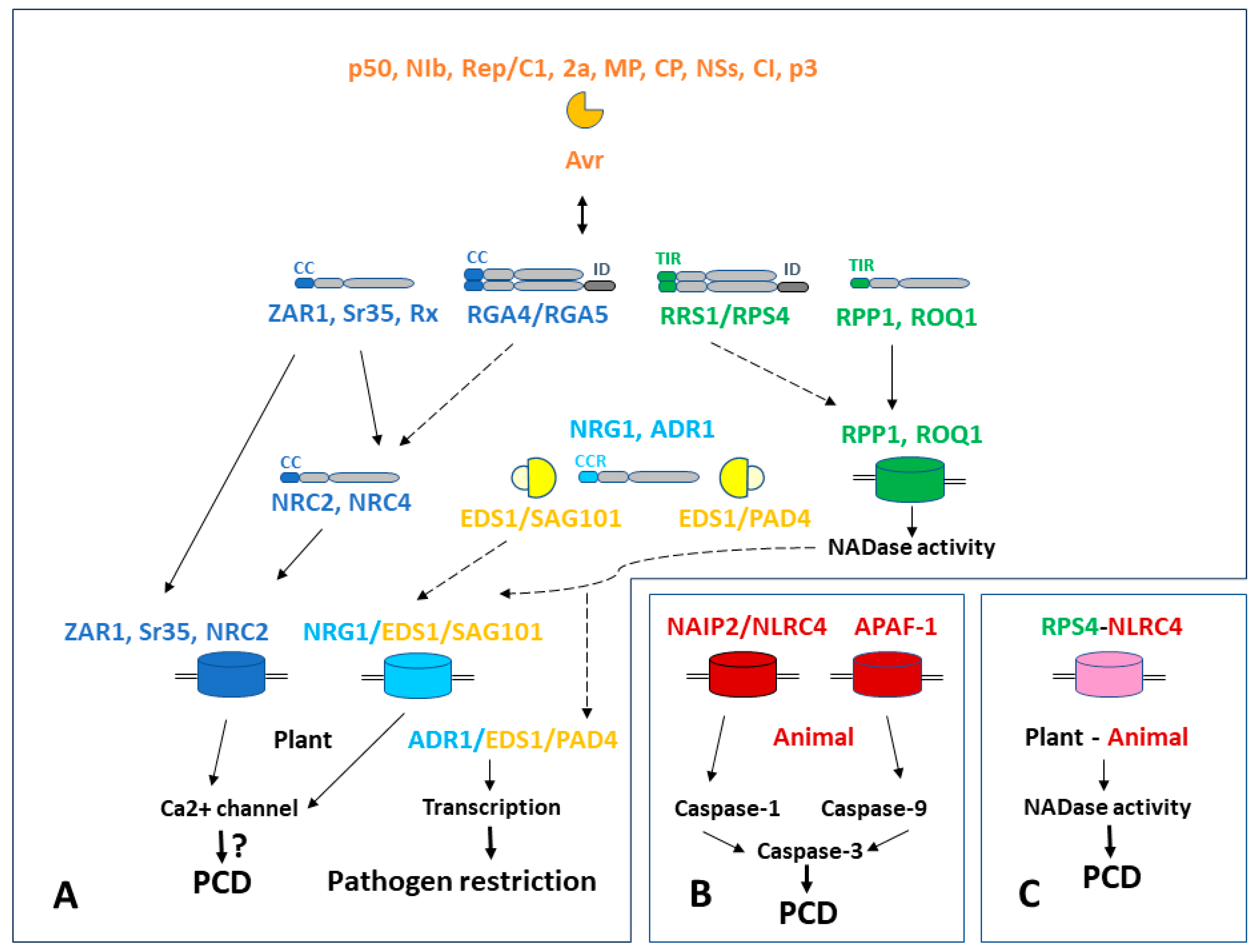
3. NLR Signaling Leading to Hypersensitive Response in Plants and Formation of Resistosomes
3.1. Plant Cell Death Signal Transmission Network
3.2. Resistosomes in Plants
3.3. Metazoan Apoptosomes/Inflammasomes
4. Common Features of Antiviral Resistance across the Kingdoms
5. Conclusions
6. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kesavardhana, S.; Malireddi, R.K.S.; Kanneganti, T.D. Caspases in Cell Death, Inflammation, and Pyroptosis. Annu. Rev. Immunol. 2020, 38, 567–595. [Google Scholar] [CrossRef] [PubMed]
- Distéfano, A.M.; Martin, M.V.; Córdoba, J.P.; Bellido, A.M.; D’Ippólito, S.; Colman, S.L.; Soto, D.; Roldán, J.A.; Bartoli, C.G.; Zabaleta, E.J.; et al. Heat stress induces ferroptosis-like cell death in plants. J. Cell Biol. 2017, 216, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Shen, Y.; Chen, C.; Sui, X.; Yang, J.; Wang, L.; Zhou, J. The crosstalk between autophagy and ferroptosis: What can we learn to target drug resistance in cancer? Cancer Biol. Med. 2019, 16, 630–646. [Google Scholar] [CrossRef] [PubMed]
- Distéfano, A.M.; López, G.A.; Setzes, N.; Marchetti, F.; Cainzos, M.; Cascallares, M.; Zabaleta, E.; Pagnussat, G.C. Ferroptosis in plants: Triggers, proposed mechanisms, and the role of iron in modulating cell death. J. Exp. Bot. 2021, 72, 2125–2135. [Google Scholar] [CrossRef]
- Wang, P.; Wang, T.; Han, J.; Li, M.; Zhao, Y.; Su, T.; Ma, C. Plant Autophagy: An Intricate Process Controlled by Various Signaling Pathways. Front. Plant Sci. 2021, 12, 754982. [Google Scholar] [CrossRef]
- Salguero-Linares, J.; Coll, N.S. Plant proteases in the control of the hypersensitive response. J. Exp. Bot. 2019, 70, 2087–2095. [Google Scholar] [CrossRef]
- Balakireva, A.V.; Zamyatnin, A.A., Jr. Cutting Out the Gaps Between Proteases and Programmed Cell Death. Front. Plant Sci. 2019, 10, 704. [Google Scholar] [CrossRef]
- Huh, S.U. Evolutionary Diversity and Function of Metacaspases in Plants: Similar to but Not Caspases. Int. J. Mol. Sci. 2022, 23, 4588. [Google Scholar] [CrossRef]
- Ebeed, H.T.; El-Helely, A.A. Programmed Cell Death in Plants: Insights into Developmental and Stress-Induced Cell Death. Curr. Protein Pept. Sci. 2021, 22, 873–889. [Google Scholar] [CrossRef]
- Huang, C. From Player to Pawn: Viral Avirulence Factors Involved in Plant Immunity. Viruses 2021, 13, 688. [Google Scholar] [CrossRef]
- Erickson, F.L.; Holzberg, S.; Calderon-Urrea, A.; Handley, V.; Axtell, M.; Corr, C.; Baker, B. The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 1999, 18, 67–75. [Google Scholar] [CrossRef]
- Kang, B.C.; Yeam, I.; Jahn, M.M. Genetics of plant virus resistance. Annu. Rev. Phytopathol. 2005, 43, 581–621. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Czymmek, K.; Tallóczy, Z.; Levine, B.; Dinesh-Kumar, S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005, 121, 567–577. [Google Scholar] [CrossRef]
- Ueda, H.; Yamaguchi, Y.; Sano, H. Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol. Biol. 2006, 61, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Solovieva, A.D.; Frolova, O.Y.; Solovyev, A.G.; Morozov, S.Y.; Zamyatnin, A.A., Jr. Effect of mitochondria-targeted antioxidant SkQ1 on programmed cell death induced by viral proteins in tobacco plants. Biochemistry 2013, 78, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Hong, Q.; Li, Y.; Li, Q.; Wang, M. Autophagy contributes to regulate the ROS levels and PCD progress in TMV-infected tomatoes. Plant Sci. 2018, 269, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Konakalla, N.C.; Nitin, M.; Kaldis, A.; Masarapu, H.; Carpentier, S.; Voloudakis, A. dsRNA Molecules from the Tobacco Mosaic Virus p126 Gene Counteract TMV-Induced Proteome Changes at an Early Stage of Infection. Front. Plant Sci. 2021, 12, 663707. [Google Scholar] [CrossRef]
- Caplan, J.L.; Mamillapalli, P.; Burch-Smith, T.M.; Czymmek, K.; Dinesh-Kumar, S.P. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 2008, 132, 449–462. [Google Scholar] [CrossRef]
- Padmanabhan, M.S.; Ma, S.; Burch-Smith, T.M.; Czymmek, K.; Huijser, P.; Dinesh-Kumar, S.P. Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog. 2013, 9, e1003235. [Google Scholar] [CrossRef]
- Jacob, P.; Kim, N.H.; Wu, F.; El-Kasmi, F.; Chi, Y.; Walton, W.G.; Furzer, O.J.; Lietzan, A.D.; Sunil, S.; Kempthorn, K.; et al. Plant “helper” immune receptors are Ca2+-permeable nonselective cation channels. Science 2021, 373, 420–425. [Google Scholar] [CrossRef]
- Li, Y.; Li, Q.; Hong, Q.; Lin, Y.; Mao, W.; Zhou, S. Reactive oxygen species triggering systemic programmed cell death process via elevation of nuclear calcium ion level in tomatoes resisting tobacco mosaic virus. Plant Sci. 2018, 270, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.B.; Lee, H.Y.; Seo, S.; Lee, J.H.; Choi, D. RNA-dependent RNA polymerase (NIb) of the potyviruses is an avirulence factor for the broad-spectrum resistance gene Pvr4 in Capsicum annuum cv. CM334. PLoS ONE 2015, 10, e0119639. [Google Scholar] [CrossRef]
