Androgens Modulate Bcl-2 Agonist of Cell Death (BAD) Expression and Function in Breast Cancer Cells
Abstract
1. Introduction
2. Results
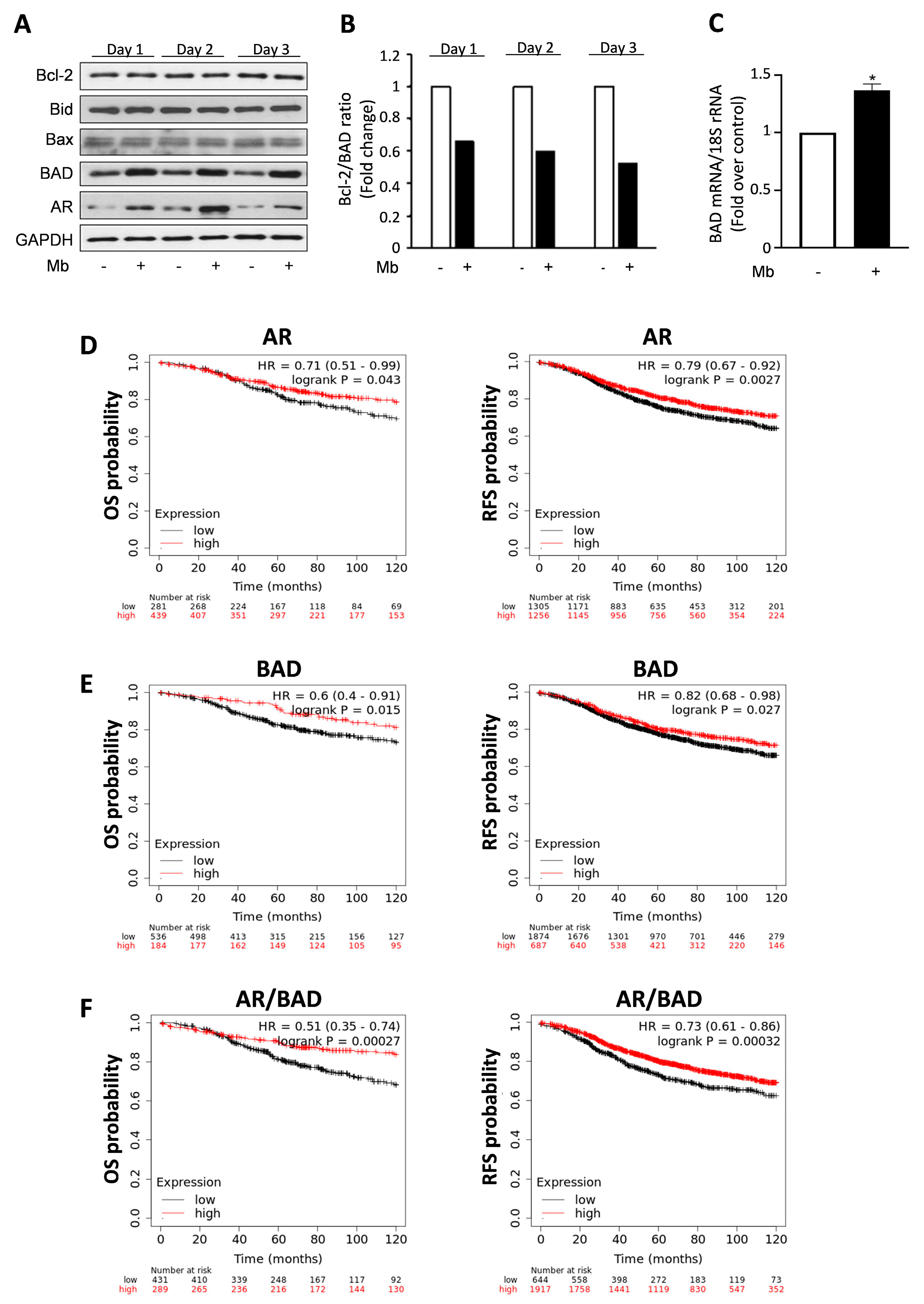
2.1. Androgens Increase BAD Expression, Which Affects Breast Cancer Patients’ Survival
2.2. Androgens Influence BAD Cellular Compartmentalization in Breast Cancer Cells
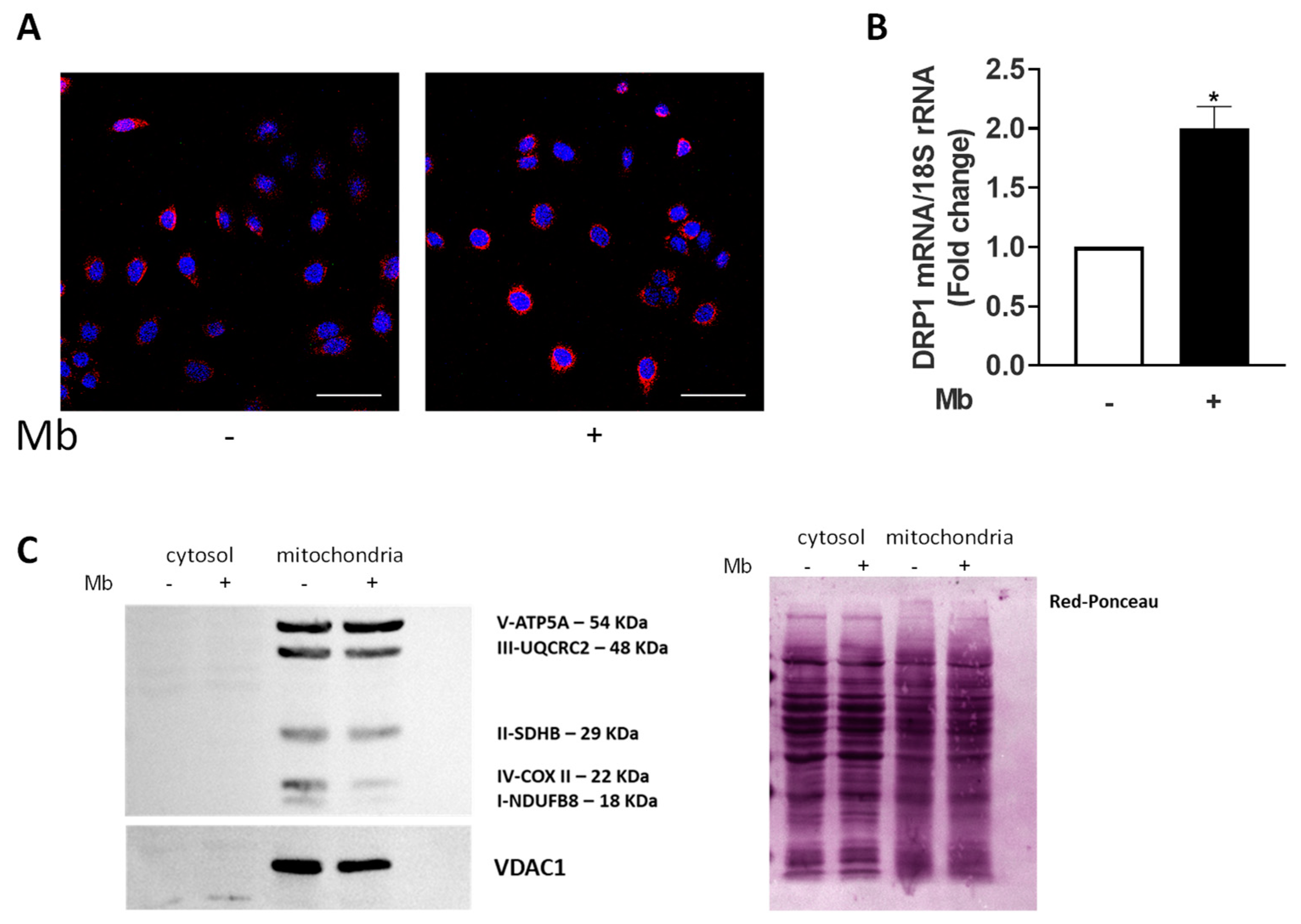
2.3. AR Activation Affects Mitochondria Functions
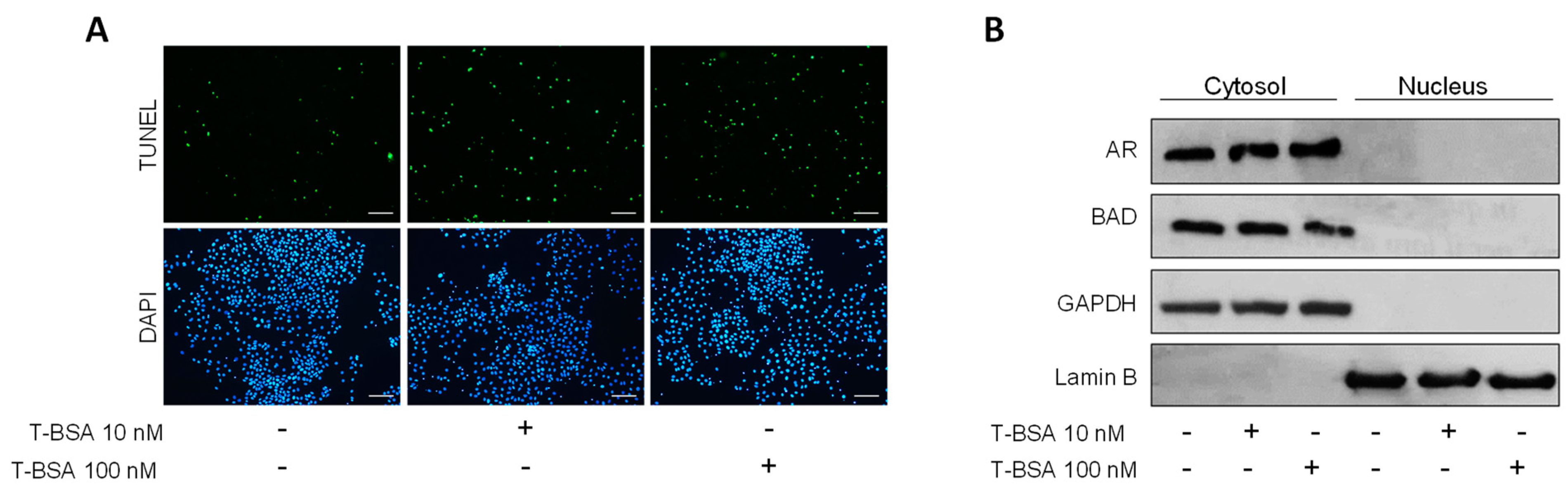
2.4. BSA-Conjugated Testosterone Induces Apoptosis without Influencing BAD Nuclear Translocation
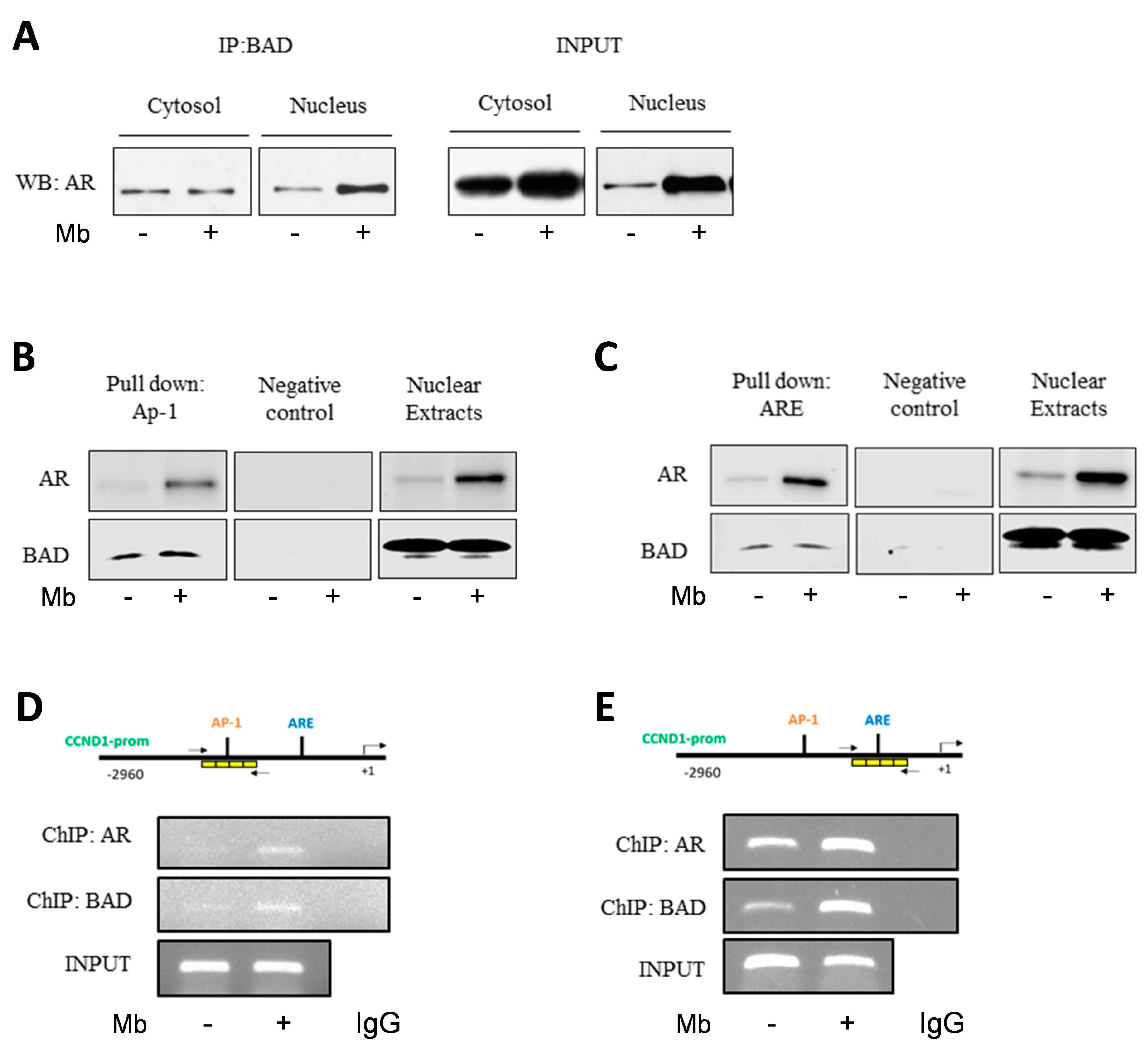
2.5. Mibolerone Induces the Formation of an AR/BAD Complex and Influences BAD Recruitment at the AP-1 and ARE Sites on the Cyclin D1 Gene Promoter
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Cell Cultures
4.3. Western Blot Analysis
4.4. Total RNA Extraction, Reverse Transcription Polymerase PCR and Real-Time RT-PCR Assay
4.5. Immunoprecipitation
4.6. Chromatin Immunoprecipitation (ChIP) Assay and PCR/Real-Time PCR ChIP
4.7. TUNEL Assay
4.8. DNA Affinity Precipitation Assay (DAPA)
4.9. Immunofluorescence Assay
4.10. Mitotracker Red CMXRos Staining
4.11. Kaplan–Meier Analysis
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.E.; Feigelson, H.S. Hormonal carcinogenesis. Carcinogenesis 2000, 21, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Yager, J.D.; Davidson, N.E. Estrogen carcinogenesis in breast cancer. N. Engl. J. Med. 2006, 354, 270–282. [Google Scholar] [CrossRef]
- Anderson, K.N.; Schwab, R.B.; Martinez, M.E. Reproductive risk factors and breast cancer subtypes: A review of the literature. Breast Cancer Res. Treat. 2014, 144, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hammond, M.E.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; BADve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M.; et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2010, 28, 2784–2795. [Google Scholar] [CrossRef]
- Wagner, R.K.; Gorlich, L.; Jungblut, P.W. Dihydrotestosterone receptor in human mammary cancer. Acta Endocrinol. Suppl. 1973, 173, 65. [Google Scholar] [CrossRef]
- Horwitz, K.B.; Zava, D.T.; Thilagar, A.K.; Jensen, E.M.; McGuire, W.L. Steroid receptor analyses of nine human breast cancer cell lines. Cancer Res. 1978, 38, 2434–2437. [Google Scholar]
- Allegra, J.C.; Lippman, M.E.; Thompson, E.B.; Simon, R.; Barlock, A.; Green, L.; Huff, K.K.; Do, H.M.; Aitken, S.C. Distribution, frequency, and quantitative analysis of estrogen, progesterone, androgen, and glucocorticoid receptors in human breast cancer. Cancer Res. 1979, 39, 1447–1454. [Google Scholar]
- Chiodo, C.; Morelli, C.; Cavaliere, F.; Sisci, D.; Lanzino, M. The Other Side of the Coin: May Androgens Have a Role in Breast Cancer Risk? Int. J. Mol. Sci. 2021, 23, 424. [Google Scholar] [CrossRef]
- You, C.P.; Leung, M.H.; Tsang, W.C.; Khoo, U.S.; Tsoi, H. Androgen Receptor as an Emerging Feasible Biomarker for Breast Cancer. Biomolecules 2022, 12, 72. [Google Scholar] [CrossRef]
- Caswell-Jin, J.L.; Curtis, C. Androgen receptor agonists as breast cancer therapeutics. Nat. Med. 2021, 27, 198–199. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.; Tarulli, G.; Portman, N.; Hickey, T.E.; Tilley, W.D.; Palmieri, C. Pushing estrogen receptor around in breast cancer. Endocr.-Relat. Cancer 2016, 23, T227–T241. [Google Scholar] [CrossRef]
