Genetic and Epigenetic Features of Uveal Melanoma—An Overview and Clinical Implications
Abstract
1. Introduction
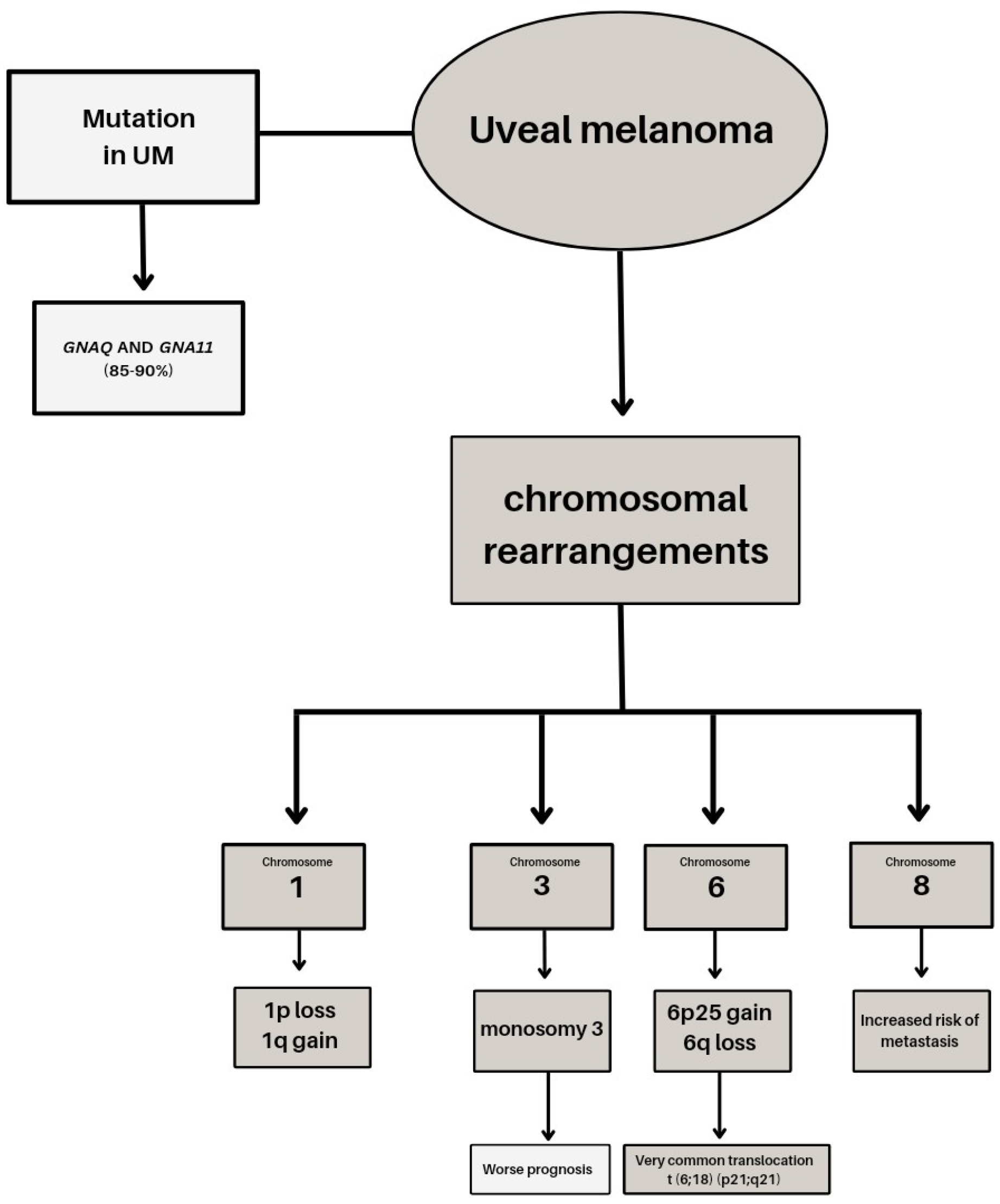
2. Genetic Basis of Uveal Melanoma
3. miRNA and Uveal Melanoma
| miRNAs | Sample Type | Role | Regulation Action | Target | Reference |
|---|---|---|---|---|---|
| miR-9 | cells with highly and poorly invasive potential | tumor suppressor | suppresses cell migration and invasion | downregulation NF-κB1 | [75] |
| miR-17-3p | UM patients tissues and cell lines | tumor suppressor | inhibiting cell proliferation, migration and invasion | decreased MDM2 expression | [76] |
| miR-20a | cells and melanocytes, and tumor tissue samples for patients | oncogenic | increases cell growth, migration and invasion activities | not validated | [89] |
| miR-21 | cell lines and tissue samples | oncogenic | promoted proliferation, migration, and invasion | inhibited expression p53 | [81] |
| miR-34a | cells lines and melanocytes | tumor suppressor | inhibiting cell proliferation, migration and invasion | downregulation c-Met; decreased LGR4 | [63,64] |
| miR-34b/c | cells and melanocytes from patients, the uveal stromal tissues | tumor suppressor | reduction in cell growth and migration | downregulation c-Met | [65] |
| miR-122 | cell lines and patients’ tissues | tumor suppressor | impaired cell proliferation and migration | reduced expression ADAM10 and c-Met | [66] |
| miR-124a | cell culture and tumor specimens | tumor suppressor | inhibited cell growth, migration and invasion | downregulation CDK4, CDK6, cyclin D2 and EZH2 | [69] |
| miR-137 | cells lines from patients | tumor suppressor | decrease cell growth | downregulated MITF and CDK6 | [67] |
| miR-142-3p | cells and uveal melanocytes from patients | tumor suppressor | inhibited cell proliferation, migration and invasiveness | reduced CDC25C, TGFβR1, GNAQ, WASL, and RAC1 | [77] |
| miR-144 | cell lines and patients tissues | tumor suppressor | impaired cell proliferation and migration | reduced expression ADAM10 and c-Met | [66] |
| miR-145 | cells with highly and low invasive potential and patients’ tissues | tumor suppressor | reduce proliferation, migration and invasion | downregulation CDC42 | [73] |
| miR-155 | cell culture and tumor specimens | oncogenic | increase cell growth and invasion | inhibited NDFIP1 | [82] |
| miR-181 | cell lines | oncogenic | promoted cell cycle progression | inhibited CTDSPL | [83] |
| miR-182 | cells lines from patients | tumor suppressor | decrease cell growth, migration, and invasiveness | downregulation MITF, BCL2 and cyclin D2 | [68] |
| miR-205 | cells with highly and low invasive potential and patients’ tissues | tumor suppressor | reduce proliferation, migration and invasion | downregulation CDC42 | [73] |
| miR-216a-5p | cell lines and human embryonic kidney cell line | tumor suppressor | reduce cell proliferation | inhibit HK2 expression | [78] |
| miR-222 | cell lines | oncogenic | increases proliferation and migration | decreased PI3K, Akt, MMP-9 | [87] |
| miR-224-5p | cell lines and human embryonic kidney cell line and patients’ tissues | tumor suppressor | inhibited capacities of proliferation, invasion and migration | decreased PIK3R3 and AKT3 | [79] |
| miR-296-3-p | choroidal tissues | tumor suppressor | inhibiting cell proliferation, cell cycle progression, migration | targeting of MMP-2 and MMP-9 in combination with FOXCUT | [90] |
| miR-367 | tissue specimens, cell culture | oncogenic | promote cell proliferation and migration | reduced PTEN | [84] |
| miR-454 | tumor samples from patients | oncogenic | promote cell proliferation, colony formation, invasion and induction | reduced PTEN | [85] |
| miR-652 | cell lines and tissues samples from patients | oncogenic | increases proliferation and migration | decrease HOXA9 | [91] |
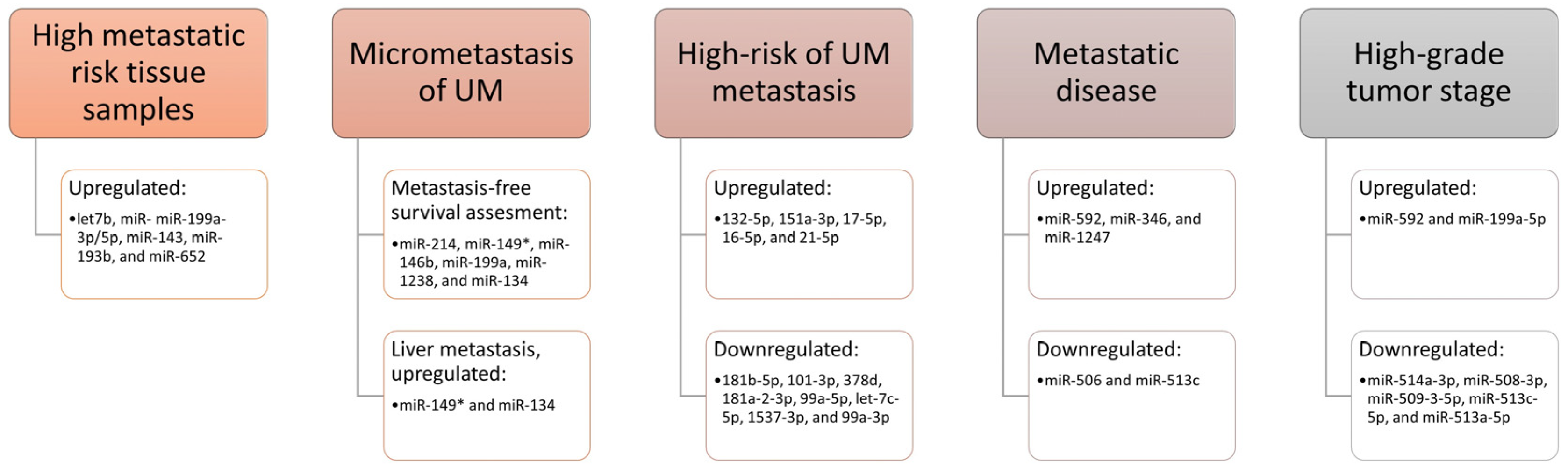
miRNAs as Biomarkers
4. Uveal Melanoma and DNA Methylation
5. Uveal Melanoma Biomarkers in Body Fluids
5.1. Liver Function Tests as Uveal Melanoma Biomarkers
5.2. UM-Serum Biomarkers
5.3. Ocular Fluid Proteins as Biomarkers in Uveal Melanoma
6. Current Challenges and Clinical Implications
6.1. Metastatic Risk Assessment
6.2. Tumor Tissue Analysis
6.3. Novel Treatment Options for Metastatic Uveal Melanoma
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chang, A.E.; Karnell, L.H.; Menck, H.R. The National Cancer Data Base Report on cutaneous and noncutaneous melanoma: A summary of 84,836 cases from the past decade. Cancer 1998, 83, 1664–1678. [Google Scholar] [CrossRef]
- Shields, C.L.; Kaliki, S.; Furuta, M.; Mashayekhi, A.; Shields, J.A. Clinical spectrum and prognosis of uveal melanoma based on age at presentation in 8033 cases. Retina 2012, 32, 1363–1372. [Google Scholar] [CrossRef]
- Kaštelan, S.; Antunica, A.G.; Beketić-Orešković, L.; Bakija, I.; Bogadi, M. Uveal mealnoma: Clinical features and diagnostic procedures. Libr. Oncol. 2017, 45, 81–88. [Google Scholar]