- Tran, P.T.; Choi, H.; Choi, D.; Kim, K.H. Molecular characterization of Pvr9 that confers a hypersensitive response to Pepper mottle virus (a potyvirus) in Nicotiana benthamiana. Virology 2015, 481, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Yu, J.; Kim, K.H. Pepper Mottle Virus and Its Host Interactions: Current State of Knowledge. Viruses 2021, 13, 1930. [Google Scholar] [CrossRef]
- Gong, Y.N.; Tang, R.Q.; Zhang, Y.; Peng, J.; Xian, O.; Zhang, Z.H.; Zhang, S.B.; Zhang, D.Y.; Liu, H.; Luo, X.W.; et al. The NIa-Protease Protein Encoded by the Pepper Mottle Virus Is a Pathogenicity Determinant and Releases DNA Methylation of Nicotiana benthamiana. Front Microbiol. 2020, 11, 102. [Google Scholar] [CrossRef] [PubMed]
- Ozeki, J.; Takahashi, S.; Komatsu, K.; Kagiwada, S.; Yamashita, K.; Mori, T.; Hirata, H.; Yamaji, Y.; Ugaki, M.; Namba, S. A single amino acid in the RNA-dependent RNA polymerase of Plantago asiatica mosaic virus contributes to systemic necrosis. Arch. Virol. 2006, 151, 2067–2075. [Google Scholar] [CrossRef]
- Komatsu, K.; Hammond, J. Plantago asiatica mosaic virus: An emerging plant virus causing necrosis in lilies and a new model RNA virus for molecular research. Mol. Plant Pathol. 2022, 23, 1401–1414. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Ohnishi, J.; Saito, A.; Ohyama, A.; Nunome, T.; Miyatake, K.; Fukuoka, H. An NB-LRR gene, TYNBS1, is responsible for resistance mediated by the Ty-2 Begomovirus resistance locus of tomato. Theor. Appl. Genet. 2018, 131, 1345–1362. [Google Scholar] [CrossRef]
- Shen, X.; Yan, Z.; Wang, X.; Wang, Y.; Arens, M.; Du, Y.; Visser, R.G.F.; Kormelink, R.; Bai, Y.; Wolters, A.A. The NLR Protein Encoded by the Resistance Gene Ty-2 Is Triggered by the Replication-Associated Protein Rep/C1 of Tomato Yellow Leaf Curl Virus. Front. Plant Sci. 2020, 11, 545306. [Google Scholar] [CrossRef]
- Lucioli, A.; Berardi, A.; Gatti, F.; Tavazza, R.; Pizzichini, D.; Tavazza, M. Tomato yellow leaf curl Sardinia virus-resistant tomato plants expressing the multifunctional N-terminal domain of the replication-associated protein show transcriptional changes resembling stress-related responses. Mol. Plant Pathol. 2014, 15, 31–43. [Google Scholar] [CrossRef]
- Lucioli, A.; Perla, C.; Berardi, A.; Gatti, F.; Spanò, L.; Tavazza, M. Transcriptomics of tomato plants infected with TYLCSV or expressing the central TYLCSV Rep protein domain uncover changes impacting pathogen response and senescence. Plant Physiol. Biochem. 2016, 103, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.S.; Rojas, M.R.; Lee, J.Y.; Lee, S.W.; Jeon, J.S.; Ronald, P.; Lucas, W.J.; Gilbertson, R.L. A viral resistance gene from common bean functions across plant families and is up-regulated in a non-virus-specific manner. Proc. Natl. Acad. Sci. USA 2006, 103, 11856–11861. [Google Scholar] [CrossRef] [PubMed]
- Bendahmane, A.; Köhn, B.A.; Dedi, C.; Baulcombe, D.C. The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. Plant J. 1995, 8, 933–941. [Google Scholar] [CrossRef]
- Goulden, M.G.; Baulcombe, D.C. Functionally Homologous Host Components Recognize Potato Virus X in Gomphrena globosa and Potato. Plant Cell 1993, 5, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Bendahmane, A.; Kanyuka, K.; Baulcombe, D.C. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 1999, 11, 781–792. [Google Scholar] [CrossRef]
- Tameling, W.I.; Baulcombe, D.C. Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell 2007, 19, 1682–1694. [Google Scholar] [CrossRef]
- Walling, L.L. Extreme resistance: The GLK-Rx1 alliance. J. Biol. Chem. 2018, 293, 3234–3235. [Google Scholar] [CrossRef]
- Zhao, Y.; DelGrosso, L.; Yigit, E.; Dempsey, D.A.; Klessig, D.F.; Wobbe, K.K. The amino terminus of the coat protein of Turnip crinkle virus is the AVR factor recognized by resistant arabidopsis. Mol. Plant Microbe Interact. 2000, 13, 1015–1018. [Google Scholar] [CrossRef]
- Cooley, M.B.; Pathirana, S.; Wu, H.J.; Kachroo, P.; Klessig, D.F. Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 2000, 12, 663–676. [Google Scholar] [CrossRef]
- Chandra-Shekara, A.C.; Navarre, D.; Kachroo, A.; Kang, H.G.; Klessig, D.; Kachroo, P. Signaling requirements and role of salicylic acid in HRT- and rrt-mediated resistance to turnip crinkle virus in Arabidopsis. Plant J. 2004, 40, 47–59. [Google Scholar] [CrossRef]
- Falk, A.; Feys, B.J.; Frost, L.N.; Jones, J.D.; Daniels, M.J.; Parker, J.E. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 1999, 96, 3292–3297. [Google Scholar] [CrossRef] [PubMed]
- Jirage, D.; Tootle, T.L.; Reuber, T.L.; Frost, L.N.; Feys, B.J.; Parker, J.E.; Ausubel, F.M.; Glazebrook, J. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 1999, 96, 13583–13588. [Google Scholar] [CrossRef] [PubMed]
