Structural Analysis of Plasmodium falciparum Hexokinase Provides Novel Information about Catalysis Due to a Plasmodium-Specific Insertion
Abstract
1. Introduction
2. Results
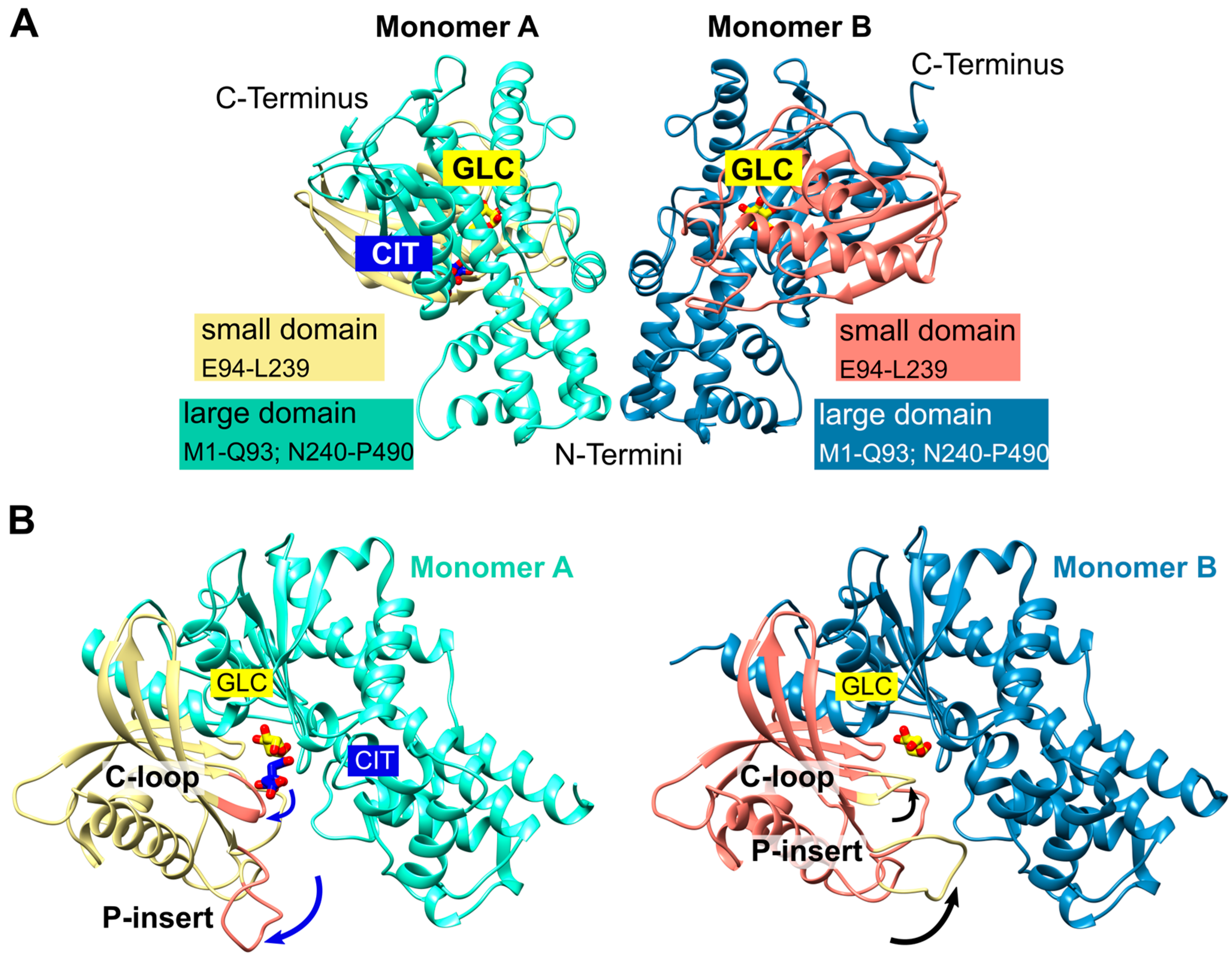
2.1. Overall Structure of P. falciparum Hexokinase
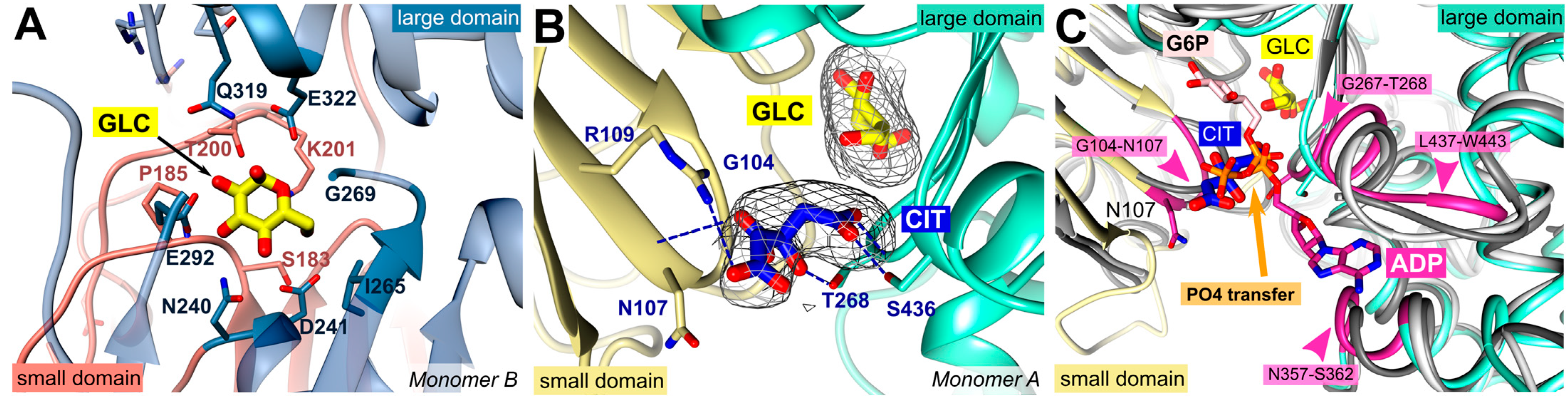
2.2. Glucose and Citrate Were Bound to the Active Site
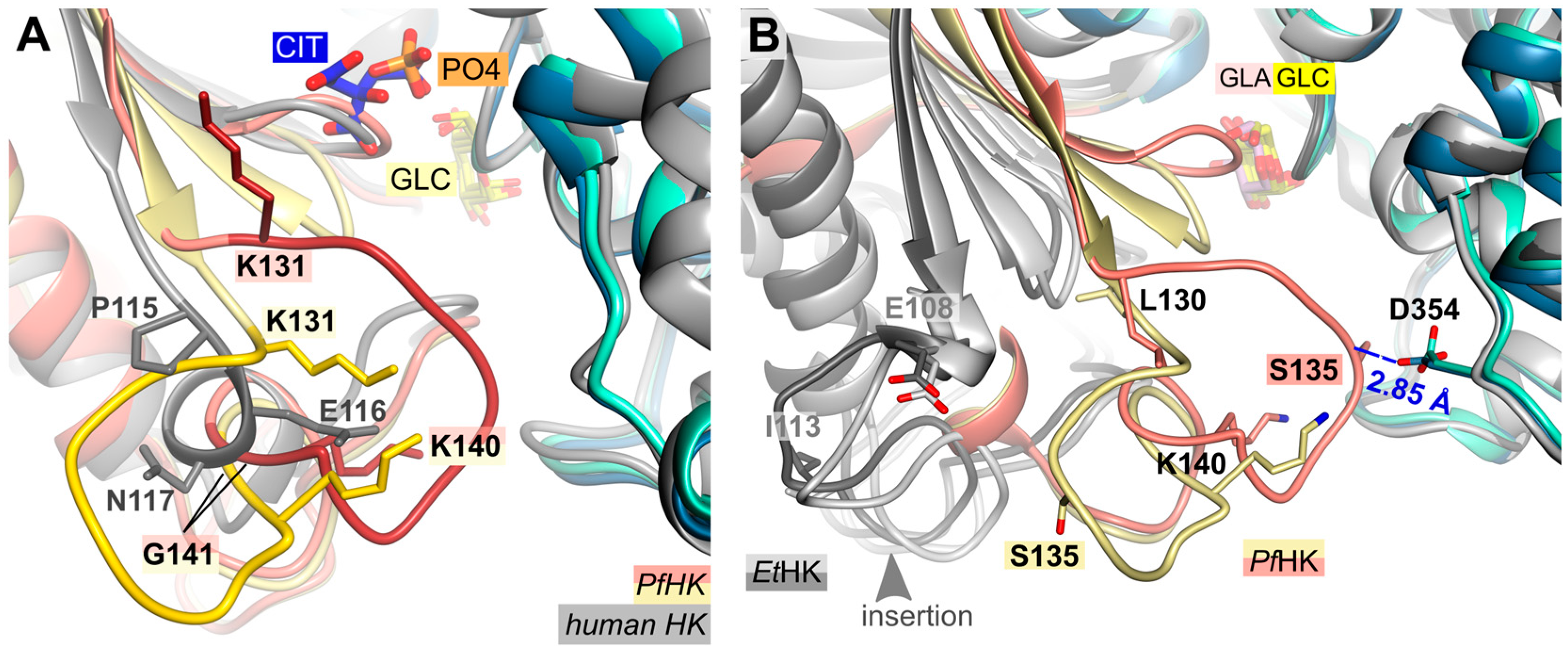
2.3. A Plasmodium-Specific Insertion (P-Insert) Lined the Active Site