- Damato, E.M.; Damato, B.E. Detection and time to treatment of uveal melanoma in the United Kingdom: An evaluation of 2384 patients. Ophthalmology 2012, 119, 1582–1589. [Google Scholar] [CrossRef]
- Andreoli, M.T.; Mieler, W.F.; Leiderman, Y.I. Epidemiological trends in uveal melanoma. Br. J. Ophthalmol. 2015, 99, 1550–1553. [Google Scholar] [CrossRef] [PubMed]
- Virgili, G.; Gatta, G.; Ciccolallo, L.; Capocaccia, R.; Biggeri, A.; Crocetti, E.; Lutz, J.M.; Paci, E. Incidence of uveal melanoma in Europe. Ophthalmology 2007, 114, 2309–2315. [Google Scholar] [CrossRef]
- Vajdic, C.M.; Kricker, A.; Giblin, M.; McKenzie, J.; Aitken, J.; Giles, G.G.; Armstrong, B.K. Incidence of ocular melanoma in Australia from 1990 to 1998. Int. J. Cancer 2003, 105, 117–122. [Google Scholar] [CrossRef]
- Margo, C.E.; Mulla, Z.; Billiris, K. Incidence of surgically treated uveal melanoma by race and ethnicity. Ophthalmology 1998, 105, 1087–1090. [Google Scholar] [CrossRef] [PubMed]
- Tarlan, B.; Kıratlı, H. Uveal melanoma: Current trends in diagnosis and management. Turk Oftalmoloiji Derg. 2016, 46, 123–137. [Google Scholar] [CrossRef]
- Nayman, T.; Bostan, C.; Logan, P.; Burnier, M.N. Uveal melanoma risk factors: A systematic review of meta-analyses. Curr. Eye Res. 2017, 42, 1085–1093. [Google Scholar] [CrossRef]
- Hammer, H.; Oláh, J.; Tóth-Molnár, E. Dysplastic nevi are a risk factor for uveal melanoma. Eur. J. Ophthalmol. 1996, 6, 472–474. [Google Scholar] [CrossRef]
- Shields, C.L.; Kaliki, S.; Livesey, M.; Walker, B.; Garoon, R.; Bucci, M.; Feinstein, E.; Pesch, A.; Gonzalez, C.; Lally, S.E.; et al. Association of ocular and oculodermal melanocytosis with the rate of uveal melanoma metastasis analysis of 7872 consecutive eyes. JAMA Ophthalmol. 2013, 131, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Weis, E.; Shah, C.P.; Lajous, M.; Shields, J.; Shields, C. The association between host susceptibility factors and uveal melanoma. Arch. Ophthalmol. 2006, 124, 54. [Google Scholar] [CrossRef]
- Harbour, J.W.; Onken, M.D.; Roberson, E.D.O.; Duan, S.; Cao, L.; Worley, L.A.; Council, M.L.; Matatall, K.A.; Helms, C.; Bowcock, A.M. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010, 330, 1410–1413. [Google Scholar] [CrossRef] [PubMed]
- Kastelan, S.; Zimak, D.M.; Ivankovic, M.; Markovic, I.; Antunica, A.G. Liver metastasis in uveal melanoma-treatment options and clinical outcome. Front. Biosci. Landmark 2022, 27, 72. [Google Scholar] [CrossRef]
- Damato, B.; Eleuteri, A.; Taktak, A.F.G.; Coupland, S.E. Estimating prognosis for survival after treatment of choroidal melanoma. Prog. Retin. Eye Res. 2011, 30, 285–295. [Google Scholar] [CrossRef]
- Rodriguez-Vidal, C.; Fernandez-Diaz, D.; Fernandez-Marta, B.; Lago-Baameiro, N.; Pardo, M.; Silva, P.; Paniagua, L.; Blanco-Teijeiro, M.J.; Piñeiro, A.; Bande, M. Treatment of metastatic uveal melanoma: Systematic review. Cancers 2020, 12, 2557. [Google Scholar] [CrossRef]
- Damato, B. Treatment of primary intraocular melanoma. Expert Rev. Anticancer Ther. 2006, 6, 493–506. [Google Scholar] [CrossRef]
- Kaštelan, S.; Gverović Antunica, A.; Beketić Oresković, L.; Kasun, B.; Hat, K. Uveal melanoma: An overview of management and prognosis. Libr. Oncol. 2018, 46, 95–104. [Google Scholar] [CrossRef]
- Garg, G.; Finger, P.T.; Kivelä, T.T.; Simpson, R.; Gallie, B.L.; Saakyan, S.; Amiryan, A.G.; Valskiy, V.; Chin, K.J.; Semenova, K.; et al. Patients presenting with metastases: Stage IV uveal melanoma, an international study. Br. J. Ophthalmol. 2022, 106, 510–517. [Google Scholar] [CrossRef]
- Lamas, N.J.; Martel, A.; Nahon-Estève, S.; Goffinet, S.; Macocco, A.; Bertolotto, C.; Lassalle, S.; Hofman, P. Prognostic biomarkers in uveal melanoma: The status quo, recent advances and future directions. Cancers 2022, 14, 96. [Google Scholar] [CrossRef]
- Kaštelan, S.; Antunica, A.G.; Oresković, L.B.; Pelčić, G.; Kasun, E.; Hat, K. Immunotherapy for uveal melanoma—Current knowledge and perspectives. Curr. Med. Chem. 2020, 27, 1350–1366. [Google Scholar] [CrossRef]
- Sussman, T.A.; Funchain, P.; Singh, A. clinical trials in metastatic uveal melanoma: Current status. Ocul. Oncol. Pathol. 2020, 6, 381–387. [Google Scholar] [CrossRef]
- Barbagallo, C.; Stella, M.; Broggi, G.; Russo, A.; Caltabiano, R.; Ragusa, M. Genetics and RNA regulation of uveal melanoma. Cancers 2023, 15, 775. [Google Scholar] [CrossRef] [PubMed]
- Kaliki, S.; Shields, C.L.; Shields, J.A. Uveal melanoma: Estimating prognosis. Indian J. Ophthalmol. 2015, 63, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Oliva, M.; Rullan, A.J.; Piulats, J.M. Uveal melanoma as a target for immune-therapy. Ann. Transl. Med. 2016, 4, 172. [Google Scholar] [CrossRef]
- Rowcroft, A.; Loveday, B.P.T.; Thomson, B.N.J.; Banting, S.; Knowles, B. Systematic review of liver directed therapy for uveal melanoma hepatic metastases. Hpb 2020, 22, 497–505. [Google Scholar] [CrossRef]
- Buder, K.; Gesierich, A.; Gelbrich, G.; Goebeler, M. Systemic treatment of metastatic uveal melanoma: Review of literature and future perspectives. Cancer Med. 2013, 2, 674–686. [Google Scholar] [CrossRef]