- Feys, B.J.; Moisan, L.J.; Newman, M.A.; Parker, J.E. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001, 20, 5400–5411. [Google Scholar] [CrossRef] [PubMed]
- Valkonen, J.P. Elucidation of virus-host interactions to enhance resistance breeding for control of virus diseases in potato. Breed. Sci. 2015, 65, 69–76. [Google Scholar] [CrossRef]
- Grech-Baran, M.; Witek, K.; Szajko, K.; Witek, A.I.; Morgiewicz, K.; Wasilewicz-Flis, I.; Jakuczun, H.; Marczewski, W.; Jones, J.D.G.; Hennig, J. Extreme resistance to Potato virus Y in potato carrying the Rysto gene is mediated by a TIR-NLR immune receptor. Plant Biotechnol. J. 2020, 18, 655–667. [Google Scholar] [CrossRef]
- Ross, B.T.; Zidack, N.K.; Flenniken, M.L. Extreme Resistance to Viruses in Potato and Soybean. Front. Plant Sci. 2021, 12, 658981. [Google Scholar] [CrossRef]
- Baebler, Š.; Coll, A.; Gruden, K. Plant Molecular Responses to Potato Virus Y: A Continuum of Outcomes from Sensitivity and Tolerance to Resistance. Viruses 2020, 12, 217. [Google Scholar] [CrossRef]
- Takahashi, H.; Suzuki, M.; Natsuaki, K.; Shigyo, T.; Hino, K.; Teraoka, T.; Hosokawa, D.; Ehara, Y. Mapping the virus and host genes involved in the resistance response in cucumber mosaic virus-Infected Arabidopsis thaliana. Plant Cell Physiol. 2001, 42, 340–347. [Google Scholar] [CrossRef]
- Takahashi, H.; Miller, J.; Nozaki, Y.; Takeda, M.; Shah, J.; Hase, S.; Ikegami, M.; Ehara, Y.; Dinesh-Kumar, S.P. RCY1, an Arabidopsis thaliana RPP8/HRT family resistance gene, conferring resistance to cucumber mosaic virus requires salicylic acid, ethylene and a novel signal transduction mechanism. Plant J. 2002, 32, 655–667. [Google Scholar] [CrossRef]
- Lanfermeijer, F.C.; Dijkhuis, J.; Sturre, M.J.; de Haan, P.; Hille, J. Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-22 from Lycopersicon esculentum. Plant Mol. Biol. 2003, 52, 1037–1049. [Google Scholar] [CrossRef][Green Version]
- Chen, T.; Liu, D.; Niu, X.; Wang, J.; Qian, L.; Han, L.; Liu, N.; Zhao, J.; Hong, Y.; Liu, Y. Antiviral Resistance Protein Tm-22 Functions on the Plasma Membrane. Plant Physiol. 2017, 173, 2399–2410. [Google Scholar] [CrossRef]
- Ishibashi, K.; Kezuka, Y.; Kobayashi, C.; Kato, M.; Inoue, T.; Nonaka, T.; Ishikawa, M.; Matsumura, H.; Katoh, E. Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1. Proc. Natl. Acad. Sci. USA 2014, 111, E3486–E3495. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.Y.; Ma, H.Y.; Wang, L.; Tettey, C.; Zhao, M.S.; Geng, C.; Tian, Y.P.; Li, X.D. Identification of genetic determinants of tomato brown rugose fruit virus that enable infection of plants harbouring the Tm-22 resistance gene. Mol. Plant Pathol. 2021, 22, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Ghazala, W.; Varrelmann, M. Tobacco rattle virus 29K movement protein is the elicitor of extreme and hypersensitive-like resistance in two cultivars of Solanum tuberosum. Mol. Plant Microbe Interact. 2007, 20, 1396–1405. [Google Scholar] [CrossRef]
- Peiró, A.; Cañizares, M.C.; Rubio, L.; López, C.; Moriones, E.; Aramburu, J.; Sánchez-Navarro, J. The movement protein (NSm) of Tomato spotted wilt virus is the avirulence determinant in the tomato Sw-5 gene-based resistance. Mol. Plant Pathol. 2014, 15, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Hallwass, M.; de Oliveira, A.S.; de Campos Dianese, E.; Lohuis, D.; Boiteux, L.S.; Inoue-Nagata, A.K.; Resende, R.O.; Kormelink, R. The Tomato spotted wilt virus cell-to-cell movement protein (NSM) triggers a hypersensitive response in Sw-5-containing resistant tomato lines and in Nicotiana benthamiana transformed with the functional Sw-5b resistance gene copy. Mol. Plant Pathol. 2014, 15, 871–880. [Google Scholar] [CrossRef]
- Zhu, M.; Jiang, L.; Bai, B.; Zhao, W.; Chen, X.; Li, J.; Liu, Y.; Chen, Z.; Wang, B.; Wang, C.; et al. The intracellular immune receptor Sw-5b confers broad-spectrum resistance to Tospoviruses through recognition of a conserved 21-amino acid viral effector epitope. Plant Cell 2017, 29, 2214–2232. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Liu, Y.; Yu, H.; Yuan, C.; Zeng, J.; Zhao, L.; Tong, Z.; Tao, X. Non-Structural Protein NSm of Tomato Spotted Wilt Virus Is an Avirulence Factor Recognized by Resistance Genes of Tobacco and Tomato via Different Elicitor Active Sites. Viruses 2018, 10, 660. [Google Scholar] [CrossRef]
- Palanichelvam, K.; Cole, A.B.; Shababi, M.; Schoelz, J.E. Agroinfiltration of Cauliflower mosaic virus gene VI elicits hypersensitive response in Nicotiana species. Mol. Plant Microbe Interact. 2000, 13, 1275–1279. [Google Scholar] [CrossRef]
- Cole, A.B.; Király, L.; Ross, K.; Schoelz, J.E. Uncoupling resistance from cell death in the hypersensitive response of Nicotiana species to cauliflower mosaic virus infection. Mol. Plant Microbe Interact. 2001, 14, 31–41. [Google Scholar] [CrossRef]
