Genetic Architecture of Ischaemic Strokes after COVID-19 Shows Similarities with Large Vessel Strokes
Abstract
:1. Introduction
2. Results
2.1. Local Genetic Covariance Estimation
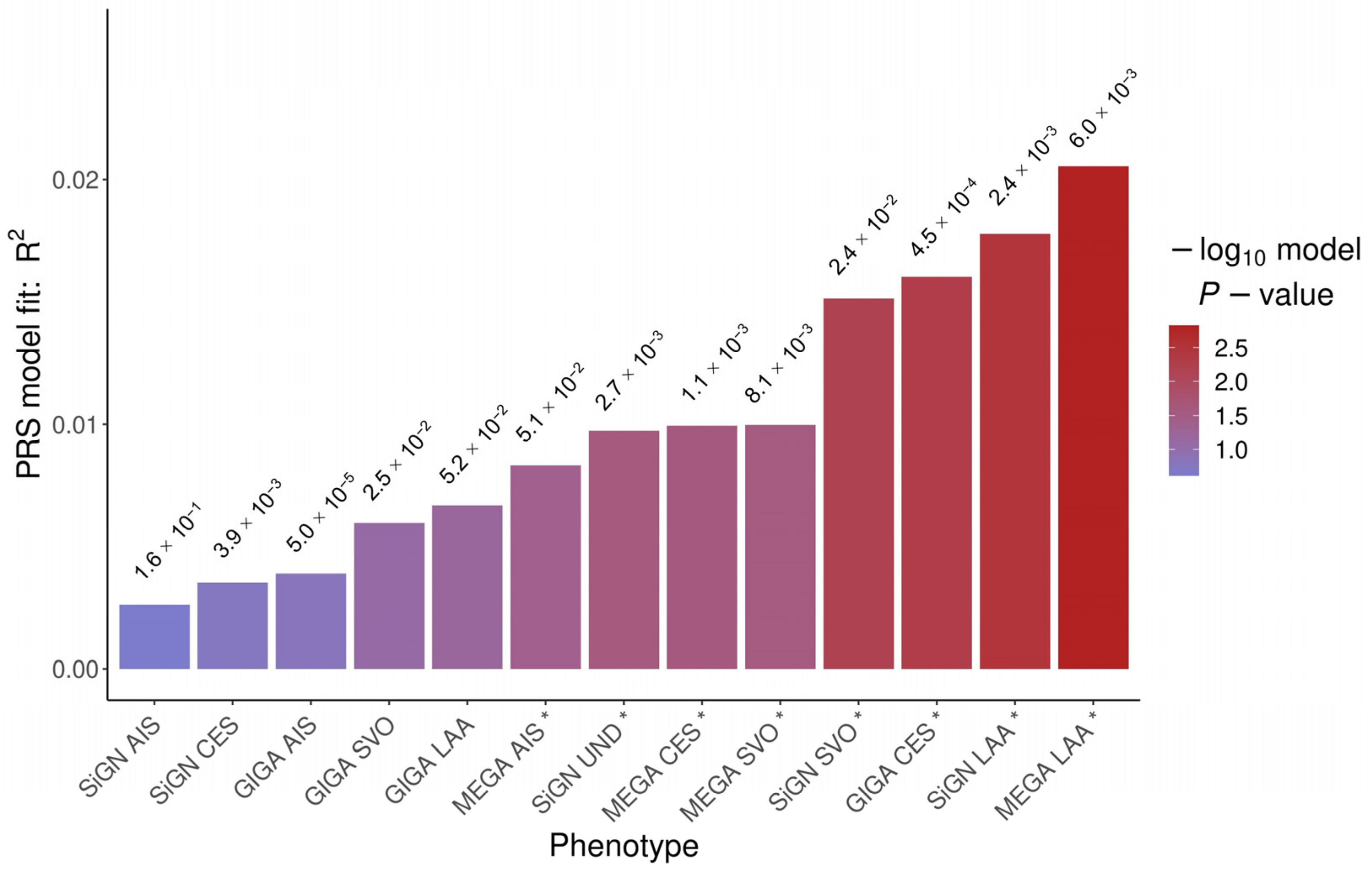
2.2. Polygenic Risk Score
3. Discussion
4. Materials and Methods
4.1. Cohorts’ Description
4.2. Genotyping
4.3. Genotyped Data Quality Controls
4.4. Genome-Wide Association Analysis
4.5. Local Genetic Covariance Estimation
4.6. Polygenic Risk Score
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klok, F.A.; Kruip, M.J.H.A.; van der Meer, N.J.M.; Arbous, M.S.; Gommers, D.; Kant, K.M.; Kaptein, F.H.J.; van Paassen, J.; Stals, M.A.M.; Huisman, M.V.; et al. Confirmation of the High Cumulative Incidence of Thrombotic Complications in Critically Ill ICU Patients with COVID-19: An Updated Analysis. Thromb. Res. 2020, 191, 148–150. [Google Scholar] [CrossRef]
- Mao, L.; Jin, H.; Wang, M.; Hu, Y.; Chen, S.; He, Q.; Chang, J.; Hong, C.; Zhou, Y.; Wang, D.; et al. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020, 77, 683–690. [Google Scholar] [CrossRef]
- Rothstein, A.; Oldridge, O.; Schwennesen, H.; Do, D.; Cucchiara, B.L. Acute Cerebrovascular Events in Hospitalized COVID-19 Patients. Stroke 2020, 51, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Lodigiani, C.; Iapichino, G.; Carenzo, L.; Cecconi, M.; Ferrazzi, P.; Sebastian, T.; Kucher, N.; Studt, J.D.; Sacco, C.; Alexia, B.; et al. Venous and Arterial Thromboembolic Complications in COVID-19 Patients Admitted to an Academic Hospital in Milan, Italy. Thromb. Res. 2020, 191, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Yaghi, S.; Ishida, K.; Torres, J.; Mac Grory, B.; Raz, E.; Humbert, K.; Henninger, N.; Trivedi, T.; Lillemoe, K.; Alam, S.; et al. SARS-CoV-2 and Stroke in a New York Healthcare System. Stroke 2020, 51, 2002–2011. [Google Scholar] [CrossRef] [PubMed]
- Feigin, V.L.; Brainin, M.; Norrving, B.; Martins, S.; Sacco, R.L.; Hacke, W.; Fisher, M.; Pandian, J.; Lindsay, P. World Stroke Organization (WSO): Global Stroke Fact Sheet 2022. Int. J. Stroke 2022, 17, 18–29. [Google Scholar] [CrossRef]
- Stamm, B.; Huang, D.; Royan, R.; Lee, J.; Marquez, J.; Desai, M. Pathomechanisms and Treatment Implications for Stroke in COVID-19: A Review of the Literature. Life 2022, 12, 207. [Google Scholar] [CrossRef]
- Alotaibi, B.A.; Aldali, J.A.; Aldali, H.J.; Meo, S.A.; Alasiri, G.A.; Elsokkary, E.M.; Alotaibi, N.D.; Alotaibi, F. The Risk Factors for Acute Cerebrovascular Accident (Stroke) in Patients with Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2). Viruses 2023, 15, 1140. [Google Scholar] [CrossRef]