- Lea, O.A.; Kvinnsland, S.; Thorsen, T. Improved measurement of androgen receptors in human breast cancer. Cancer Res. 1989, 49 24 Pt 1, 7162–7167. [Google Scholar]
- Moinfar, F.; Okcu, M.; Tsybrovskyy, O.; Regitnig, P.; Lax, S.F.; Weybora, W.; Ratschek, M.; Tavassoli, F.A.; Denk, H. Androgen receptors frequently are expressed in breast carcinomas: Potential relevance to new therapeutic strategies. Cancer 2003, 98, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Cimino-Mathews, A.; Hicks, J.L.; Illei, P.B.; Halushka, M.K.; Fetting, J.H.; De Marzo, A.M.; Park, B.H.; Argani, P. Androgen receptor expression is usually maintained in initial surgically resected breast cancer metastases but is often lost in end-stage metastases found at autopsy. Hum. Pathol. 2012, 43, 1003–1011. [Google Scholar] [CrossRef]
- Honma, N.; Horii, R.; Iwase, T.; Saji, S.; Younes, M.; Ito, Y.; Akiyama, F. Clinical importance of androgen receptor in breast cancer patients treated with adjuvant tamoxifen monotherapy. Breast Cancer 2012, 20, 323–330. [Google Scholar] [CrossRef]
- Ren, Q.; Zhang, L.; Ruoff, R.; Ha, S.; Wang, J.; Jain, S.; Reuter, V.; Gerald, W.; Giri, D.D.; Melamed, J.; et al. Expression of androgen receptor and its phosphorylated forms in breast cancer progression. Cancer 2013, 119, 2532–2540. [Google Scholar] [CrossRef]
- Gonzalez-Angulo, A.M.; Stemke-Hale, K.; Palla, S.L.; Carey, M.; Agarwal, R.; Meric-Berstam, F.; Traina, T.A.; Hudis, C.; Hortobagyi, G.N.; Gerald, W.L.; et al. Androgen receptor levels and association with PIK3CA mutations and prognosis in breast cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 2472–2478. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.A.; Buchanan, G.; Ricciardelli, C.; Bianco-Miotto, T.; Centenera, M.M.; Harris, J.M.; Jindal, S.; Segara, D.; Jia, L.; Moore, N.L.; et al. Androgen receptor inhibits estrogen receptor-alpha activity and is prognostic in breast cancer. Cancer Res. 2009, 69, 6131–6140. [Google Scholar] [CrossRef]
- Micello, D.; Marando, A.; Sahnane, N.; Riva, C.; Capella, C.; Sessa, F. Androgen receptor is frequently expressed in HER2-positive, ER/PR-negative breast cancers. Virchows Arch. Int. J. Pathol. 2010, 457, 467–476. [Google Scholar] [CrossRef]
- Niemeier, L.A.; Dabbs, D.J.; Beriwal, S.; Striebel, J.M.; Bhargava, R. Androgen receptor in breast cancer: Expression in estrogen receptor-positive tumors and in estrogen receptor-negative tumors with apocrine differentiation. Mod. Pathol. 2010, 23, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Koo, J.; Park, H.S.; Kim, J.H.; Choi, S.Y.; Lee, J.H.; Park, B.W.; Lee, K.S. Expression of androgen receptors in primary breast cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2010, 21, 488–492. [Google Scholar] [CrossRef]
- Collins, L.C.; Cole, K.S.; Marotti, J.D.; Hu, R.; Schnitt, S.J.; Tamimi, R.M. Androgen receptor expression in breast cancer in relation to molecular phenotype: Results from the Nurses’ Health Study. Mod. Pathol. 2011, 24, 924–931. [Google Scholar] [CrossRef]
- Hu, R.; Dawood, S.; Holmes, M.D.; Collins, L.C.; Schnitt, S.J.; Cole, K.; Marotti, J.D.; Hankinson, S.E.; Colditz, G.A.; Tamimi, R.M. Androgen receptor expression and breast cancer survival in postmenopausal women. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 1867–1874. [Google Scholar] [CrossRef] [PubMed]