- Harries, P.A.; Palanichelvam, K.; Yu, W.; Schoelz, J.E.; Nelson, R.S. The cauliflower mosaic virus protein P6 forms motile inclusions that traffic along actin microfilaments and stabilize microtubules. Plant Physiol. 2009, 149, 1005–1016. [Google Scholar] [CrossRef]
- Love, A.J.; Geri, C.; Laird, J.; Carr, C.; Yun, B.; Loake, G.J.; Tada, Y.; Sadanandom, A.; Milner, J.J. Cauliflower mosaic virus protein P6 inhibits signaling responses to salicylic acid and regulates innate immunity. PLoS ONE 2012, 7, e47535. [Google Scholar] [CrossRef]
- Cawly, J.; Cole, A.B.; Király, L.; Qiu, W.; Schoelz, J.E. The plant gene CCD1 selectively blocks cell death during the hypersensitive response to Cauliflower mosaic virus infection. Mol. Plant Microbe Interact. 2005, 18, 212–219. [Google Scholar] [CrossRef]
- de Ronde, D.; Butterbach, P.; Lohuis, D.; Hedil, M.; van Lent, J.W.; Kormelink, R. Tsw gene-based resistance is triggered by a functional RNA silencing suppressor protein of the Tomato spotted wilt virus. Mol. Plant Pathol. 2013, 14, 405–415. [Google Scholar] [CrossRef]
- van Grinsven, I.L.; Martin, E.C.; Petrescu, A.J.; Kormelink, R. Tsw—A case study on structure-function puzzles in plant NLRs with unusually large LRR domains. Front. Plant Sci. 2022, 13, 983693. [Google Scholar] [CrossRef]
- Schnettler, E.; Hemmes, H.; Huismann, R.; Goldbach, R.; Prins, M.; Kormelink, R. Diverging affinity of tospovirus RNA silencing suppressor proteins, NSs, for various RNA duplex molecules. J. Virol. 2010, 84, 11542–11554. [Google Scholar] [CrossRef]
- Margaria, P.; Bosco, L.; Vallino, M.; Ciuffo, M.; Mautino, G.C.; Tavella, L.; Turina, M. The NSs protein of tomato spotted wilt virus is required for persistent infection and transmission by Frankliniella occidentalis. J. Virol. 2014, 88, 5788–5802. [Google Scholar] [CrossRef]
- de Ronde, D.; Pasquier, A.; Ying, S.; Butterbach, P.; Lohuis, D.; Kormelink, R. Analysis of Tomato spotted wilt virus NSs protein indicates the importance of the N-terminal domain for avirulence and RNA silencing suppression. Mol. Plant Pathol. 2014, 15, 185–195. [Google Scholar] [CrossRef]
- Almási, A.; Nemes, K.; Csömör, Z.; Tóbiás, I.; Palkovics, L.; Salánki, K. A single point mutation in Tomato spotted wilt virus NSs protein is sufficient to overcome Tsw-gene-mediated resistance in pepper. J. Gen. Virol. 2017, 98, 1521–1525. [Google Scholar] [CrossRef]
- Sharma, N.; Sahu, P.P.; Prasad, A.; Muthamilarasan, M.; Waseem, M.; Khan, Y.; Thakur, J.K.; Chakraborty, S.; Prasad, M. The Sw5a gene confers resistance to ToLCNDV and triggers an HR response after direct AC4 effector recognition. Proc. Natl. Acad. Sci. USA 2021, 118, e2101833118. [Google Scholar] [CrossRef]
- Moriones, E.; Praveen, S.; Chakraborty, S. Tomato Leaf Curl New Delhi Virus: An Emerging Virus Complex Threatening Vegetable and Fiber Crops. Viruses 2017, 9, 264. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Wang, L.; Jin, T.; Nie, Y.; Liu, H.; Qiu, Y.; Yang, Y.; Li, B.; Zhang, J.; Wang, D.; et al. A cell wall-localized NLR confers resistance to Soybean mosaic virus by recognizing viral-encoded cylindrical inclusion protein. Mol. Plant. 2021, 14, 1881–1900. [Google Scholar] [CrossRef] [PubMed]
- Eggenberger, A.L.; Hajimorad, M.R.; Hill, J.H. Gain of virulence on Rsv1-genotype soybean by an avirulent Soybean mosaic virus requires concurrent mutations in both P3 and HC-Pro. Mol. Plant Microbe Interact. 2008, 21, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Lee, S.H.; Kim, K.H. Strain-specific cylindrical inclusion protein of soybean mosaic virus elicits extreme resistance and a lethal systemic hypersensitive response in two resistant soybean cultivars. Mol. Plant Microbe Interact. 2009, 22, 1151–1159. [Google Scholar] [CrossRef]
- Chowda-Reddy, R.V.; Sun, H.; Chen, H.; Poysa, V.; Ling, H.; Gijzen, M.; Wang, A. Mutations in the P3 protein of Soybean mosaic virus G2 isolates determine virulence on Rsv4-genotype soybean. Mol. Plant Microbe Interact. 2011, 24, 37–43. [Google Scholar] [CrossRef]
- Khatabi, B.; Wen, R.H.; Hajimorad, M.R. Fitness penalty in susceptible host is associated with virulence of Soybean mosaic virus on Rsv1-genotype soybean: A consequence of perturbation of HC-Pro and not P3. Mol. Plant Pathol. 2013, 14, 885–897. [Google Scholar] [CrossRef]
- Martin, R.; Qi, T.; Zhang, H.; Liu, F.; King, M.; Toth, C.; Nogales, E.; Staskawicz, B.J. Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science 2020, 370, eabd9993. [Google Scholar] [CrossRef]
- Duxbury, Z.; Wu, C.H.; Ding, P. A Comparative Overview of the Intracellular Guardians of Plants and Animals: NLRs in Innate Immunity and Beyond. Annu. Rev. Plant Biol. 2021, 72, 155–184. [Google Scholar] [CrossRef]