- Merkler, A.E.; Parikh, N.S.; Mir, S.; Gupta, A.; Kamel, H.; Lin, E.; Lantos, J.; Schenck, E.J.; Goyal, P.; Bruce, S.S.; et al. Risk of Ischemic Stroke in Patients With Coronavirus Disease 2019 (COVID-19) vs Patients With Influenza. JAMA Neurol. 2020, 77, 1366–1372. [Google Scholar] [CrossRef]
- Marto, J.P.; Strambo, D.; Ntaios, G.; Nguyen, T.N.; Herzig, R.; Czlonkowska, A.; Demeestere, J.; Mansour, O.Y.; Salerno, A.; Wegener, S.; et al. Safety and Outcome of Revascularization Treatment in Patients With Acute Ischemic Stroke and COVID-19: The Global COVID-19 Stroke Registry. Neurology 2023, 100, E739–E750. [Google Scholar] [CrossRef]
- Zuin, M.; Mazzitelli, M.; Rigatelli, G.; Bilato, C.; Cattelan, A.M. Risk of Ischemic Stroke in Patients Recovered from COVID-19 Infection: A Systematic Review and Meta-Analysis. Eur. Stroke J. 2023. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.W.; Park, S.H.; Kim, N.; Kim, W.J.; Park, J.H.; Ko, Y.; Yang, M.H.; Jang, M.S.; Han, M.K.; Jung, C.; et al. Trial of ORG 10172 in Acute Stroke Treatment (TOAST) Classification and Vascular Territory of Ischemic Stroke Lesions Diagnosed by Diffusion-Weighted Imaging. J. Am. Heart Assoc. 2014, 3, e001119. [Google Scholar] [CrossRef] [PubMed]
- Parodi, L.; Myserlis, E.P.; Chung, J.; Georgakis, M.K.; Mayerhofer, E.; Henry, J.; Montgomery, B.E.; Moy, M.; Xu, H.; Malik, R.; et al. Shared Genetic Background between SARS-CoV-2 Infection and Large Artery Stroke. Int. J. Stroke 2022, 17, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Zuber, V.; Cameron, A.; Myserlis, E.P.; Bottolo, L.; Fernandez-Cadenas, I.; Burgess, S.; Anderson, C.D.; Dawson, J.; Gill, D. Leveraging Genetic Data to Elucidate the Relationship Between COVID-19 and Ischemic Stroke. J. Am. Heart Assoc. 2021, 10, e022433. [Google Scholar] [CrossRef] [PubMed]
- Martí-Fàbregas, J.; Guisado-Alonso, D.; Delgado-Mederos, R.; Martínez-Domeño, A.; Prats-Sánchez, L.; Guasch-Jiménez, M.; Cardona, P.; Núñez-Guillén, A.; Requena, M.; Rubiera, M.; et al. Impact of COVID-19 Infection on the Outcome of Patients With Ischemic Stroke. Stroke 2021, 52, 3908. [Google Scholar] [CrossRef]
- Ramos-Araque, M.E.; Siegler, J.E.; Ribo, M.; Requena, M.; López, C.; de Lera, M.; Arenillas, J.F.; Pérez, I.H.; Gómez-Vicente, B.; Talavera, B.; et al. Stroke Etiologies in Patients with COVID-19: The SVIN COVID-19 Multinational Registry. BMC Neurol. 2021, 21, 43. [Google Scholar] [CrossRef]
- Bahouth, M.N.; Venkatesan, A. Acute Viral Illnesses and Ischemic Stroke: Pathophysiological Considerations in the Era of the COVID-19 Pandemic. Stroke 2021, 52, 1885. [Google Scholar] [CrossRef]