- Wang, Z.; Xie, Y.; Wang, J.; Cheng, Y.; Fleishman, J.; Chen, Z.; Chen, Y. Tebentafusp: A novel drug for the treatment of metastatic uveal melanoma. Drugs Today 2023, 59, 179–193. [Google Scholar] [CrossRef]
- Montazeri, K.; Pattanayak, V.; Sullivan, R.J. Tebentafusp in the treatment of metastatic uveal melanoma: Patient selection and special considerations. Drug Des. Devel. Ther. 2023, 17, 333–339. [Google Scholar] [CrossRef]
- Fallico, M.; Raciti, G.; Longo, A.; Reibaldi, M.; Bonfiglio, V.; Russo, A.; Caltabiano, R.; Gattuso, G.; Falzone, L.; Avitabile, T. Current molecular and clinical insights into uveal melanoma (review). Int. J. Oncol. 2021, 58, 1–22. [Google Scholar] [CrossRef]
- Shain, A.H.; Bagger, M.M.; Yu, R.; Chang, D.; Liu, S.; Vemula, S.; Weier, J.F.; Wadt, K.; Heegaard, S.; Bastian, B.C.; et al. The Genetic evolution of metastatic uveal melanoma. Nat. Genet. 2019, 51, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Kilic, E.; Van Gils, W.; Lodder, E.; Beverloo, H.B.; Van Til, M.E.; Mooy, C.M.; Paridaens, D.; De Klein, A.; Luyten, G.P.M. Clinical and cytogenetic analyses in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3703–3707. [Google Scholar] [CrossRef]
- Harbour, J.W. Molecular prognostic testing and individualized patient care in uveal melanoma. Am. J. Ophthalmol. 2009, 148, 823–829.e1. [Google Scholar] [CrossRef]
- Mudhar, H.S.; Parsons, M.A.; Sisley, K.; Rundle, P.; Singh, A.; Rennie, I.G. A Critical appraisal of the prognostic and predictive factors for uveal malignant melanoma. Histopathology 2004, 45, 1–12. [Google Scholar] [CrossRef]
- Sisley, K.; Doherty, R.; Cross, N.A. What hope for the future? GNAQ and uveal melanoma. Br. J. Ophthalmol. 2011, 95, 620–623. [Google Scholar] [CrossRef]
- Van Raamsdonk, C.D.; Griewank, K.G.; Crosby, M.B.; Garrido, M.C.; Vemula, S.; Wiesner, T.; Obenauf, A.C.; Wackernagel, W.; Green, G.; Bouvier, N.; et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010, 363, 2191–2199. [Google Scholar] [CrossRef]
- Onken, M.D.; Worley, L.A.; Long, M.D.; Duan, S.; Council, M.L.; Bowcock, A.M.; Harbour, J.W. Oncogenic Mutations in GNAQ occur early in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2008, 49, 5230–5234. [Google Scholar] [CrossRef]
- Bauer, J.; Kilic, E.; Vaarwater, J.; Bastian, B.C.; Garbe, C.; De Klein, A. oncogenic GNAQ mutations are not correlated with disease-free survival in uveal melanoma. Br. J. Cancer 2009, 101, 813–815. [Google Scholar] [CrossRef] [PubMed]
- Koopmans, A.E.; Verdijk, R.M.; Brouwer, R.W.W.; Van Den Bosch, T.P.P.; Van Den Berg, M.M.P.; Vaarwater, J.; Kockx, C.E.M.; Paridaens, D.; Naus, N.C.; Nellist, M.; et al. Clinical significance of immunohistochemistry for detection of BAP1 mutations in uveal melanoma. Mod. Pathol. 2014, 27, 1321–1330. [Google Scholar] [CrossRef]
- Jensen, D.E.; Proctor, M.; Marquis, S.T.; Gardner, H.P.; Ha, S.I.; Chodosh, L.A.; Ishov, A.M.; Tommerup, N.; Vissing, H.; Sekido, Y.; et al. BAP1: A novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 1998, 16, 1097–1112. [Google Scholar] [CrossRef]
- Karlsson, J.; Nilsson, L.M.; Mitra, S.; Alsén, S.; Shelke, G.V.; Sah, V.R.; Forsberg, E.M.V.; Stierner, U.; All-Eriksson, C.; Einarsdottir, B.; et al. Molecular profiling of driver events in metastatic uveal melanoma. Nat. Commun. 2020, 11, 1894. [Google Scholar] [CrossRef]
- Riechardt, A.I.; Kilic, E.; Joussen, A.M. The genetics of uveal melanoma: Overview and clinical relevance. Klin. Monbl. Augenheilkd. 2021, 238, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Harbour, J.W.; Roberson, E.D.O.; Anbunathan, H.; Onken, M.D.; Worley, L.A.; Bowcock, A.M. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet. 2013, 45, 133–135. [Google Scholar] [CrossRef] [PubMed]
- Alsafadi, S.; Houy, A.; Battistella, A.; Popova, T.; Wassef, M.; Henry, E.; Tirode, F.; Constantinou, A.; Piperno-Neumann, S.; Roman-Roman, S.; et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 2016, 7, 10615. [Google Scholar] [CrossRef]
- Li, Y.; Shi, J.; Yang, J.; Ge, S.; Zhang, J.; Jia, R.; Fan, X. Uveal melanoma: Progress in molecular biology and therapeutics. Ther. Adv. Med. Oncol. 2020, 12, 175883592096585. [Google Scholar] [CrossRef]
- Johnson, C.P.; Kim, I.K.; Esmaeli, B.; Amin-Mansour, A.; Treacy, D.J.; Carter, S.L.; Hodis, E.; Wagle, N.; Seepo, S.; Yu, X.; et al. Systematic Genomic and translational efficiency studies of uveal melanoma. PLoS ONE 2017, 12, e0178189. [Google Scholar] [CrossRef]
- Koopmans, A.E.; Ober, K.; Dubbink, H.J.; Paridaens, D.; Naus, N.C.; Belunek, S.; Krist, B.; Post, E.; Zwarthoff, E.C.; De Klein, A.; et al. Prevalence and implications of TERT Promoter mutation in uveal and conjunctival melanoma and in benign and premalignant conjunctival melanocytic lesions. Investig. Ophthalmol. Vis. Sci. 2014, 55, 6024–6030. [Google Scholar] [CrossRef]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly recurrent TERT promoter mutations in human melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef]