- Ramírez-Zavaleta, C.Y.; García-Barrera, L.J.; Rodríguez-Verástegui, L.L.; Arrieta-Flores, D.; Gregorio-Jorge, J. An Overview of PRR- and NLR-Mediated Immunities: Conserved Signaling Components across the Plant Kingdom That Communicate Both Pathways. Int. J. Mol. Sci. 2022, 23, 12974. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Ding, P.; Jones, J.D.G. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 2022, 34, 1447–1478. [Google Scholar] [CrossRef]
- Wróblewski, T.; Spiridon, L.; Martin, E.C.; Petrescu, A.J.; Cavanaugh, K.; Truco, M.J.; Xu, H.; Gozdowski, D.; Pawłowski, K.; Michelmore, R.W.; et al. Genome-wide functional analyses of plant coiled-coil NLR-type pathogen receptors reveal essential roles of their N-terminal domain in oligomerization, networking, and immunity. PLoS Biol. 2018, 16, e2005821. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Pennill, L.A.; Ning, J.; Lee, S.W.; Ramalingam, J.; Webb, C.A.; Zhao, B.; Sun, Q.; Nelson, J.C.; Leach, J.E.; et al. Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res. 2002, 12, 1871–1884. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.D.; Wu, R.; Guttman, D.S.; Desveaux, D. Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet. 2010, 6, e1000894. [Google Scholar] [CrossRef]
- Schultink, A.; Qi, T.; Bally, J.; Staskawicz, B. Using forward genetics in Nicotiana benthamiana to uncover the immune signaling pathway mediating recognition of the Xanthomonas perforans effector XopJ4. New Phytol. 2019, 221, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Moffett, P.; Farnham, G.; Peart, J.; Baulcombe, D.C. Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 2002, 21, 4511–4519. [Google Scholar] [CrossRef]
- Botella, M.A.; Parker, J.E.; Frost, L.N.; Bittner-Eddy, P.D.; Beynon, J.L.; Daniels, M.J.; Holub, E.B.; Jones, J.D. Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 1998, 10, 1847–1860. [Google Scholar] [CrossRef]
- Wu, C.H.; Abd-El-Haliem, A.; Bozkurt, T.O.; Belhaj, K.; Terauchi, R.; Vossen, J.H.; Kamoun, S. NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA 2017, 114, 8113–8118. [Google Scholar] [CrossRef]
- Wu, C.H.; Derevnina, L.; Kamoun, S. Receptor networks underpin plant immunity. Science 2018, 360, 1300–1301. [Google Scholar] [CrossRef]
- Wang, J.; Hu, M.; Wang, J.; Qi, J.; Han, Z.; Wang, G.; Qi, Y.; Wang, H.W.; Zhou, J.M.; Chai, J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 2019, 364, eaav5870. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Hu, M.; Wu, S.; Qi, J.; Wang, G.; Han, Z.; Qi, Y.; Gao, N.; Wang, H.W.; et al. Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 2019, 364, eaav5868. [Google Scholar] [CrossRef]
- Ma, S.; Lapin, D.; Liu, L.; Sun, Y.; Song, W.; Zhang, X.; Logemann, E.; Yu, D.; Wang, J.; Jirschitzka, J.; et al. Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 2020, 370, eabe3069. [Google Scholar] [CrossRef] [PubMed]
- Förderer, A.; Li, E.; Lawson, A.W.; Deng, Y.N.; Sun, Y.; Logemann, E.; Zhang, X.; Wen, J.; Han, Z.; Chang, J.; et al. A wheat resistosome defines common principles of immune receptor channels. Nature 2022, 610, 532–539. [Google Scholar] [CrossRef]
- Adachi, H.; Contreras, M.P.; Harant, A.; Wu, C.H.; Derevnina, L.; Sakai, T.; Duggan, C.; Moratto, E.; Bozkurt, T.O.; Maqbool, A.; et al. N-terminal motif in NLR immune receptors is functionally conserved across distantly related plant species. eLife 2019, 8, e49956. [Google Scholar] [CrossRef] [PubMed]
- Swiderski, M.R.; Birker, D.; Jones, J.D. The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol. Plant Microbe Interact. 2009, 22, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Collier, S.M.; Hamel, L.P.; Moffett, P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant Microbe Interact. 2011, 24, 918–931. [Google Scholar] [CrossRef]
- Cesari, S.; Moore, J.; Chen, C.; Webb, D.; Periyannan, S.; Mago, R.; Bernoux, M.; Lagudah, E.S.; Dodds, P.N. Cytosolic activation of cell death and stem rust resistance by cereal MLA-family CC-NLR proteins. Proc. Natl. Acad. Sci. USA 2016, 113, 10204–10209. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wu, X.; Sun, K.; Gao, Z. Structure and function analysis of a CC-NBS-LRR protein AT1G12290. Biochem. Biophys. Res. Commun. 2021, 534, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, T.; Cheng, W.; Spiridon, L.N.; Töller, A.; Lukasik, E.; Saijo, Y.; Liu, P.; Shen, Q.H.; Micluta, M.A.; Somssich, I.E.; et al. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 2011, 9, 187–199. [Google Scholar] [CrossRef]
- Bernoux, M.; Ve, T.; Williams, S.; Warren, C.; Hatters, D.; Valkov, E.; Zhang, X.; Ellis, J.G.; Kobe, B.; Dodds, P.N. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 2011, 9, 200–211. [Google Scholar] [CrossRef]
- Rairdan, G.J.; Collier, S.M.; Sacco, M.A.; Baldwin, T.T.; Boettrich, T.; Moffett, P. The coiled-coil and nucleotide binding domains of the Potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 2008, 20, 739–751. [Google Scholar] [CrossRef]