- Słomka, A.; Kowalewski, M.; Żekanowska, E. Coronavirus Disease 2019 (COVID–19): A Short Review on Hematological Manifestations. Pathogens 2020, 9, 493. [Google Scholar] [CrossRef]
- Sánchez, K.E.; Rosenberg, G.A. Shared Inflammatory Pathology of Stroke and COVID-19. Int. J. Mol. Sci. 2022, 23, 5150. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, X.; Chen, Z.; Duan, J.; Hashimoto, K.; Yang, L.; Liu, C.; Yang, C. Nervous System Involvement after Infection with COVID-19 and Other Coronaviruses. Brain Behav. Immun. 2020, 87, 18. [Google Scholar] [CrossRef]
- Rosand, J.; Mitchell, B.D.; Ay, H.; de Bakker, P.I.W.; Gwinn, K.; Kittner, S.J.; Lindgren, A.; Meschia, J.F.; Pulit, S.L.; Sudlow, C.L.M.; et al. Loci Associated with Ischaemic Stroke and Its Subtypes (SiGN): A Genome-Wide Association Study. Lancet Neurol. 2016, 15, 174–184. [Google Scholar] [CrossRef]
- Malik, R.; Rannikmäe, K.; Traylor, M.; Georgakis, M.K.; Sargurupremraj, M.; Markus, H.S.; Hopewell, J.C.; Debette, S.; Sudlow, C.L.M.; Dichgans, M. Genome-wide Meta-analysis Identifies 3 Novel Loci Associated with Stroke. Ann. Neurol. 2018, 84, 934. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Malik, R.; Hachiya, T.; Jürgenson, T.; Namba, S.; Posner, D.C.; Kamanu, F.K.; Koido, M.; le Grand, Q.; Shi, M.; et al. Stroke Genetics Informs Drug Discovery and Risk Prediction across Ancestries. Nature 2022, 611, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Machiela, M.J.; Chanock, S.J. LDlink: A Web-Based Application for Exploring Population-Specific Haplotype Structure and Linking Correlated Alleles of Possible Functional Variants. Bioinformatics 2015, 31, 3555–3557. [Google Scholar] [CrossRef]
- Altshuler, D.M.; Durbin, R.M.; Abecasis, G.R.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; Flicek, P.; Gabriel, S.B.; et al. An Integrated Map of Genetic Variation from 1,092 Human Genomes. Nature 2012, 491, 56–65. [Google Scholar] [CrossRef]
- Malik, R.; Chauhan, G.; Traylor, M.; Sargurupremraj, M.; Okada, Y.; Mishra, A.; Rutten-Jacobs, L.; Giese, A.K.; Van Der Laan, S.W.; Gretarsdottir, S.; et al. Multiancestry Genome-Wide Association Study of 520,000 Subjects Identifies 32 Loci Associated with Stroke and Stroke Subtypes. Nat. Genet. 2018, 50, 524–537. [Google Scholar] [CrossRef]
- Pruim, R.J.; Welch, R.P.; Sanna, S.; Teslovich, T.M.; Chines, P.S.; Gliedt, T.P.; Boehnke, M.; Abecasis, G.R.; Willer, C.J.; Frishman, D. LocusZoom: Regional Visualization of Genome-Wide Association Scan Results. Bioinformatics 2010, 26, 2336–2337. [Google Scholar] [CrossRef]