- Wu, X.; Huang, C.; He, X.; Tian, Y.; Zhou, D.; He, Y.; Liu, X.; Li, J. Feedback regulation of telomerase reverse transcriptase: New insight into the evolving field of telomerase in cancer. Cell. Signal. 2013, 25, 2462–2468. [Google Scholar] [CrossRef]
- Derrien, A.C.; Rodrigues, M.; Eeckhoutte, A.; Dayot, S.; Houy, A.; Mobuchon, L.; Gardrat, S.; Lequin, D.; Ballet, S.; Pierron, G.; et al. Germline MBD4 mutations and predisposition to uveal melanoma. J. Natl. Cancer Inst. 2021, 113, 80–87. [Google Scholar] [CrossRef]
- Li, Y.; Jia, R.; Ge, S. Role of epigenetics in uveal melanoma. Int. J. Biol. Sci. 2017, 13, 426–433. [Google Scholar] [CrossRef]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L.; et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 2017, 32, 204–220.e15. [Google Scholar] [CrossRef] [PubMed]
- Jurmeister, P.; Wrede, N.; Hoffmann, I.; Vollbrecht, C.; Heim, D.; Hummel, M.; Wolkenstein, P.; Koch, I.; Heynol, V.; Schmitt, W.D.; et al. Mucosal melanomas of different anatomic sites share a common global DNA methylation profile with cutaneous melanoma but show location-dependent patterns of genetic and epigenetic alterations. J. Pathol. 2022, 256, 61–70. [Google Scholar] [CrossRef]
- Harbour, J.W. A prognostic test to predict the risk of metastasis in uveal melanoma based on a 15-gene expression profile. Methods Mol. Biol. 2014, 1102, 427–440. [Google Scholar] [PubMed]
- Field, M.G.; Decatur, C.L.; Kurtenbach, S.; Gezgin, G.; van der Velden, P.A.; Jager, M.J.; Kozak, K.N.; Harbour, J.W. PRAME as an independent biomarker for metastasis in uveal melanoma. Clin. Cancer Res. 2016, 22, 1234–1242. [Google Scholar] [CrossRef]
- Fabian, I.D.; Thaung, C.; AlHarby, L.; Sisley, K.; Mudhar, H.S.; Doherty, R.E.; Stacey, A.W.; Arora, A.K.; Cohen, V.M.L.; Sagoo, M.S. Late solitary extraocular recurrence from previously resected iris melanoma. Am. J. Ophthalmol. 2017, 181, 97–105. [Google Scholar] [CrossRef]
- Mensink, H.W.; Vaarwater, J.; de Keizer, R.J.W.; de Wolff-Rouendaal, D.; Mooy, C.M.; de Klein, A.; Paridaens, D. Chromosomal aberrations in iris melanomas. Br. J. Ophthalmol. 2011, 95, 424–428. [Google Scholar] [CrossRef][Green Version]
- Henriquez, F.; Janssen, C.; Kemp, E.G.; Roberts, F. The T1799A BRAF mutation is present in iris melanoma. Investig. Ophthalmol. Vis. Sci. 2007, 48, 4897–4900. [Google Scholar] [CrossRef] [PubMed]
- Scholz, S.L.; Möller, I.; Reis, H.; Süßkind, D.; Van De Nes, J.A.P.; Leonardelli, S.; Schilling, B.; Livingstone, E.; Schimming, T.; Paschen, A.; et al. Frequent GNAQ, GNA11, and EIF1AX mutations in iris melanoma. Investig. Ophthalmol. Vis. Sci. 2017, 58, 3464–3470. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.H.; Burge, C.B.; Bartel, D.P. Most mammalian MRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of microRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Zhou, X.; Chen, X.; Hu, D.N.; Da Dong, X.; Wang, J.; Lu, F.; Tu, L.L.; Qu, J. MicroRNA-34a inhibits uveal melanoma cell proliferation and migration through downregulation of c-Met. Investig. Ophthalmol. Vis. Sci. 2009, 50, 1559–1565. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Han, S.; Yang, L.; Chen, S.; Chen, J.; Ma, N.; Tang, J.; Chen, X.; Chen, F.; Da, X.; et al. The interplay of microRNA-34a, LGR4, EMT-associated factors, and MMP2 in regulating uveal melanoma cells. Invest. Ophthalmol. Vis. Sci. 2019, 60, 4503–4510. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Lou, D. MicroRNA-34b/c suppresses uveal melanoma cell proliferation and migration through multiple targets. Mol. Vis. 2012, 18, 537–546. [Google Scholar]
- Amaro, A.; Croce, M.; Ferrini, S.; Barisione, G.; Gualco, M.; Perri, P.; Pfe, U.; Jager, M.J.; Coupland, S.E.; Mosci, C.; et al. Potential onco-suppressive role of miR122 and miR144 in uveal melanoma through ADAM10 and C-Met inhibition. Cancers 2020, 12, 1468. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, J.; Shen, H.; Lu, J.; Li, C.; Hu, D.N.; da Dong, X.; Yan, D.; Tu, L. Epigenetics, microRNAs, and carcinogenesis: Functional role of microRNA-137 in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2011, 52, 1193–1199. [Google Scholar] [CrossRef]
- Yan, D.; da Dong, X.; Chen, X.; Yao, S.; Wang, L.; Wang, J.; Wang, C.; Hu, D.N.; Qu, J.; Tu, L.L. Role of microRNA-182 in posterior uveal melanoma: Regulation of tumor development through MITF, BCL2 and cyclin D2. PLoS ONE 2012, 7, e40967. [Google Scholar] [CrossRef]
- Chen, X.; He, D.; Da Dong, X.; Dong, F.; Wang, J.; Wang, L.; Tang, J.; Hu, D.N.; Yan, D.; Tu, L.L. MicroRNA-124a is epigenetically regulated and acts as a tumor suppressor by controlling multiple targets in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2013, 54, 2248–2256. [Google Scholar] [CrossRef]
- Ewen, M.E.; Sluss, H.K.; Sherr, C.J.; Matsushime, H.; Kato, J.; Livingston, D.M. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell 1993, 73, 487–497. [Google Scholar] [CrossRef]