- Loibl, S.; Müller, B.M.; von Minckwitz, G.; Schwabe, M.; Roller, M.; Darb-Esfahani, S.; Ataseven, B.; du Bois, A.; Fissler-Eckhoff, A.; Gerber, B.; et al. Androgen receptor expression in primary breast cancer and its predictive and prognostic value in patients treated with neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2011, 130, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Niu, Y.; Liu, N.; Zhang, J.Z.; Liu, T.J.; Zhang, R.J.; Wang, S.L.; Ding, X.M.; Xiao, X.Q. Expression of androgen receptor in breast cancer and its significance as a prognostic factor. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2011, 22, 1288–1294. [Google Scholar] [CrossRef] [PubMed]
- Hickey, T.E.; Robinson, J.L.L.; Carroll, J.S.; Tilley, W.D. Minireview: The androgen receptor in breast tissues: Growth inhibitor, tumor suppressor, oncogene? Mol. Endocrinol. 2012, 26, 1252–1267. [Google Scholar] [CrossRef]
- Tsang, J.Y.S.; Ni, Y.-B.; Chan, S.-K.; Shao, M.-M.; Law, B.K.B.; Tan, P.H.; Tse, G.M. Androgen receptor expression shows distinctive significance in ER positive and negative breast cancers. Ann. Surg. Oncol. 2014, 21, 2218–2228. [Google Scholar] [CrossRef]
- Ricciardelli, C.; Bianco-Miotto, T.; Jindal, S.; Butler, L.M.; Leung, S.; McNeil, C.M.; O’Toole, S.A.; Ebrahimie, E.; Millar, E.K.A.; Sakko, A.J.; et al. The Magnitude of Androgen Receptor Positivity in Breast Cancer Is Critical for Reliable Prediction of Disease Outcome. Clin. Cancer Res. 2018, 24, 2328–2341. [Google Scholar] [CrossRef]
- Jiang, H.-S.; Kuang, X.-Y.; Sun, W.-L.; Xu, Y.; Zheng, Y.-Z.; Liu, Y.-R.; Lang, G.-T.; Qiao, F.; Hu, X.; Shao, Z.-M. Androgen receptor expression predicts different clinical outcomes for breast cancer patients stratified by hormone receptor status. Oncotarget 2016, 7, 41285–41293. [Google Scholar] [CrossRef]
- Hickey, T.E.; Selth, L.A.; Chia, K.M.; Laven-Law, G.; Milioli, H.H.; Roden, D.; Jindal, S.; Hui, M.; Finlay-Schultz, J.; Ebrahimie, E.; et al. The androgen receptor is a tumor suppressor in estrogen receptor-positive breast cancer. Nat. Med. 2021, 27, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.A.; Ingman, W.V.; Tilley, W.D.; Butler, L.M. Differential effects of exogenous androgen and an androgen receptor antagonist in the peri- and postpubertal murine mammary gland. Endocrinology 2011, 152, 3728–3737. [Google Scholar] [CrossRef] [PubMed]
- Andò, S.; De Amicis, F.; Rago, V.; Carpino, A.; Maggiolini, M.; Panno, M.; Lanzino, M. Breast cancer: From estrogen to androgen receptor. Mol. Cell Endocrinol. 2002, 193, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Lanzino, M.; De Amicis, F.; McPhaul, M.J.; Marsico, S.; Panno, M.L.; Ando, S. Endogenous coactivator ARA70 interacts with estrogen receptor alpha (ERalpha) and modulates the functional ERalpha/androgen receptor interplay in MCF-7 cells. J. Biol. Chem. 2005, 280, 20421–20430. [Google Scholar] [CrossRef] [PubMed]
- Lanzino, M.; Maris, P.; Sirianni, R.; Barone, I.; Casaburi, I.; Chimento, A.; Giordano, C.; Morelli, C.; Sisci, D.; Rizza, P.; et al. DAX-1, as an androgen-target gene, inhibits aromatase expression: A novel mechanism blocking estrogen-dependent breast cancer cell proliferation. Cell Death Dis. 2013, 4, e724. [Google Scholar] [CrossRef]