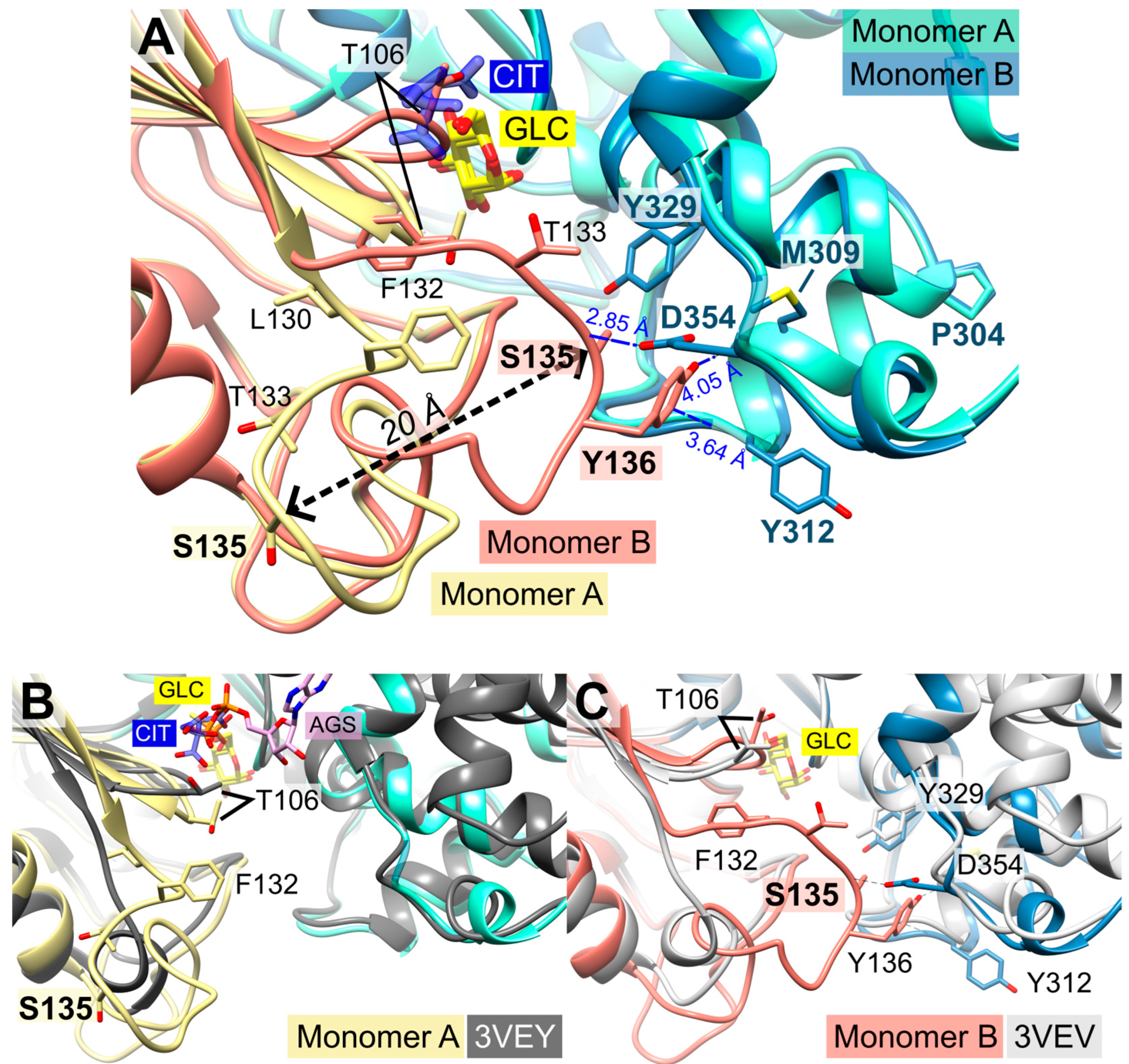
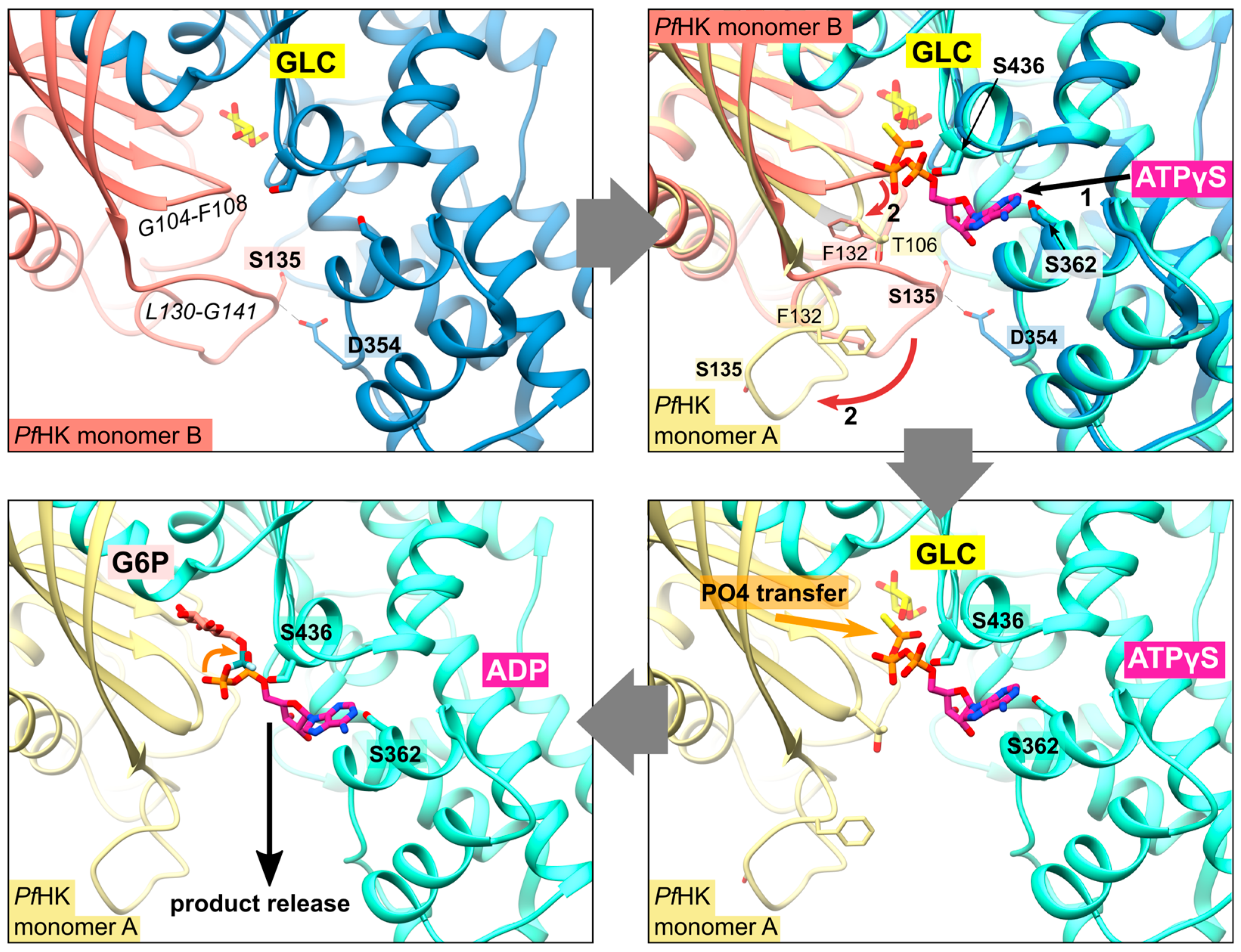
2.4. Does Catalysis Affect the Conformation of the C-Loop and the P-Insert?
2.5. Structural Comparisons Suggest a Plasmodium-Specific Mechanism
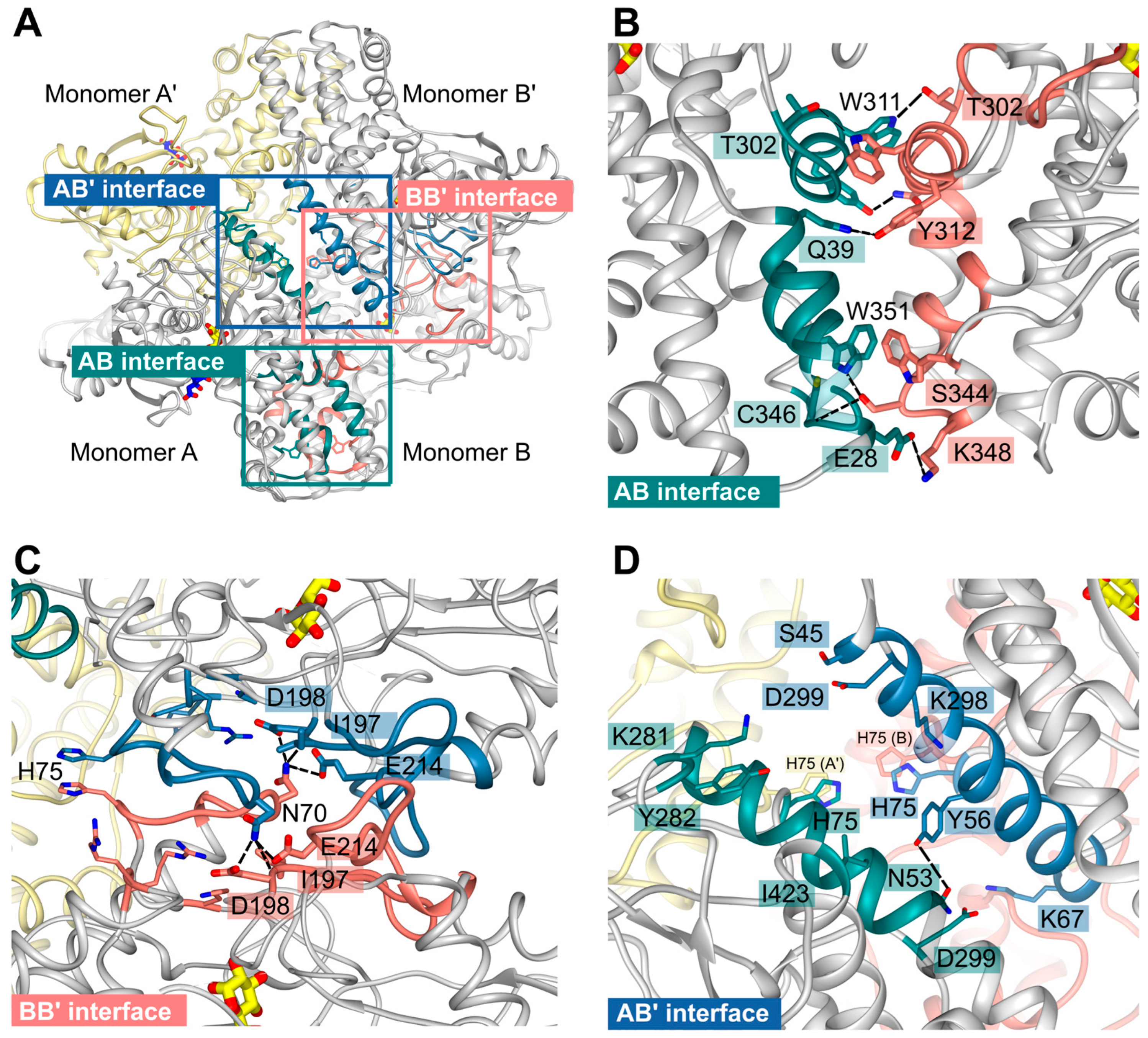
2.6. Oligomerization of P. falciparum Hexokinase
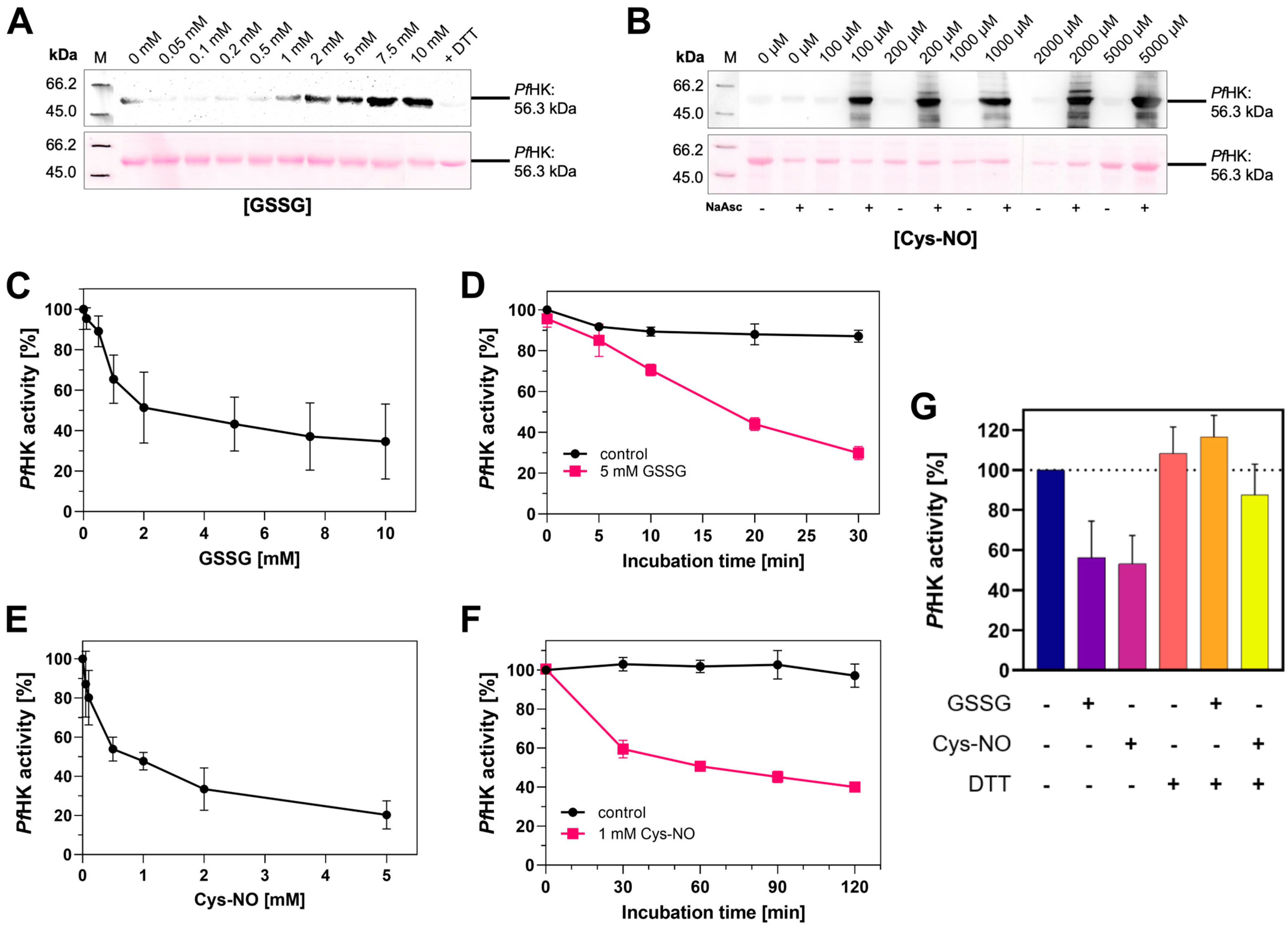
2.7. Oxidative Posttranslational Modifications Inhibit the Enzymatic Activity of PfHK