- Harbour, J.W.; Luo, R.X.; Dei Santi, A.; Postigo, A.A.; Dean, D.C. Cdk phosphorylation triggers sequential intramolecular interactions that progressively Block Rb Functions as Cells Move through G1. Cell 1999, 98, 859–869. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, I.M.; Halvorsen, O.J.; Collett, K.; Stefansson, I.M.; Straume, O.; Haukaas, S.A.; Salvesen, H.B.; Otte, A.P.; Akslen, L.A. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, Prostate, and Breast. J. Clin. Oncol. 2006, 24, 268–273. [Google Scholar] [CrossRef]
- Li, Y.; Luo, J.T.; Liu, Y.M.; Wei, W. Bin miRNA-145/miRNA-205 inhibits proliferation and invasion of uveal melanoma cells by targeting NPR1/CDC42. Int. J. Ophthalmol. 2020, 13, 718–724. [Google Scholar] [CrossRef]
- Del Mar Maldonado, M.; Dharmawardhane, S. Targeting Rac and Cdc42 GT pases in cancer. Cancer Res. 2018, 78, 3101–3111. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Sun, Q.; Chen, J.; Li, J.; Zeng, Y.; Zhai, S.; Li, P.; Wang, B.; Wang, X. MicroRNA-9 suppresses uveal melanoma cell migration and invasion through the NF-ΚB1 pathway. Oncol. Rep. 2012, 28, 961–968. [Google Scholar] [CrossRef]
- Wu, S.; Chen, H.; Han, N.; Zhang, C.; Yan, H. Long noncoding RNA PVT1 silencing prevents the development of uveal melanoma by impairing microRNA-17-3p–dependent MDM2 upregulation. Investig. Ophthalmol. Vis. Sci. 2019, 60, 4904–4914. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Dong, J.; Zhao, Y.; Peng, X.; Tang, J.; Chen, X.; Wang, L.; Hu, D.N.; Reinach, P.S.; Qu, J.; et al. MiR-142-3p Suppresses uveal melanoma by targeting CDC25C, TGFβR1, GNAQ, WASL, and RAC1. Cancer Manag. Res. 2019, 11, 4729–4742. [Google Scholar] [CrossRef]
- Liu, Y.; Huo, Y.; Wang, D.; Tai, Y.; Li, J.; Pang, D.; Zhang, Y.; Zhao, W.; Du, N.; Huang, Y. MiR-216a-5p/Hexokinase 2 axis regulates uveal melanoma growth through modulation of warburg effect. Biochem. Biophys. Res. Commun. 2018, 501, 885–892. [Google Scholar] [CrossRef]
- Li, J.; Liu, X.; Li, C.; Wang, W. MiR-224-5p inhibits proliferation, migration, and invasion by targeting PIK3R3/AKT3 in uveal melanoma. J. Cell. Biochem. 2019, 120, 12412–12421. [Google Scholar] [CrossRef]
- Okayama, H.; Schetter, A.J.; Harris, C.C. MicroRNAs and inflammation in the pathogenesis and progression of colon cancer. Dig. Dis. 2012, 30, 9–15. [Google Scholar] [CrossRef]
- Wang, Y.C.; Yang, X.; Bin Wei, W.; Xu, X.L. Role of microRNA-21 in uveal melanoma cell invasion and metastasis by regulating P53 and its downstream protein. Int. J. Ophthalmol. 2018, 11, 1258–1268. [Google Scholar] [CrossRef]
- Peng, J.; Liu, H.; Liu, C. MiR-155 promotes uveal melanoma cell proliferation and invasion by regulating NDFIP1 expression. Technol. Cancer Res. Treat. 2017, 16, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; He, X.; Li, F.; Pan, H.; Huang, X.; Wen, X.; Zhang, H.; Li, B.; Ge, S.; Xu, X.; et al. The miR-181 family promotes cell cycle by targeting CTDSPL, a phosphatase-like tumor suppressor in uveal melanoma. J. Exp. Clin. Cancer Res. 2018, 37, 1–13. [Google Scholar] [CrossRef]
- Ling, J.W.; Lu, P.R.; Zhang, Y.B.; Jiang, S.; Zhang, Z.C. MiR-367 promotes uveal melanoma cell proliferation and migration by regulating PTEN. Genet. Mol. Res. 2017, 16, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, Q.; Gao, X.; Shi, D.; Mi, S.; Han, Q. MicroRNA-454 functions as an oncogene by regulating pten in uveal melanoma. FEBS Lett. 2015, 589, 2791–2796. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.H.; Yang, Y.; Zhou, X.P.; Craig, E.L.; Davidorf, F.H.; Eng, C. High frequency of submicroscopic hemizygous deletion is a major mechanism of loss of expression of PTEN in uveal melanoma. J. Clin. Oncol. 2006, 24, 288–295. [Google Scholar] [CrossRef]
- Cheng, Y.; Cheng, T.; Zhao, Y.; Qu, Y. HMGA1 exacerbates tumor progression by activating miR-222 through PI3K/Akt/MMP-9 signaling pathway in uveal melanoma. Cell. Signal. 2019, 63, 109386. [Google Scholar] [CrossRef]
- Souri, Z.; Wierenga, A.P.A.; Kiliç, E.; Brosens, E.; Böhringer, S.; Kroes, W.G.M.; Verdijk, R.M.; van der Velden, P.A.; Luyten, G.P.M.; Jager, M.J. MiRNAs correlate with HLA expression in uveal melanoma: Both up-and Downregulation Are Related to Monosomy 3. Cancers 2021, 13, 4020. [Google Scholar] [CrossRef]
- Zhou, J.; Jiang, J.; Wang, S.; Xia, X. Oncogenic role of microrNA-20a in human uveal melanoma. Mol. Med. Rep. 2016, 14, 1560–1566. [Google Scholar] [CrossRef]
- Wang, X.; Hu, Y.; Cui, J.; Zhou, Y.; Chen, L. Coordinated targeting of MMP-2/MMP-9 by miR-296-3p/FOXCUT exerts tumor-suppressing effects in choroidal malignant melanoma. Mol. Cell. Biochem. 2018, 445, 25–33. [Google Scholar] [CrossRef]
- Xia, Z.; Yang, C.; Yang, X.; Wu, S.; Feng, Z.; Qu, L.; Chen, X.; Liu, L.; Ma, Y. MiR-652 promotes proliferation and migration of uveal melanoma cells by targeting HOXA9. Med. Sci. Monit. 2019, 25, 8722–8732. [Google Scholar] [CrossRef]
- Worley, L.A.; Long, M.D.; Onken, M.D.; Harbour, J.W. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res. 2008, 18, 184–190. [Google Scholar] [CrossRef]