- Mountjoy, E.; Schmidt, E.M.; Carmona, M.; Schwartzentruber, J.; Peat, G.; Miranda, A.; Fumis, L.; Hayhurst, J.; Buniello, A.; Karim, M.A.; et al. An Open Approach to Systematically Prioritize Causal Variants and Genes at All Published Human GWAS Trait-Associated Loci. Nat. Genet. 2021, 53, 1527–1533. [Google Scholar] [CrossRef]
- Isabel, C.; Calvet, D.; Mas, J.L. Stroke Prevention. Presse Med. 2016, 45, e457–e471. [Google Scholar] [CrossRef]
- Nannoni, S.; de Groot, R.; Bell, S.; Markus, H.S. Stroke in COVID-19: A Systematic Review and Meta-Analysis. Int. J. Stroke 2021, 16, 137–149. [Google Scholar] [CrossRef]
- Franco, D.; Sedmera, D.; Lozano-Velasco, E. Multiple Roles of Pitx2 in Cardiac Development and Disease. J. Cardiovasc. Dev. Dis. 2017, 4, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Hu, X.; Hao, J.; Guo, L.; Zhang, W.; Liu, J.; Jin, T.; Gao, D.; Zhi, J. Effect of PITX2 Genetic Variants on the Susceptibility to Stroke in the Chinese Han Population. Infect. Genet. Evol. 2022, 98, 105201. [Google Scholar] [CrossRef] [PubMed]
- Zou, R.; Zhang, D.; Lv, L.; Shi, W.; Song, Z.; Yi, B.; Lai, B.; Chen, Q.; Yang, S.; Hua, P. Bioinformatic Gene Analysis for Potential Biomarkers and Therapeutic Targets of Atrial Fibrillation-Related Stroke. J. Transl. Med. 2019, 17, 45. [Google Scholar] [CrossRef]
- Steimle, J.D.; Grisanti Canozo, F.J.; Park, M.; Kadow, Z.A.; Samee, M.A.H.; Martin, J.F. Decoding the PITX2-Controlled Genetic Network in Atrial Fibrillation. JCI Insight 2022, 7, e158895. [Google Scholar] [CrossRef]
- Bhatla, A.; Mayer, M.M.; Adusumalli, S.; Hyman, M.C.; Oh, E.; Tierney, A.; Moss, J.; Chahal, A.A.; Anesi, G.; Denduluri, S.; et al. COVID-19 and Cardiac Arrhythmias. Heart Rhythm. 2020, 17, 1439. [Google Scholar] [CrossRef]
- Gawałko, M.; Kapłon-Cieślicka, A.; Hohl, M.; Dobrev, D.; Linz, D. COVID-19 Associated Atrial Fibrillation: Incidence, Putative Mechanisms and Potential Clinical Implications. Int. J. Cardiol. Heart Vasc. 2020, 30, 100631. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Pedersen, L.C. Anticoagulant Heparan Sulfate: Structural Specificity and Biosynthesis. Appl. Microbiol. Biotechnol. 2007, 74, 263. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.K.; Price, D.J.; Pratt, T. FGF8 Morphogen Gradients Are Differentially Regulated by Heparan Sulphotransferases Hs2st and Hs6st1 in the Developing Brain. Biol. Open 2017, 6, 1933. [Google Scholar] [CrossRef]
- Liaqat, K.; Hussain, S.; Bilal, M.; Nasir, A.; Acharya, A.; Ali, R.H.; Nawaz, S.; Umair, M.; Schrauwen, I.; Ahmad, W.; et al. Further Evidence of Involvement of TMEM132E in Autosomal Recessive Nonsyndromic Hearing Impairment. J. Hum. Genet. 2020, 65, 187. [Google Scholar] [CrossRef]