- Feehan, J.M.; Castel, B.; Bentham, A.R.; Jones, J.D. Plant NLRs get by with a little help from their friends. Curr. Opin. Plant Biol. 2020, 56, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Derevnina, L.; Kamoun, S. NLR singletons, pairs, and networks: Evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 2019, 50, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Derevnina, L.; Contreras, M.P.; Adachi, H.; Upson, J.; Vergara Cruces, A.; Xie, R.; Skłenar, J.; Menke, F.L.H.; Mugford, S.T.; MacLean, D.; et al. Plant pathogens convergently evolved to counteract redundant nodes of an NLR immune receptor network. PLoS Biol. 2021, 19, e3001136. [Google Scholar] [CrossRef]
- Contreras, M.P.; Pai, H.; Tumtas, Y.; Duggan, C.; Yuen, E.L.H.; Cruces, A.V.; Kourelis, J.; Ahn, H.K.; Lee, K.T.; Wu, C.H.; et al. Sensor NLR immune proteins activate oligomerization of their NRC helpers in response to plant pathogens. EMBO J. 2023, 42, e111519. [Google Scholar] [CrossRef]
- Oh, S.K.; Kwon, S.Y.; Choi, D. Rpi-blb2-Mediated Hypersensitive Cell Death Caused by Phytophthora infestans AVRblb2 Requires SGT1, but not EDS1, NDR1, Salicylic Acid-, Jasmonic Acid-, or Ethylene-Mediated Signaling. Plant Pathol. J. 2014, 30, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Duggan, C.; Moratto, E.; Savage, Z.; Hamilton, E.; Adachi, H.; Wu, C.H.; Leary, A.Y.; Tumtas, Y.; Rothery, S.M.; Maqbool, A.; et al. Dynamic localization of a helper NLR at the plant-pathogen interface underpins pathogen recognition. Proc. Natl. Acad. Sci. USA 2021, 118, e2104997118. [Google Scholar] [CrossRef]
- Adachi, H.; Sakai, T.; Harant, A.; Pai, H.; Honda, K.; Toghani, A.; Claeys, J.; Duggan, C.; Bozkurt, T.O.; Wu, C.H.; et al. An atypical NLR protein modulates the NRC immune receptor network in Nicotiana benthamiana. PLoS Genet. 2023, 19, e1010500. [Google Scholar] [CrossRef] [PubMed]
- Lapin, D.; Bhandari, D.D.; Parker, J.E. Origins and Immunity Networking Functions of EDS1 Family Proteins. Annu. Rev. Phytopathol. 2020, 58, 253–276. [Google Scholar] [CrossRef] [PubMed]
- Lapin, D.; Johanndrees, O.; Wu, Z.; Li, X.; Parker, J.E. Molecular innovations in plant TIR-based immunity signaling. Plant Cell 2022, 34, 1479–1496. [Google Scholar] [CrossRef]
- Saile, S.C.; Jacob, P.; Castel, B.; Jubic, L.M.; Salas-Gonzáles, I.; Bäcker, M.; Jones, J.D.G.; Dangl, J.L.; El Kasmi, F. Two unequally redundant “helper” immune receptor families mediate Arabidopsis thaliana intracellular “sensor” immune receptor functions. PLoS Biol. 2020, 18, e3000783. [Google Scholar] [CrossRef]
- Sun, X.; Lapin, D.; Feehan, J.M.; Stolze, S.C.; Kramer, K.; Dongus, J.A.; Rzemieniewski, J.; Blanvillain-Baufumé, S.; Harzen, A.; Bautor, J.; et al. Pathogen effector recognition-dependent association of NRG1 with EDS1 and SAG101 in TNL receptor immunity. Nat. Commun. 2021, 12, 3335. [Google Scholar] [CrossRef] [PubMed]
- Feehan, J.M.; Wang, J.; Sun, X.; Choi, J.; Ahn, H.K.; Ngou, B.P.M.; Parker, J.E.; Jones, J.D.G. Oligomerization of a plant helper NLR requires cell-surface and intracellular immune receptor activation. Proc. Natl. Acad. Sci. USA 2023, 120, e2210406120. [Google Scholar] [CrossRef] [PubMed]
- Peart, J.R.; Mestre, P.; Lu, R.; Malcuit, I.; Baulcombe, D.C. NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr. Biol. 2005, 15, 968–973. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Seong, K.; Thomazella, D.P.T.; Kim, J.R.; Pham, J.; Seo, E.; Cho, M.J.; Schultink, A.; Staskawicz, B.J. NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 2018, 115, E10979–E10987. [Google Scholar] [CrossRef] [PubMed]
- Lapin, D.; Kovacova, V.; Sun, X.; Dongus, J.A.; Bhandari, D.; von Born, P.; Bautor, J.; Guarneri, N.; Rzemieniewski, J.; Stuttmann, J.; et al. A Coevolved EDS1-SAG101-NRG1 Module Mediates Cell Death Signaling by TIR-Domain Immune Receptors. Plant Cell 2019, 31, 2430–2455. [Google Scholar] [CrossRef]
- Voss, M.; Toelzer, C.; Bhandari, D.D.; Parker, J.E.; Niefind, K. Arabidopsis immunity regulator EDS1 in a PAD4/SAG101-unbound form is a monomer with an inherently inactive conformation. J. Struct. Biol. 2019, 208, 107390. [Google Scholar] [CrossRef]
- Dongus, J.A.; Parker, J.E. EDS1 signalling: At the nexus of intracellular and surface receptor immunity. Curr. Opin. Plant Biol. 2021, 62, 102039. [Google Scholar] [CrossRef]
- Wu, Z.; Tian, L.; Liu, X.; Zhang, Y.; Li, X. TIR signal promotes interactions between lipase-like proteins and ADR1-L1 receptor and ADR1-L1 oligomerization. Plant Physiol. 2021, 187, 681–686. [Google Scholar] [CrossRef]
- Huang, S.; Jia, A.; Song, W.; Hessler, G.; Meng, Y.; Sun, Y.; Xu, L.; Laessle, H.; Jirschitzka, J.; Ma, S.; et al. Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity. Science 2022, 377, eabq3297. [Google Scholar] [CrossRef]