- Venkatesan, N.; Kanwar, J.; Deepa, P.R.; Khetan, V.; Crowley, T.M.; Raguraman, R.; Sugneswari, G.; Rishi, P.; Natarajan, V.; Biswas, J.; et al. Clinico-pathological association of delineated miRNAs in uveal melanoma with monosomy 3/disomy 3 chromosomal aberrations. PLoS ONE 2016, 11, e0146128. [Google Scholar] [CrossRef]
- Smit, K.N.; Chang, J.; Derks, K.; Vaarwater, J.; Brands, T.; Verdijk, R.M.; Wiemer, E.A.C.; Mensink, H.W.; Pothof, J.; de Klein, A.; et al. Aberrant microRNA expression and its implications for uveal melanoma metastasis. Cancers 2019, 11, 815. [Google Scholar] [CrossRef]
- Wróblewska, J.P.; Lach, M.S.; Ustaszewski, A.; Kulcenty, K.; Ibbs, M.; Jagiełło, I.; Suchorska, W.M.; Marszałek, A. The Potential role of selected miRNA in uveal melanoma primary tumors as early biomarkers of disease progression. Genes 2020, 11, 271. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Romano, G.L.; Salemi, R.; Bucolo, C.; Tomasello, B.; Lupo, G.; Anfuso, C.D.; Spandidos, D.A.; Libra, M.; Candido, S. Prognostic significance of deregulated microRNAs in uveal melanomas. Mol. Med. Rep. 2019, 19, 2599–2610. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Zhang, Y.; Ling, F.; Wang, L.; Sheng, X.; Qin, L.; Zhao, X. Identification of a nine-miRNA signature for the prognosis of uveal melanoma. Exp. Eye Res. 2019, 180, 242–249. [Google Scholar] [CrossRef]
- Vashishtha, A.; Lee, T.J.; Sharma, A.; Wallbillich, J.J. Research paper changes in microRNA expression associated with metastasis and survival in patients with uveal melanoma. Oncotarget 2020, 11, 1435–1447. [Google Scholar] [CrossRef][Green Version]
- Venza, I.; Visalli, M.; Beninati, C.; Benfatto, S.; Teti, D.; Venza, M. IL-10Rα expression is post-transcriptionally regulated by miR-15a, miR-185, and miR-211 in melanoma. BMC Med. Genomics 2015, 8, 81. [Google Scholar] [CrossRef]
- Triozzi, P.L.; Achberger, S.; Aldrich, W.; Singh, A.D.; Grane, R.; Borden, E.C. The association of blood angioregulatory microRNA levels with circulating endothelial cells and angiogenic proteins in patients receiving dacarbazine and interferon. J. Transl. Med. 2012, 10, 241. [Google Scholar] [CrossRef] [PubMed]
- Triozzi, P.L.; Achberger, S.; Aldrich, W.; Crabb, J.W.; Saunthararajah, Y.; Singh, A.D. Association of tumor and plasma microRNA expression with tumor monosomy-3 in patients with uveal melanoma. Clin. Epigenetics 2016, 8, 35. [Google Scholar] [CrossRef]
- Stark, M.S.; Gray, E.S.; Isaacs, T.; Chen, F.K.; Millward, M.; McEvoy, A.; Zaenker, P.; Ziman, M.; Soyer, H.P.; Glasson, W.J.; et al. A panel of circulating micrornas detects uveal melanoma with high precision. Transl. Vis. Sci. Technol. 2019, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Caltabiano, R.; Longo, A.; Avitabile, T.; Franco, L.M.; Bonfiglio, V.; Puzzo, L.; Reibaldi, M. Increased levels of mirNA-146a in serum and histologic samples of patients with uveal melanoma. Front. Pharmacol. 2016, 7, 424. [Google Scholar] [CrossRef]
- Yang, Z.K.; Yang, J.Y.; Xu, Z.Z.; Yu, W.H. DNA methylation and uveal melanoma. Chin. Med. J. 2018, 131, 845–851. [Google Scholar] [CrossRef]
- Bakhoum, M.F.; Curtis, E.J.; Goldbaum, M.H.; Mischel, P.S. BAP1 methylation: A prognostic marker of uveal melanoma metastasis. NPJ Precis. Onc. 2021, 5, 89. [Google Scholar] [CrossRef]
- Smit, K.N.; Boers, R.; Vaarwater, J.; Boers, J.; Brands, T.; Mensink, H.; Verdijk, R.M.; van IJcken, W.F.J.; Gribnau, J.; de Klein, A.; et al. Genome-wide aberrant methylation in primary metastatic UM and their matched metastases. Sci. Rep. 2022, 12, 42. [Google Scholar] [CrossRef]
- Ness, C.; Katta, K.; Garred, Ø.; Kumar, T.; Olstad, O.K.; Petrovski, G.; Moe, M.C.; Noer, A. Integrated differential DNA methylation and gene expression of formalin-fixed paraffin-embedded uveal melanoma specimens identifies genes associated with early metastasis and poor prognosis. Exp. Eye Res. 2021, 203, 108426. [Google Scholar] [CrossRef]
- Soltysova, A.; Dvorska, D.; Kajabova, V.H.; Pecimonova, M.; Cepcekova, K.; Ficek, A.; Demkova, L.; Buocikova, V.; Babal, P.; Juras, I.; et al. DNA methylation aberrancy is a reliable prognostic tool in uveal melanoma. Res. Square 2023. [Google Scholar] [CrossRef]
- Mouriaux, F.; Diorio, C.; Bergeron, D.; Berchi, C.; Rousseau, A. Liver function testing is not helpful for early diagnosis of metastatic uveal melanoma. Ophthalmology 2012, 119, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Eskelin, S.; Pyrhönen, S.; Hahka-Kemppinen, M.; Tuomaala, S.; Kivelä, T. A prognostic model and staging for metastatic uveal melanoma. Cancer 2003, 97, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Patel, M. Characterization of computed tomography scan abnormalities in patients with biopsy-proven hepatic metastases from uveal melanoma. Arch. Ophthalmol. 2011, 129, 1576. [Google Scholar] [CrossRef][Green Version]
- Eskelin, S.; Pyrhonen, S.; Summanen, P.; Hahka-Kemppinen, M.; Kivelä, T.; Schachat, A.P. Tumor doubling times in metastatic malignant melanoma of the uvea: Tumor progression before and after treatment. Evid.-Based Eye Care 2001, 2, 36–37. [Google Scholar] [CrossRef]