- Li, J.; Zhao, X.; Xin, Q.; Shan, S.; Jiang, B.; Jin, Y.; Yuan, H.; Dai, P.; Xiao, R.; Zhang, Q.; et al. Whole-Exome Sequencing Identifies a Variant in TMEM132E Causing Autosomal-Recessive Nonsyndromic Hearing Loss DFNB99. Hum. Mutat. 2015, 36, 98–105. [Google Scholar] [CrossRef]
- Wang, Y.; Herzig, G.; Molano, C.; Liu, A. Differential Expression of the Tmem132 Family Genes in the Developing Mouse Nervous System. Gene Expr. Patterns 2022, 45, 119257. [Google Scholar] [CrossRef] [PubMed]
- Lane, J.M.; Liang, J.; Vlasac, I.; Anderson, S.G.; Bechtold, D.A.; Bowden, J.; Emsley, R.; Gill, S.; Little, M.A.; Luik, A.I.; et al. Genome-Wide Association Analyses of Sleep Disturbance Traits Identify New Loci and Highlight Shared Genetics with Neuropsychiatric and Metabolic Traits. Nat. Genet. 2017, 49, 274. [Google Scholar] [CrossRef] [PubMed]
- Sklar, P.; Smoller, J.W.; Fan, J.; Ferreira, M.A.R.; Perlis, R.H.; Chambert, K.; Nimgaonkar, V.L.; McQueen, M.B.; Faraone, S.V.; Kirby, A.; et al. Whole-Genome Association Study of Bipolar Disorder. Mol. Psychiatry 2008, 13, 558. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, N.O.; Buttenschøn, H.N.; Hedemand, A.; Dahl, H.A.; Kristensen, A.S.; Clementsen, B.; Woldbye, D.P.D.; Koefoed, P.; Erhardt, A.; Kruse, T.A.; et al. Are TMEM Genes Potential Candidate Genes for Panic Disorder? Psychiatr. Genet. 2014, 24, 37–41. [Google Scholar] [CrossRef]
- Sanchez-Pulido, L.; Ponting, C.P. TMEM132: An Ancient Architecture of Cohesin and Immunoglobulin Domains Define a New Family of Neural Adhesion Molecules. Bioinformatics 2018, 34, 721. [Google Scholar] [CrossRef]
- Roder, K.; Kabakov, A.; Moshal, K.S.; Murphy, K.R.; Xie, A.; Dudley, S.; Turan, N.N.; Lu, Y.; MacRae, C.A.; Koren, G. Trafficking of the Human Ether-a-Go-Go-Related Gene (HERG) Potassium Channel Is Regulated by the Ubiquitin Ligase Rififylin (RFFL). J. Biol. Chem. 2019, 294, 351–360. [Google Scholar] [CrossRef]
- Lu, J.; Auduong, L.; White, E.S.; Yue, X. Up-Regulation of Heparan Sulfate 6-O-Sulfation in Idiopathic Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 2014, 50, 106–114. [Google Scholar] [CrossRef]
- Taniguchi, S.; Ito, Y.; Kiritani, H.; Maruo, A.; Sakai, R.; Ono, Y.; Fukuda, R.; Okiyoneda, T. The Ubiquitin Ligase RNF34 Participates in the Peripheral Quality Control of CFTR (RNF34 Role in CFTR PeriQC). Front. Mol. Biosci. 2022, 9, 840649. [Google Scholar] [CrossRef]
- Mashima, R.; Okuyama, T.; Ohira, M. Physiology and Pathophysiology of Heparan Sulfate in Animal Models: Its Biosynthesis and Degradation. Int. J. Mol. Sci. 2022, 23, 1963. [Google Scholar] [CrossRef]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043. [Google Scholar] [CrossRef]