- Lanzino, M.; Sisci, D.; Morelli, C.; Garofalo, C.; Catalano, S.; Casaburi, I.; Capparelli, C.; Giordano, C.; Giordano, F.; Maggiolini, M.; et al. Inhibition of cyclin D1 expression by androgen receptor in breast cancer cells--identification of a novel androgen response element. Nucleic Acids Res. 2010, 38, 5351–5365. [Google Scholar] [CrossRef]
- Casaburi, I.; Cesario, M.G.; Donà, A.; Rizza, P.; Aquila, S.; Avena, P.; Lanzino, M.; Pellegrino, M.; Vivacqua, A.; Tucci, P.; et al. Androgens downregulate miR-21 expression in breast cancer cells underlining the protective role of androgen receptor. Oncotarget 2016, 7, 12651–12661. [Google Scholar] [CrossRef]
- Rizza, P.; Barone, I.; Zito, D.; Giordano, F.; Lanzino, M.; De Amicis, F.; Mauro, L.; Sisci, D.; Catalano, S.; Wright, K.D.; et al. Estrogen receptor beta as a novel target of androgen receptor action in breast cancer cell lines. Breast Cancer Res. 2014, 16, R21. [Google Scholar] [CrossRef]
- Vasiliou, S.K.; Filippou, P.S.; Clotet-Freixas, S.; Soosaipillai, A.; Batruch, I.; Viktor Tsianos, F.; Konvalinka, A.; Diamandis, E.P. Transcriptome profiling and proteomic validation reveals targets of the androgen receptor signaling in the BT-474 breast cancer cell line. Clin. Proteom. 2022, 19, 14. [Google Scholar] [CrossRef]
- Hickey, T.E.; Dwyer, A.R.; Tilley, W.D. Arming androgen receptors to oppose oncogenic estrogen receptor activity in breast cancer. Br. J. Cancer 2021, 125, 1599–1601. [Google Scholar] [CrossRef]
- Greeve, M.A.; Allan, R.K.; Harvey, J.M.; Bentel, J.M. Inhibition of MCF-7 breast cancer cell proliferation by 5alpha-dihydrotestosterone; a role for p21(Cip1/Waf1). J. Mol. Endocrinol. 2004, 32, 793–810. [Google Scholar] [CrossRef]
- Wang, Y.; Romigh, T.; He, X.; Tan, M.-H.; Orloff, M.S.; Silverman, R.H.; Heston, W.D.; Eng, C. Differential regulation of PTEN expression by androgen receptor in prostate and breast cancers. Oncogene 2011, 30, 4327–4338. [Google Scholar] [CrossRef]
- Thompson, C.B. Apoptosis in the pathogenesis and treatment of disease. Science 1995, 267, 1456–1462. [Google Scholar] [CrossRef] [PubMed]
- Fernald, K.; Kurokawa, M. Evading apoptosis in cancer. Trends Cell Biol. 2013, 23, 620–633. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.C.; O’Reilly, L.A.; Strasser, A.; Cory, S. The anti-apoptosis function of Bcl-2 can be genetically separated from its inhibitory effect on cell cycle entry. EMBO J. 1997, 16, 4628–4638. [Google Scholar] [CrossRef]
- Yeretssian, G.; Correa, R.G.; Doiron, K.; Fitzgerald, P.; Dillon, C.P.; Green, D.R.; Reed, J.C.; Saleh, M. Non-apoptotic role of BID in inflammation and innate immunity. Nature 2011, 474, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Danial, N.N.; Gramm, C.F.; Scorrano, L.; Zhang, C.Y.; Krauss, S.; Ranger, A.M.; Datta, S.R.; Greenberg, M.E.; Licklider, L.J.; Lowell, B.B.; et al. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature 2003, 424, 952–956. [Google Scholar] [CrossRef]
- Lowman, X.H.; McDonnell, M.A.; Kosloske, A.; Odumade, O.A.; Jenness, C.; Karim, C.B.; Jemmerson, R.; Kelekar, A. The proapoptotic function of Noxa in human leukemia cells is regulated by the kinase Cdk5 and by glucose. Mol. Cell 2010, 40, 823–833. [Google Scholar] [CrossRef]