- Ma, Y.; Guo, H.; Hu, L.; Martinez, P.P.; Moschou, P.N.; Cevik, V.; Ding, P.; Duxbury, Z.; Sarris, P.F.; Jones, J.D.G. Distinct modes of derepression of an Arabidopsis immune receptor complex by two different bacterial effectors. Proc. Natl. Acad. Sci. USA 2018, 115, 10218–10227. [Google Scholar] [CrossRef]
- Césari, S.; Kanzaki, H.; Fujiwara, T.; Bernoux, M.; Chalvon, V.; Kawano, Y.; Shimamoto, K.; Dodds, P.; Terauchi, R.; Kroj, T. The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 2014, 33, 1941–1959. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.L.; Kim, S.T.; Castel, B.; Charoennit, N.; Chae, E. Genetics of autoimmunity in plants: An evolutionary genetics perspective. New Phytol. 2021, 229, 1215–1233. [Google Scholar] [CrossRef] [PubMed]
- Kourelis, J.; Adachi, H. Activation and Regulation of NLR Immune Receptor Networks. Plant Cell Physiol. 2022, 63, 1366–1377. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, J.P.; Seddon, A.E.; Moghe, G.D.; Simenc, M.C.; Shiu, S.H. Characteristics of Plant Essential Genes Allow for within- and between-Species Prediction of Lethal Mutant Phenotypes. Plant Cell 2015, 27, 2133–2147. [Google Scholar] [CrossRef] [PubMed]
- Riedl, S.J.; Li, W.; Chao, Y.; Schwarzenbacher, R.; Shi, Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 2005, 434, 926–933. [Google Scholar] [CrossRef]
- Reubold, T.F.; Wohlgemuth, S.; Eschenburg, S. Crystal structure of full-length Apaf-1: How the death signal is relayed in the mitochondrial pathway of apoptosis. Structure 2011, 19, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Li, Y.; Hu, Q.; Bai, X.C.; Huang, W.; Yan, C.; Scheres, S.H.; Shi, Y. Atomic structure of the apoptosome: Mechanism of cytochrome c- and dATP-mediated activation of Apaf-1. Genes. Dev. 2015, 29, 2349–2361. [Google Scholar] [CrossRef]
- Hu, Z.; Yan, C.; Liu, P.; Huang, Z.; Ma, R.; Zhang, C.; Wang, R.; Zhang, Y.; Martinon, F.; Miao, D.; et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 2013, 341, 172–175. [Google Scholar] [CrossRef]
- Maekawa, S.; Ohto, U.; Shibata, T.; Miyake, K.; Shimizu, T. Crystal structure of NOD2 and its implications in human disease. Nat. Commun. 2016, 7, 11813. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, S.; Ruan, J.; Wu, J.; Tong, A.B.; Yin, Q.; Li, Y.; David, L.; Lu, A.; Wang, W.L.; et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 2015, 350, 404–409. [Google Scholar] [CrossRef]
- Bi, G.; Su, M.; Li, N.; Liang, Y.; Dang, S.; Xu, J.; Hu, M.; Wang, J.; Zou, M.; Deng, Y.; et al. The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 2021, 184, 3528–3541.e12. [Google Scholar] [CrossRef]
- Horsefield, S.; Burdett, H.; Zhang, X.; Manik, M.K.; Shi, Y.; Chen, J.; Qi, T.; Gilley, J.; Lai, J.S.; Rank, M.X.; et al. NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 2019, 365, 793–799. [Google Scholar] [CrossRef] [PubMed]
- Lukasik, E.; Takken, F.L. STANDing strong, resistance proteins instigators of plant defence. Curr. Opin. Plant Biol. 2009, 12, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Pang, Y.; Hu, Q.; Liu, Q.; Li, H.; Zhou, Y.; He, T.; Liang, Q.; Liu, Y.; Yuan, X.; et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 2010, 141, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Urbach, J.M.; Ausubel, F.M. The NBS-LRR architectures of plant R-proteins and metazoan NLRs evolved in independent events. Proc. Natl. Acad. Sci. USA 2017, 114, 1063–1068. [Google Scholar] [CrossRef]
- Hu, Z.; Chai, J. Structural Mechanisms in NLR Inflammasome Assembly and Signaling. Curr. Top. Microbiol. Immunol. 2016, 397, 23–42. [Google Scholar]
- Li, Y.; Zhou, M.; Hu, Q.; Bai, X.C.; Huang, W.; Scheres, S.H.; Shi, Y. Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme. Proc. Natl. Acad. Sci. USA 2017, 114, 1542–1547. [Google Scholar] [CrossRef]
- Wang, L.; Wu, H. Keeping the Death Protein in Check. Immunity 2019, 51, 1–2. [Google Scholar] [CrossRef]
- Wan, L.; Essuman, K.; Anderson, R.G.; Sasaki, Y.; Monteiro, F.; Chung, E.H.; Osborne Nishimura, E.; DiAntonio, A.; Milbrandt, J.; Dangl, J.L.; et al. TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 2019, 365, 799–803. [Google Scholar] [CrossRef]
- Gerdts, J.; Brace, E.J.; Sasaki, Y.; DiAntonio, A.; Milbrandt, J. SARM1 activation triggers axon degeneration locally via NAD⁺ destruction. Science 2015, 348, 453–457. [Google Scholar] [CrossRef]
- Duxbury, Z.; Wang, S.; MacKenzie, C.I.; Tenthorey, J.L.; Zhang, X.; Huh, S.U.; Hu, L.; Hill, L.; Ngou, P.M.; Ding, P.; et al. Induced proximity of a TIR signaling domain on a plant-mammalian NLR chimera activates defense in plants. Proc. Natl. Acad. Sci. USA 2020, 117, 18832–18839. [Google Scholar] [CrossRef] [PubMed]
- Pontier, D.; Mittler, R.; Lam, E. Mechanism of cell death and disease resistance induction by transgenic expression of bacterio-opsin. Plant J. 2002, 30, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, S.; Lee, J.; Gäbler, Y.; Kemmerling, B.; Haapalainen, M.L.; Li, C.M.; Wei, Z.; Keller, H.; Joosten, M.; Taira, S.; et al. Separable roles of the Pseudomonas syringae pv. phaseolicola accessory protein HrpZ1 in ion-conducting pore formation and activation of plant immunity. Plant J. 2009, 57, 706–717. [Google Scholar] [CrossRef]