- Domingues-Montanari, S.; Fernández-Cadenas, I.; del Río-Espinola, A.; Mendioroz, M.; Fernandez-Morales, J.; Corbeto, N.; Delgado, P.; Ribó, M.; Rubiera, M.; Obach, V.; et al. KCNK17 Genetic Variants in Ischemic Stroke. Atherosclerosis 2010, 208, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Riba, I.; Jarca, C.I.; Mundet, X.; Tovar, J.L.; Orfila, F.; Nafría, C.; Raga, A.; Girona, A.; Fernández-Lara, P.; Castañé, X.; et al. Cognitive Assessment Protocol Design in the ISSYS (Investigating Silent Strokes in HYpertensives: A Magnetic Resonance Imaging Study). J. Neurol. Sci. 2012, 322, 79–81. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Cadenas, I.; Mendióroz, M.; Giralt, D.; Nafria, C.; Garcia, E.; Carrera, C.; Gallego-Fabrega, C.; Domingues-Montanari, S.; Delgado, P.; Ribó, M.; et al. GRECOS Project (Genotyping Recurrence Risk of Stroke): The Use of Genetics to Predict the Vascular Recurrence After Stroke. Stroke 2017, 48, 1147–1153. [Google Scholar] [CrossRef]
- COVID-19 Host Genetics Initiative. A First Update on Mapping the Human Genetic Architecture of COVID-19. Nature 2022, 608, E1–E10. [Google Scholar] [CrossRef] [PubMed]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank Resource with Deep Phenotyping and Genomic Data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Budde, M.; Anderson-Schmidt, H.; Gade, K.; Reich-Erkelenz, D.; Adorjan, K.; Kalman, J.L.; Senner, F.; Papiol, S.; Andlauer, T.F.M.; Comes, A.L.; et al. A Longitudinal Approach to Biological Psychiatric Research: The PsyCourse Study. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2019, 180, 89–102. [Google Scholar] [CrossRef]
- Das, S.; Forer, L.; Schönherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-Generation Genotype Imputation Service and Methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef]
- Jiang, L.; Zheng, Z.; Qi, T.; Kemper, K.E.; Wray, N.R.; Visscher, P.M.; Yang, J. A Resource-Efficient Tool for Mixed Model Association Analysis of Large-Scale Data. Nat. Genet. 2019, 51, 1749–1755. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, Q.; Ye, Y.; Huang, K.; Liu, W.; Wu, Y.; Zhong, X.; Li, B.; Yu, Z.; Travers, B.G.; et al. SUPERGNOVA: Local Genetic Correlation Analysis Reveals Heterogeneous Etiologic Sharing of Complex Traits. Genome Biol. 2021, 22, 262. [Google Scholar] [CrossRef]
- Choi, S.W.; O’Reilly, P.F. PRSice-2: Polygenic Risk Score software for biobank-scale data. GigaScience 2019, 8, 7. [Google Scholar] [CrossRef]
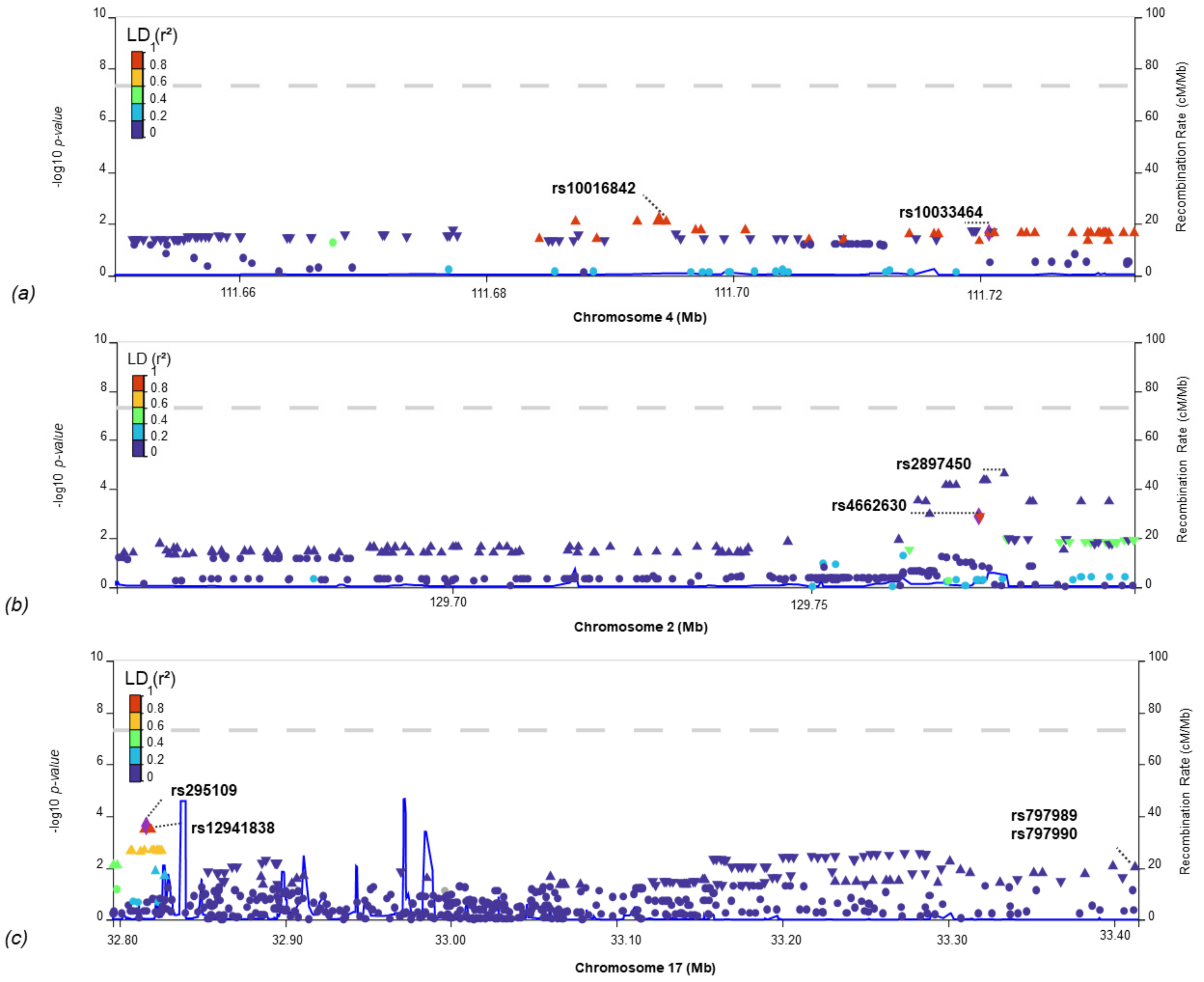
| Phenotype | Chr | Start | End | Corr | p-Value | SNVs |
|---|---|---|---|---|---|---|
| GIGA-LAA | 2 | 129,312,188 | 129,864,416 | 0.65 | 1.4 × 10−2 | 198 |