- Barak, V.; Frenkel, S.; Kalickman, I.; Maniotis, A.; Folberg, R.; Pe’Er, J. Serum markers to detect metastatic uveal melanoma. J. Anticancer Res 2007, 27, 1897–1900. [Google Scholar]
- Barak, V.; Kaiserman, I.; Frenkel, S.; Hendler, K.; Kalickman, I.; Pe’er, J. The dynamics of serum tumor markers in predicting metastatic uveal melanoma (part 1). Anticancer Res. 2011, 31, 345–349. [Google Scholar] [PubMed]
- Haritoglou, I.; Wolf, A.; Maier, T.; Haritoglou, C.; Hein, R.; Schaller, U.C. Osteopontin and “melanoma inhibitory activity”: Comparison of two serological tumor markers in metastatic uveal melanoma patients. Ophthalmologica 2009, 223, 239–243. [Google Scholar] [CrossRef]
- Missotten, G.S.; Korse, C.M.; Van Dehn, C.; Linders, T.C.; Keunen, J.E.; Jager, M.J.; Bonfrer, J.M. S-100B Protein and melanoma inhibitory activity protein in uveal melanoma screening: A comparison with liver function tests. Tumor Biol. 2007, 28, 63–69. [Google Scholar] [CrossRef]
- Wróblewska, J.P.; Lach, M.S.; Kulcenty, K.; Galus, Ł.; Suchorska, W.M.; Rösel, D.; Brábek, J.; Marszałek, A. The analysis of inflammation-related proteins in a cargo of exosomes derived from the serum of uveal melanoma patients reveals potential biomarkers of disease progression. Cancers 2021, 13, 3334. [Google Scholar] [CrossRef]
- Frenkel, S.; Zloto, O.; Pe’er, J.; Barak, V. Insulin-like growth factor-1 as a predictive biomarker for metastatic uveal melanoma in humans. Investig. Ophthalmol. Vis. Sci. 2013, 54, 490–493. [Google Scholar] [CrossRef] [PubMed]
- Husain, B.; Kirchberger, M.C.; Erdmann, M.; Schüpferling, S.; Abolhassani, A.R.; Fröhlich, W.; Berking, C.; Heinzerling, L. Inflammatory markers in autoimmunity induced by checkpoint inhibitors. J. Cancer Res. Clin. Oncol. 2021, 147, 1623–1630. [Google Scholar] [CrossRef]
- Sunderland, D.; Sapra, A. Physiology, Aqueous Humor Circulation. In Study Guide StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK553209/ (accessed on 1 May 2023).
- Goel, M. Aqueous humor dynamics: A review. Open Ophthalmol. J. 2010, 4, 52–59. [Google Scholar] [CrossRef]
- Cheng, Y.; Feng, J.; Zhu, X.; Liang, J. Cytokines Concentrations in aqueous humor of eyes with uveal melanoma. Medicine 2019, 98, e14030. [Google Scholar] [CrossRef] [PubMed]
- Midena, E.; Parrozzani, R.; Midena, G.; Trainiti, S.; Marchione, G.; Cosmo, E.; Londei, D.; Frizziero, L. In vivo intraocular biomarkers: Changes of aqueous humor cytokines and chemokines in patients affected by uveal melanoma. Medicine 2020, 99, e22091. [Google Scholar] [CrossRef]
- Nagarkatti-Gude, N.; Bronkhorst, I.H.G.; van Duinen, S.G.; Luyten, G.P.M.; Jager, M.J. Cytokines and Chemokines in the vitreous fluid of eyes with uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2012, 53, 6748–6755. [Google Scholar] [CrossRef] [PubMed]
- Usui, Y.; Tsubota, K.; Agawa, T.; Ueda, S.; Umazume, K.; Okunuki, Y.; Kezuka, T.; Yamakawa, N.; Goto, H. Aqueous immune mediators in malignant uveal melanomas in comparison to benign pigmented intraocular tumors. Graefe’s Arch. Clin. Exp. Ophthalmol. 2017, 255, 393–399. [Google Scholar] [CrossRef]
- Dunavoelgyi, R.; Funk, M.; Sacu, S.; Georgopoulos, M.; Zlabinger, G.; Zehetmayer, M.; Schmidt-Erfurth, U. Intraocular activation of angiogenic and inflammatory pathways in uveal melanoma. Retina 2012, 32, 1373–1384. [Google Scholar] [CrossRef]
- Wierenga, A.P.A.; Gezgin, G.; van Beelen, E.; Eikmans, M.; Spruyt-Gerritse, M.; Brouwer, N.J.; Versluis, M.; Verdijk, R.M.; van Duinen, S.G.; Marinkovic, M.; et al. Soluble HLA in the aqueous humour of uveal melanoma is associated with unfavourable tumour characteristics. Cancers 2019, 11, 1202. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.X.; Liu, Y.M.; Yang, L.; Xuan, Y.; Wei, W. Bin elevated VEGF-A & PLGF concentration in aqueous humor of patients with uveal melanoma following iodine-125 plaque radiotherapy. Int. J. Ophthalmol. 2020, 13, 599–605. [Google Scholar] [CrossRef]
- Lee, C.S.; Jun, I.H.; Kim, T.I.; Byeon, S.H.; Koh, H.J.; Lee, S.C. Expression of 12 cytokines in aqueous humour of uveal melanoma before and after combined ruthenium-106 brachytherapy and transpupillary thermotherapy. Acta Ophthalmol. 2012, 90, 314–320. [Google Scholar] [CrossRef]
- Ten Voorde, A.M.W.; Wierenga, A.P.A.; Nell, R.J.; van der Velden, P.A.; Luyten, G.P.M.; Verdijk, R.M.; Jager, M.J. In uveal melanoma, angiopoietin-2 but not angiopoietin-1 is increased in high-risk tumors, providing a potential druggable target. Cancers 2021, 13, 3986. [Google Scholar] [CrossRef]
- Dikovskaya, M.A.; Russkikh, G.S.; Loktev, K.V.; Johnston, T.P.; Gevorgyan, M.M.; Voronina, N.P.; Chernykh, V.V.; Trunov, A.N.; Korolenko, T.A. Cystatin C and cystatin SN as possible soluble tumor markers in malignant uveal melanoma. Radiol. Oncol. 2021, 56, 83–91. [Google Scholar] [CrossRef]
- Kujala, E.; Mäkitie, T.; Kivelä, T. Very long-term prognosis of patients with malignant uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2003, 44, 4651–4659. [Google Scholar] [CrossRef]