- Mann, J.; Githaka, J.M.; Buckland, T.W.; Yang, N.; Montpetit, R.; Patel, N.; Li, L.; Baksh, S.; Godbout, R.; Lemieux, H.; et al. Non-canonical BAD activity regulates breast cancer cell and tumor growth via 14-3-3 binding and mitochondrial metabolism. Oncogene 2019, 38, 3325–3339. [Google Scholar] [CrossRef]
- Fernando, R.; Foster, J.S.; Bible, A.; Strom, A.; Pestell, R.G.; Rao, M.; Saxton, A.; Baek, S.J.; Yamaguchi, K.; Donnell, R.; et al. Breast cancer cell proliferation is inhibited by BAD: Regulation of cyclin D1. J. Biol. Chem. 2007, 282, 28864–28873. [Google Scholar] [CrossRef]
- Cekanova, M.; Fernando, R.I.; Siriwardhana, N.; Sukhthankar, M.; de la Parra, C.; Woraratphoka, J.; Malone, C.; Ström, A.; Baek, S.J.; Wade, P.A.; et al. BCL-2 family protein, BAD is down-regulated in breast cancer and inhibits cell invasion. Exp. Cell Res. 2015, 331, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zha, J.; Harada, H.; Yang, E.; Jockel, J.; Korsmeyer, S.J. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L). Cell 1996, 87, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.R.; Ranger, A.M.; Lin, M.Z.; Sturgill, J.F.; Ma, Y.C.; Cowan, C.W.; Dikkes, P.; Korsmeyer, S.J.; Greenberg, M.E. Survival factor-mediated BAD phosphorylation raises the mitochondrial threshold for apoptosis. Dev. Cell 2002, 3, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Fu, J.; Lu, S.X.; Liu, L.L.; Luo, R.Z.; Yun, J.P.; Zhang, C.Z. Decrease of Bcl-xL/Bcl-2-associated death promoter in hepatocellular carcinoma indicates poor prognosis. Am. J. Cancer Res. 2015, 5, 1805–1813. [Google Scholar]
- Sinicrope, F.A.; Rego, R.L.; Foster, N.R.; Thibodeau, S.N.; Alberts, S.R.; Windschitl, H.E.; Sargent, D.J. Proapoptotic BAD and Bid protein expression predict survival in stages II and III colon cancers. Clin. Cancer Res. 2008, 14, 4128–4133. [Google Scholar] [CrossRef]
- Yancey, D.; Nelson, K.C.; Baiz, D.; Hassan, S.; Flores, A.; Pullikuth, A.; Karpova, Y.; Axanova, L.; Moore, V.; Sui, G.; et al. BAD dephosphorylation and decreased expression of MCL-1 induce rapid apoptosis in prostate cancer cells. PLoS ONE 2013, 8, e74561. [Google Scholar] [CrossRef]
- Craik, A.C.; Veldhoen, R.A.; Czernick, M.; Buckland, T.W.; Kyselytzia, K.; Ghosh, S.; Lai, R.; Damaraju, S.; Underhill, D.A.; Mackey, J.R.; et al. The BH3-only protein BAD confers breast cancer taxane sensitivity through a nonapoptotic mechanism. Oncogene 2010, 29, 5381–5391. [Google Scholar] [CrossRef]
- Cannings, E.; Kirkegaard, T.; Tovey, S.M.; Dunne, B.; Cooke, T.G.; Bartlett, J.M.S. BAD expression predicts outcome in patients treated with tamoxifen. Breast Cancer Res. Treat. 2007, 102, 173–179. [Google Scholar] [CrossRef]
- Yu, B.; Sun, X.; Shen, H.-Y.; Gao, F.; Fan, Y.-M.; Sun, Z.-J. Expression of the apoptosis-related genes BCL-2 and BAD in human breast carcinoma and their associated relationship with chemosensitivity. J. Exp. Clin. Cancer Res. 2010, 29, 107. [Google Scholar] [CrossRef]