| MEGA-LAA | 0.63 | 3.1 × 10−2 | 200 | |||
| SiGN-LAA | 0.99 | 6.0 × 10−4 | 200 | |||
| GIGA-LAA | 17 | 32,677,947 | 33,614,452 | 0.92 | 9.4 × 10−3 | 442 |
| MEGA-LAA | 0.91 | 4.6 × 10−3 | 442 | |||
| SiGN-LAA | 0.88 | 3.6 × 10−2 | 442 | |||
| GIGA-CES | 4 | 109,980,374 | 112,204,254 | −0.87 | 2.1 × 10−3 | 697 |
| MEGA-CES | −0.87 | 4.0 × 10−3 | 697 | |||
| SiGN-CES | −0.86 | 6.8 × 10−3 | 697 |
| rsID | Chr | BP | A1 | A2 | Trait1 | Trait2 | p-Value. Trait1 | B.Trait1 | SE.Trait1 | p-value. Trait2 | B.Trait2 | SE.Trait2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10033464 | 4 | 111,720,761 | T | G | GIGA_CES | IS-COV | 1.4 × 10−2 | 0.12 | 0.03 | 2.3 × 10−2 | 0.04 | 0.02 |
| rs4662630 | 2 | 129,773,352 | C | T | SiGN_LAA | IS-COV | 2.7 × 10−2 | −0.08 | 0.04 | 1.3 × 10−3 | −0.04 | 0.01 |
| rs12941838 | 17 | 32,819,326 | A | G | GIGA_LAA | IS-COV | 1.8 × 10−2 | 0.07 | 0.03 | 3.6 × 10−4 | 0.05 | 0.01 |
| rs797989 | 17 | 33,414,758 | A | C | MEGA_LAA | IS-COV | 3.8 × 10−2 | 0.05 | 0.03 | 1.0 × 10−2 | 0.03 | 0.01 |
| PRSice-2 | PLINK 2.0 | |||||
|---|---|---|---|---|---|---|
| PRS | Threshold | Num_SNV | R2 | p-Value | R2 | p-Value |
| MEGA_LAA * | 6.0 × 10−3 | 4004 | 2.1 × 10−2 | 1.5 × 10−3 | 3.4 × 10−3 | 2.2 × 10−2 |
| SiGN_LAA * | 2.4 × 10−3 | 1305 | 1.8 × 10−2 | 3.2 × 10−3 | 4.7 × 10−3 | 7.1 × 10−3 |
| GIGA_CES | 4.5 × 10−4 | 582 | 1.6 × 10−2 | 5.0 × 10−3 | 1.3 × 10−3 | 1.6 × 10−1 |
| SiGN_SVO * | 2.4 × 10−2 | 9276 | 1.5 × 10−2 | 6.3 × 10−3 | 3.0 × 10−3 | 3.4 × 10−2 |
| MEGA_SVO | 8.1 × 10−3 | 5068 | 1.0 × 10−2 | 2.7 × 10−2 | 1.9 × 10−3 | 8.9 × 10−2 |
| MEGA_CES | 1.1 × 10−3 | 1089 | 9.9 × 10−3 | 2.5 × 10−2 | 9.7 × 10−4 | 2.2 × 10−1 |
| SiGN_UND | 2.7 × 10−3 | 1453 | 9.7 × 10−3 | 2.6 × 10−2 | 2.5 × 10−3 | 5.2 × 10−2 |
| MEGA_AIS | 5.1 × 10−2 | 19,869 | 8.3 × 10−3 | 4.0 × 10−2 | 1.9 × 10−3 | 9.1 × 10−2 |
| GIGA_LAA | 5.2 × 10−2 | 21,350 | 6.7 × 10−3 | 6.2 × 10−2 | 5.4 × 10−4 | 3.6 × 10−1 |
| GIGA_SVO | 2.5 × 10−2 | 12,262 | 6.0 × 10−3 | 8.2 × 10−2 | 2.1 × 10−3 | 7.4 × 10−2 |
| GIGA_AIS | 5.0 × 10−5 | 267 | 3.9 × 10−3 | 1.6 × 10−1 | 4.9 × 10−4 | 3.9 × 10−1 |
| SiGN_CES | 3.9 × 10−3 | 2067 | 3.5 × 10−3 | 1.8 × 10−1 | 3.1 × 10−6 | 9.5 × 10−1 |
| SiGN_AIS | 1.6 × 10−1 | 39,071 | 2.6 × 10−3 | 2.4 × 10−1 | 4.8 × 10−4 | 3.9 × 10−1 |
| Cases (N = 73) | Controls (N = 701) | Overall (N = 774) | |
|---|---|---|---|
| Age | |||
| Mean (SD) | 70.6 (13.0) | 65.6 (9.34) | 66.1 (9.84) |
| Median [Min, Max] | 73.0 [36.0, 90.0] | 67.0 [23.0, 90.0] | 67.0 [23.0, 90.0] |
| Sex | |||
| Female | 24 (32.9%) | 352 (50.2%) | 376 (48.6%) |
| Male | 49 (67.1%) | 349 (49.8%) | 398 (51.4%) |
| Cases (N = 73) | |||