- Remick, B.C.; Gaidt, M.M.; Vance, R.E. Effector-Triggered Immunity. Annu. Rev. Immunol. 2023, 41, 453–481. [Google Scholar] [CrossRef] [PubMed]
- Tenthorey, J.L.; Haloupek, N.; López-Blanco, J.R.; Grob, P.; Adamson, E.; Hartenian, E.; Lind, N.A.; Bourgeois, N.M.; Chacón, P.; Nogales, E.; et al. The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion. Science 2017, 358, 888–893. [Google Scholar] [CrossRef] [PubMed]
- Tenthorey, J.L.; Kofoed, E.M.; Daugherty, M.D.; Malik, H.S.; Vance, R.E. Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol. Cell 2014, 54, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Krasileva, K.V.; Dahlbeck, D.; Staskawicz, B.J. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 2010, 22, 2444–2458. [Google Scholar] [CrossRef]
- Schultink, A.; Qi, T.; Lee, A.; Steinbrenner, A.D.; Staskawicz, B. Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J. 2017, 92, 787–795. [Google Scholar] [CrossRef]
- Mackey, D.; Belkhadir, Y.; Alonso, J.M.; Ecker, J.R.; Dangl, J.L. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 2003, 112, 379–389. [Google Scholar] [CrossRef]
- Redditt, T.J.; Chung, E.H.; Karimi, H.Z.; Rodibaugh, N.; Zhang, Y.; Trinidad, J.C.; Kim, J.H.; Zhou, Q.; Shen, M.; Dangl, J.L.; et al. AvrRpm1 Functions as an ADP-Ribosyl Transferase to Modify NOI Domain-Containing Proteins, Including Arabidopsis and Soybean RPM1-Interacting Protein4. Plant Cell 2019, 31, 2664–2681. [Google Scholar]
- Xu, H.; Shi, J.; Gao, H.; Liu, Y.; Yang, Z.; Shao, F.; Dong, N. The N-end rule ubiquitin ligase UBR2 mediates NLRP1B inflammasome activation by anthrax lethal toxin. EMBO J. 2019, 38, e101996. [Google Scholar] [CrossRef] [PubMed]
- Sarris, P.F.; Duxbury, Z.; Huh, S.U.; Ma, Y.; Segonzac, C.; Sklenar, J.; Derbyshire, P.; Cevik, V.; Rallapalli, G.; Saucet, S.B.; et al. A Plant Immune Receptor Detects Pathogen Effectors that Target WRKY Transcription Factors. Cell 2015, 161, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Dyrka, W.; Lamacchia, M.; Durrens, P.; Kobe, B.; Daskalov, A.; Paoletti, M.; Sherman, D.J.; Saupe, S.J. Diversity and variability of NOD-like receptors in fungi. Genome Biol. Evol. 2014, 6, 3137–3158. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, aaf6395. [Google Scholar] [CrossRef] [PubMed]
| Type of Virus Specific Proteins Dealing with Programmed Cell Death | Virus Protein | Virus Protein/Characteristics | Type NLR (Nucleotide-Binding Leucine-Rich Repeats Proteins) | Ref. |
|---|---|---|---|---|
| Replication-related | P50 Nib RdRp Rep/C1 2a | TMV PepMoV PlAMV TYLCV CMV | TIR-NBS-LRR CC-NBS-LRR CC-NBS-LRR Not reported type NLR TIR-NBS-LRR | [11,12,13,14,15,16,17,18,19,20,21] [22,23,24,25] [26,27] [28,29,30,31] [32] |
| CP | CP P38 CP CP | PVX TCV PVY CMV | CC-NBS-LRR CC-NBS-LRR TIR-NBS-LRR CC-NBS-LRR | [33,34,35,36,37] [38,39,40,41,42,43] [44,45,46,47] [48,49] |
| MP | MP MP (29K) NSM P6 | ToMV TRV TSWV CaMV | CC-NBS-LRR Not reported type NLR CC-NBS-LRR Not reported type NLR | [50,51,52,53] [54] [55,56,57,58] [59,60,61,62,63] |
| Other proteins | NSs (silencing suppressor) AC4 (silencing suppressor) C1 (cylindrical protein) P3 (multifunctional protein) HC-Pro (multifunctional protein) | TSWV ToLCNDV SMV | CC-NBS-LRR CC-NBS-LRR CC-NBS-LRR | [64,65,66,67,68,69] [70,71] [72,73,74,75,76] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanov, P.A.; Gasanova, T.V.; Repina, M.N.; Zamyatnin, A.A., Jr. Signaling and Resistosome Formation in Plant Innate Immunity to Viruses: Is There a Common Mechanism of Antiviral Resistance Conserved across Kingdoms? Int. J. Mol. Sci. 2023, 24, 13625. https://doi.org/10.3390/ijms241713625
Ivanov PA, Gasanova TV, Repina MN, Zamyatnin AA Jr. Signaling and Resistosome Formation in Plant Innate Immunity to Viruses: Is There a Common Mechanism of Antiviral Resistance Conserved across Kingdoms? International Journal of Molecular Sciences. 2023; 24(17):13625. https://doi.org/10.3390/ijms241713625
Chicago/Turabian StyleIvanov, Peter A., Tatiana V. Gasanova, Maria N. Repina, and Andrey A. Zamyatnin, Jr. 2023. "Signaling and Resistosome Formation in Plant Innate Immunity to Viruses: Is There a Common Mechanism of Antiviral Resistance Conserved across Kingdoms?" International Journal of Molecular Sciences 24, no. 17: 13625. https://doi.org/10.3390/ijms241713625
APA StyleIvanov, P. A., Gasanova, T. V., Repina, M. N., & Zamyatnin, A. A., Jr. (2023). Signaling and Resistosome Formation in Plant Innate Immunity to Viruses: Is There a Common Mechanism of Antiviral Resistance Conserved across Kingdoms? International Journal of Molecular Sciences, 24(17), 13625. https://doi.org/10.3390/ijms241713625