- Bol, K.F.; Donia, M.; Heegaard, S.; Kiilgaard, J.F.; Svane, I.M. Genetic biomarkers in melanoma of the ocular region: What the medical oncologist should know. Int. J. Mol. Sci. 2020, 21, 5231. [Google Scholar] [CrossRef]
- Schank, T.E.; Hassel, J.C. Immunotherapies for the treatment of uveal melanoma—History and future. Cancers 2019, 11, 1048. [Google Scholar] [CrossRef]
- Beasley, A.B.; Chen, F.K.; Isaacs, T.W.; Gray, E.S. Future perspectives of uveal melanoma blood based biomarkers. Br. J. Cancer 2022, 126, 1511–1528. [Google Scholar] [CrossRef]
- Gallenga, C.E.; Franco, E.; Adamo, G.G.; Violanti, S.S.; Tassinari, P.; Tognon, M.; Perri, P. Genetic basis and molecular mechanisms of uveal melanoma metastasis: A focus on prognosis. Front. Oncol. 2022, 12, 828112. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.F.B.; Marta, B.F.; Baameiro, N.L.; Santiago-Varela, M.; Silva-Rodríguez, P.; Blanco-Teijeiro, M.J.; Perez, M.P.; Ces, A.P. Blood biomarkers of uveal melanoma: Current perspectives. Clin. Ophthalmol. 2020, 14, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Y.; Lai, T.T.; Liao, W.T.; Li, C.J. Clinicopathological and prognostic significance and molecular mechanisms governing uveal melanoma. Ther. Adv. Med. Oncol. 2020, 12, 17566. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.A.; Fraile-Martínez, O.; García-Honduvilla, N.; Coca, S.; Álvarez-Mon, M.; Buján, J.; Teus, M.A. Update on uveal melanoma: Translational research from biology to clinical practice (review). Int. J. Oncol. 2020, 57, 1262–1279. [Google Scholar] [CrossRef]
- Seedor, R.S.; Orloff, M.; Sato, T. Genetic landscape and emerging therapies in uveal melanoma. Cancers 2021, 13, 5503. [Google Scholar] [CrossRef]
- Jin, E.; Burnier, J.V. Liquid biopsy in uveal melanoma: Are we there yet? Ocul. Oncol. Pathol. 2021, 7, 1–16. [Google Scholar] [CrossRef]
- Howlett, S.; Carter, T.J.; Shaw, H.M.; Nathan, P.D. Tebentafusp: A first-in-class treatment for metastatic uveal melanoma. Ther. Adv. Med. Oncol. 2023, 15, 175883592311601. [Google Scholar] [CrossRef] [PubMed]
| Sample Type | Biomarker(s) | Type of Molecules | Significancve in Diagnosis and Prognosis | References |
|---|---|---|---|---|
| aqueous humor | IL-6, IL-7, IL-8, IP-10, PGF-1, MCP-1, NGF-β, EGF, b-FGF, PDGF-AA, VEGF-A, RANTES, MIP-1α, MIP-1β, TNF-α, eotaxin | cytokines, growth factors | significantly higher in UM than cataract | [122,123,124,126] |
| aqueous humor | IL-8, MCP-1, angiogenin | cytokines, growth factors | differentiate between UM and benign ocular tumors | [125] |
| vitreous humor | Flt-3 ligand, IL-6, IL7, IL-8, IP-10, MCP-1, MIP-1α, PDGF-AA, VEGF | cytokines, growth factors | significantly higher in UM than cataract | [126] |
| aqueous humor | IP-10, MIP-1α | cytokines, growth factors | positive correlation with UM-tumor dimensions | [126] |
| vitreus humor | IP-10, MIP-1α, FLT3LG, IL-6, IL-8, MCP-1 | cytokines, growth factors | positive Correlation with UM-tumor dimensions | [126] |
| aqueous humor | sHLA | antigen | metastases, worse survival | [127] |
| aqueous humor | VEGF-A and PLGF | growth factors | positive correlation with tumor thickness and increaly expressed after the surgery | [128] |
| aqueous humor | IL-6, IL-8 and IL-1β | growth factors | increased expression after Ru-106 brachytherapy and transpupillary thermotherapy | [129] |
| aqueous humor | ANG-2 | angiognesis | metastasis | [130] |
| tears | Cystatin C | proteases inhibitor | diagnosis of UM | [131] |
| serum | LFT | enzymes and bile pigments | metastasis, but with low predictive value | [109,110,111,112] |
| serum | OPN, MIA and S-100β | tumor markers | UM hepatic metastases | [113,114,115,116] |
| exosomes | interferon-γ, IL-2, IL-22 and IL-12, Pentraxin-3, TNFSF13B and TNFSF8 | interleukins and other inflammatory-related molecules | UM metastases | [117] |
| serum | IGF-1 | growth factor | prediction of metastases | [118] |
| serum | IL-6, CRP | interleukins and inflammatory-related molecules | early detection of irAR | [119] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pašalić, D.; Nikuševa-Martić, T.; Sekovanić, A.; Kaštelan, S. Genetic and Epigenetic Features of Uveal Melanoma—An Overview and Clinical Implications. Int. J. Mol. Sci. 2023, 24, 12807. https://doi.org/10.3390/ijms241612807
Pašalić D, Nikuševa-Martić T, Sekovanić A, Kaštelan S. Genetic and Epigenetic Features of Uveal Melanoma—An Overview and Clinical Implications. International Journal of Molecular Sciences. 2023; 24(16):12807. https://doi.org/10.3390/ijms241612807
Chicago/Turabian StylePašalić, Daria, Tamara Nikuševa-Martić, Ankica Sekovanić, and Snježana Kaštelan. 2023. "Genetic and Epigenetic Features of Uveal Melanoma—An Overview and Clinical Implications" International Journal of Molecular Sciences 24, no. 16: 12807. https://doi.org/10.3390/ijms241612807
APA StylePašalić, D., Nikuševa-Martić, T., Sekovanić, A., & Kaštelan, S. (2023). Genetic and Epigenetic Features of Uveal Melanoma—An Overview and Clinical Implications. International Journal of Molecular Sciences, 24(16), 12807. https://doi.org/10.3390/ijms241612807