| Severity of COVID-19 | |||
| Hospitalized ICU | 34 (46.6%) | ||
| Hospitalized not ICU | 13 (17.8%) | ||
| Not hospitalized | 14 (19.2%) | ||
| Missing | 12 (16.4%) | ||
| TOAST | |||
| CES | 13 (17.8%) | ||
| INF | 4 (5.5%) | ||
| LAA | 10 (13.7%) | ||
| SVO | 5 (6.8%) | ||
| UND | 29 (39.7%) | ||
| Missing | 12 (16.4%) | ||
| Project | Array | Controls | Cases |
|---|---|---|---|
| CONIC | Illumina® Human Core Exome chip | 189 | |
| ISSYS | Illumina® Human Core Exome chip | 274 | |
| GRECOS | Illumina® Human Core Exome chip | 189 | |
| INMUNGEN-CoV2 | Axiom Spain Biobank Array | 49 | 45 |
| UK Biobank | Applied Biosystems UK BiLEVE Axiom and Applied Biosystems UK Biobank Axiom Array | 12 | |
| BelCovid | Illumina’s Human OmniExpress BeadChips | 12 | |
| SPGRX | Infinium Global Screening Array-24 | 2 | |
| COMRI | Infinium Global Screening Array-24 v3.0 Kit | 2 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llucià-Carol, L.; Muiño, E.; Cullell, N.; Cárcel-Márquez, J.; Lledós, M.; Gallego-Fabrega, C.; Martin-Campos, J.; Martí-Fàbregas, J.; Aguilera-Simón, A.; Planas, A.M.; et al. Genetic Architecture of Ischaemic Strokes after COVID-19 Shows Similarities with Large Vessel Strokes. Int. J. Mol. Sci. 2023, 24, 13452. https://doi.org/10.3390/ijms241713452
Llucià-Carol L, Muiño E, Cullell N, Cárcel-Márquez J, Lledós M, Gallego-Fabrega C, Martin-Campos J, Martí-Fàbregas J, Aguilera-Simón A, Planas AM, et al. Genetic Architecture of Ischaemic Strokes after COVID-19 Shows Similarities with Large Vessel Strokes. International Journal of Molecular Sciences. 2023; 24(17):13452. https://doi.org/10.3390/ijms241713452
Chicago/Turabian StyleLlucià-Carol, Laia, Elena Muiño, Natalia Cullell, Jara Cárcel-Márquez, Miquel Lledós, Cristina Gallego-Fabrega, Jesús Martin-Campos, Joan Martí-Fàbregas, Ana Aguilera-Simón, Anna M. Planas, and et al. 2023. "Genetic Architecture of Ischaemic Strokes after COVID-19 Shows Similarities with Large Vessel Strokes" International Journal of Molecular Sciences 24, no. 17: 13452. https://doi.org/10.3390/ijms241713452
APA StyleLlucià-Carol, L., Muiño, E., Cullell, N., Cárcel-Márquez, J., Lledós, M., Gallego-Fabrega, C., Martin-Campos, J., Martí-Fàbregas, J., Aguilera-Simón, A., Planas, A. M., DeDiego, M. L., de Felipe Mimbrera, A., Masjuan, J., García-Madrona, S., Segura, T., González-Villar, E., Serrano-Heras, G., Domínguez Mayoral, A., Menéndez-Valladares, P., ... Fernández-Cadenas, I. (2023). Genetic Architecture of Ischaemic Strokes after COVID-19 Shows Similarities with Large Vessel Strokes. International Journal of Molecular Sciences, 24(17), 13452. https://doi.org/10.3390/ijms241713452