- Al-Bazz, Y.O.; Underwood, J.C.; Brown, B.L.; Dobson, P.R. Prognostic significance of Akt, phospho-Akt and BAD expression in primary breast cancer. Eur. J. Cancer 2009, 45, 694–704. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, D.; Chen, B.; Zeng, J.; Wang, L.; Zhang, S.; Mo, X.; Li, W. Loss of BAD expression confers poor prognosis in non-small cell lung cancer. Med. Oncol. 2012, 29, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Troutaud, D.; Petit, B.; Bellanger, C.; Marin, B.; Gourin-Chaury, M.-P.; Petit, D.; Olivrie, A.; Feuillard, J.; Jauberteau, M.-O.; Bordessoule, D. Prognostic significance of BAD and AIF apoptotic pathways in diffuse large B-cell lymphoma. Clin. Lymphoma Myeloma Leuk. 2010, 10, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Kitada, S.; Krajewska, M.; Zhang, X.; Scudiero, D.; Zapata, J.M.; Wang, H.G.; Shabaik, A.; Tudor, G.; Krajewski, S.; Myers, T.G.; et al. Expression and location of pro-apoptotic Bcl-2 family protein BAD in normal human tissues and tumor cell lines. Am. J. Pathol. 1998, 152, 51–61. [Google Scholar] [PubMed]
- Gyorffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef]
- Landes, T.; Martinou, J.C. Mitochondrial outer membrane permeabilization during apoptosis: The role of mitochondrial fission. Biochim. Biophys. Acta 2011, 1813, 540–545. [Google Scholar] [CrossRef]
- Papadopoulou, N.; Papakonstanti, E.A.; Kallergi, G.; Alevizopoulos, K.; Stournaras, C. Membrane androgen receptor activation in prostate and breast tumor cells: Molecular signaling and clinical impact. IUBMB Life 2009, 61, 56–61. [Google Scholar] [CrossRef]
- Gu, S.; Papadopoulou, N.; Gehring, E.-M.; Nasir, O.; Dimas, K.; Bhavsar, S.K.; Föller, M.; Alevizopoulos, K.; Lang, F.; Stournaras, C. Functional membrane androgen receptors in colon tumors trigger pro-apoptotic responses in vitro and reduce drastically tumor incidence in vivo. Mol. Cancer 2009, 8, 114. [Google Scholar] [CrossRef]
- Kampa, M.; Nifli, A.-P.; Charalampopoulos, I.; Alexaki, V.-I.; Theodoropoulos, P.A.; Stathopoulos, E.N.; Gravanis, A.; Castanas, E. Opposing effects of estradiol- and testosterone-membrane binding sites on T47D breast cancer cell apoptosis. Exp. Cell Res. 2005, 307, 41–51. [Google Scholar] [CrossRef]
- Liu, G.; Honisch, S.; Liu, G.; Schmidt, S.; Pantelakos, S.; Alkahtani, S.; Toulany, M.; Lang, F.; Stournaras, C. Inhibition of SGK1 enhances mAR-induced apoptosis in MCF-7 breast cancer cells. Cancer Biol. Ther. 2015, 16, 52–59. [Google Scholar] [CrossRef]
- Pelekanou, V.; Notas, G.; Sanidas, E.; Tsapis, A.; Castanas, E.; Kampa, M. Testosterone membrane-initiated action in breast cancer cells: Interaction with the androgen signaling pathway and EPOR. Mol. Oncol. 2010, 4, 135–149. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Fouad, Y.A.; Aanei, C. Revisiting the hallmarks of cancer. Am. J. Cancer Res. 2017, 7, 1016–1036. [Google Scholar] [PubMed]
- Lopez, A.; Reyna, D.E.; Gitego, N.; Kopp, F.; Zhou, H.; Miranda-Roman, M.A.; Nordstrøm, L.U.; Narayanagari, S.-R.; Chi, P.; Vilar, E.; et al. Co-targeting of BAX and BCL-XL proteins broadly overcomes resistance to apoptosis in cancer. Nat. Commun. 2022, 13, 1199. [Google Scholar] [CrossRef]
- Frenzel, A.; Grespi, F.; Chmelewskij, W.; Villunger, A. Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis 2009, 14, 584–596. [Google Scholar] [CrossRef] [PubMed]
- Ranger, A.M.; Zha, J.; Harada, H.; Datta, S.R.; Danial, N.N.; Gilmore, A.P.; Kutok, J.L.; Le Beau, M.M.; Greenberg, M.E.; Korsmeyer, S.J. BAD-deficient mice develop diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. USA 2003, 100, 9324–9329. [Google Scholar] [CrossRef]
- Canevari, R.A.; Marchi, F.A.; Domingues, M.A.C.; de Andrade, V.P.; Caldeira, J.R.F.; Verjovski-Almeida, S.; Rogatto, S.R.; Reis, E.M. Identification of novel biomarkers associated with poor patient outcomes in invasive breast carcinoma. Tumour Biol. 2016, 37, 13855–13870. [Google Scholar] [CrossRef]
- Popgeorgiev, N.; Jabbour, L.; Gillet, G. Subcellular Localization and Dynamics of the Bcl-2 Family of Proteins. Front. Cell Dev. Biol. 2018, 6, 13. [Google Scholar] [CrossRef]
- Dumitru, R.; Gama, V.; Fagan, B.M.; Bower, J.J.; Swahari, V.; Pevny, L.H.; Deshmukh, M. Human embryonic stem cells have constitutively active Bax at the Golgi and are primed to undergo rapid apoptosis. Mol. Cell 2012, 46, 573–583. [Google Scholar] [CrossRef]
- Hosoi, K.I.; Miyata, N.; Mukai, S.; Furuki, S.; Okumoto, K.; Cheng, E.H.; Fujiki, Y. The VDAC2-BAK axis regulates peroxisomal membrane permeability. J. Cell Biol. 2017, 216, 709–722. [Google Scholar] [CrossRef]
- Kamer, I.; Sarig, R.; Zaltsman, Y.; Niv, H.; Oberkovitz, G.; Regev, L.; Haimovich, G.; Lerenthal, Y.; Marcellus, R.C.; Gross, A. Proapoptotic BID is an ATM effector in the DNA-damage response. Cell 2005, 122, 593–603. [Google Scholar] [CrossRef]
- Bonneau, B.; Prudent, J.; Popgeorgiev, N.; Gillet, G. Non-apoptotic roles of Bcl-2 family: The calcium connection. Biochim. Biophys. Acta 2013, 1833, 1755–1765. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Chen, Z.; Tang, L.H.; Fang, Y.; Shin, S.J.; Panarelli, N.C.; Chen, Y.T.; Li, Y.; Jiang, X.; Du, Y.N. Bcl-xL promotes metastasis independent of its anti-apoptotic activity. Nat. Commun. 2016, 7, 10384. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, N.; Verma, A.; Bivens, C.B.; Schwartz, Z.; Boyan, B.D. Rapid steroid hormone actions via membrane receptors. Biochim. Biophys. Acta 2016, 1863, 2289–2298. [Google Scholar] [CrossRef] [PubMed]
- Kousteni, S.; Bellido, T.; Plotkin, L.I.; O’Brien, C.A.; Bodenner, D.L.; Han, L.; Han, K.; DiGregorio, G.B.; Katzenellenbogen, J.A.; Katzenellenbogen, B.S.; et al. Nongenotropic, sex-nonspecific signaling through the estrogen or androgen receptors: Dissociation from transcriptional activity. Cell 2001, 104, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.; Han, L.; O’brien, C.A.; Kousteni, S.; Manolagas, S.C. Classical genotropic versus kinase-initiated regulation of gene transcription by the estrogen receptor alpha. Endocrinology 2006, 147, 1986–1996. [Google Scholar] [CrossRef]
- Grazzini, E.; Guillon, G.; Mouillac, B.; Zingg, H.H. Inhibition of oxytocin receptor function by direct binding of progesterone. Nature 1998, 392, 509–512. [Google Scholar] [CrossRef]
- Vertino, A.M.; Bula, C.M.; Chen, J.R.; Almeida, M.; Han, L.; Bellido, T.; Kousteni, S.; Norman, A.W.; Manolagas, S.C. Nongenotropic, anti-apoptotic signaling of 1alpha,25(OH)2-vitamin D3 and analogs through the ligand binding domain of the vitamin D receptor in osteoblasts and osteocytes. Mediation by Src, phosphatidylinositol 3-, and JNK kinases. J. Biol. Chem. 2005, 280, 14130–14137. [Google Scholar] [CrossRef]
- Hammes, S.R.; Levin, E.R. Extranuclear steroid receptors: Nature and actions. Endocr. Rev. 2007, 28, 726–741. [Google Scholar] [CrossRef]
- Pedram, A.; Razandi, M.; Sainson, R.C.; Kim, J.K.; Hughes, C.C.; Levin, E.R. A conserved mechanism for steroid receptor translocation to the plasma membrane. J. Biol. Chem. 2007, 282, 22278–22288. [Google Scholar] [CrossRef]
- Danial, N.N.; Walensky, L.D.; Zhang, C.-Y.; Choi, C.S.; Fisher, J.K.; Molina, A.J.; Datta, S.R.; Pitter, K.L.; Bird, G.H.; Wikstrom, J.D.; et al. Dual role of proapoptotic BAD in insulin secretion and beta cell survival. Nat. Med. 2008, 14, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Cassina, A.; Garcia-Haro, L.; Choi, C.S.; Osundiji, M.A.; Lane, E.A.; Huang, H.; Yildirim, M.A.; Szlyk, B.; Fisher, J.K.; Polak, K.; et al. Regulation of hepatic energy metabolism and gluconeogenesis by BAD. Cell Metab. 2014, 19, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.-C.; Leach, K.M.; Hardwick, J.M. Mitochondrial death pathways in yeast and mammalian cells. Biochim. Biophys. Acta 2008, 1783, 1272–1279. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.G.; Nam, Y.; Shin, K.J.; Yoon, S.; Park, W.S.; Joung, J.Y.; Seo, J.K.; Jang, J.; Lee, S.; Nam, D.; et al. Androgen-induced expression of DRP1 regulates mitochondrial metabolic reprogramming in prostate cancer. Cancer Lett. 2020, 471, 72–87. [Google Scholar] [CrossRef]
- Lanzino, M.; Garofalo, C.; Morelli, C.; Le Pera, M.; Casaburi, I.; McPhaul, M.J.; Surmacz, E.; Andò, S.; Sisci, D. Insulin receptor substrate 1 modulates the transcriptional activity and the stability of androgen receptor in breast cancer cells. Breast Cancer Res. Treat. 2009, 115, 297–306. [Google Scholar] [CrossRef][Green Version]

| Cell Line | Media | Supplements |
|---|---|---|
| MCF-7 | DMEM (1X) | 5% FBS, 1% Pen/Strep |
| T47D | RPMI 1640 | 10% FBS, 1% Pen/Strep, 0.2 mM L-Glutamine |
| SKBR3 | RPMI 1640 phenol red-free | 10% FBS, 1% Pen/Strep, 0.2 mM L-Glutamine, 0.2 Units/mL insulin |
| Gene | Forward | Reverse |
|---|---|---|
| BAD | 5′-GGAGGATGAGTGACGAGTTTGTG-3′ | 5′-GGGTGGAGTTTCGGGATGT-3′ |
| DRP1 | 5′-GAGAGGTAGATCCAGATGGT-3′ | 5′-CCCTTCCCATCAATACATCC-3′ |
| 18S | 5′-CGGCGACGACCCATTCGAAC-3′ | 5′-GAATCGAACCCTGATTCCCCGTC-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morelli, C.; Chiodo, C.; Nocito, M.C.; Cormace, A.; Catalano, S.; Sisci, D.; Sirianni, R.; Casaburi, I.; Andò, S.; Lanzino, M. Androgens Modulate Bcl-2 Agonist of Cell Death (BAD) Expression and Function in Breast Cancer Cells. Int. J. Mol. Sci. 2023, 24, 13464. https://doi.org/10.3390/ijms241713464
Morelli C, Chiodo C, Nocito MC, Cormace A, Catalano S, Sisci D, Sirianni R, Casaburi I, Andò S, Lanzino M. Androgens Modulate Bcl-2 Agonist of Cell Death (BAD) Expression and Function in Breast Cancer Cells. International Journal of Molecular Sciences. 2023; 24(17):13464. https://doi.org/10.3390/ijms241713464
Chicago/Turabian StyleMorelli, Catia, Chiara Chiodo, Marta Claudia Nocito, Alessandro Cormace, Stefania Catalano, Diego Sisci, Rosa Sirianni, Ivan Casaburi, Sebastiano Andò, and Marilena Lanzino. 2023. "Androgens Modulate Bcl-2 Agonist of Cell Death (BAD) Expression and Function in Breast Cancer Cells" International Journal of Molecular Sciences 24, no. 17: 13464. https://doi.org/10.3390/ijms241713464
APA StyleMorelli, C., Chiodo, C., Nocito, M. C., Cormace, A., Catalano, S., Sisci, D., Sirianni, R., Casaburi, I., Andò, S., & Lanzino, M. (2023). Androgens Modulate Bcl-2 Agonist of Cell Death (BAD) Expression and Function in Breast Cancer Cells. International Journal of Molecular Sciences, 24(17), 13464. https://doi.org/10.3390/ijms241713464