3. Discussion
4. Materials and Methods
4.1. Heterologous Expression and Purification of Recombinant PfHK
4.2. Kinetic Characterization of PfHK
4.3. Crystallization
4.4. Data Collection and Refinement
4.5. Gel Filtration Chromatography
4.6. Glutathionylation
4.7. Synthesis of S-Nitrosocysteine, Nitrosation, and Biotin-Switch Assay
4.8. Mass Spectrometry Analysis
4.9. Data Analyses and Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. World Malaria Report 2022; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Blasco, B.; Leroy, D.; Fidock, D.A. Antimalarial Drug Resistance: Linking Plasmodium Falciparum Parasite Biology to the Clinic. Nat. Med. 2017, 23, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Nsanzabana, C.; Ariey, F.; Beck, H.-P.; Ding, X.C.; Kamau, E.; Krishna, S.; Legrand, E.; Lucchi, N.; Miotto, O.; Nag, S.; et al. Molecular Assays for Antimalarial Drug Resistance Surveillance: A Target Product Profile. PLoS ONE 2018, 13, e0204347. [Google Scholar] [CrossRef] [PubMed]
- Kirk, K. Glucose Uptake in Plasmodium Falciparum-Infected Erythrocytes Is an Equilibrative Not an Active Process. Mol. Biochem. Parasitol. 1996, 82, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Moxon, C.A.; Gibbins, M.P.; McGuinness, D.; Milner, D.A.; Marti, M. New Insights into Malaria Pathogenesis. Annu. Rev. Pathol. 2020, 15, 315–343. [Google Scholar] [CrossRef]
- Roth, E.F.; Calvin, M.C.; Max-Audit, I.; Rosa, J.; Rosa, R. The Enzymes of the Glycolytic Pathway in Erythrocytes Infected with Plasmodium Falciparum Malaria Parasites. Blood 1988, 72, 1922–1925. [Google Scholar] [CrossRef]
- Harris, M.T.; Walker, D.M.; Drew, M.E.; Mitchell, W.G.; Dao, K.; Schroeder, C.E.; Flaherty, D.P.; Weiner, W.S.; Golden, J.E.; Morris, J.C. Interrogating a Hexokinase-Selected Small-Molecule Library for Inhibitors of Plasmodium Falciparum Hexokinase. Antimicrob. Agents Chemother. 2013, 57, 3731–3737. [Google Scholar] [CrossRef]
- Davis, M.I.; Patrick, S.L.; Blanding, W.M.; Dwivedi, V.; Suryadi, J.; Golden, J.E.; Coussens, N.P.; Lee, O.W.; Shen, M.; Boxer, M.B.; et al. Identification of Novel Plasmodium Falciparum Hexokinase Inhibitors with Antiparasitic Activity. Antimicrob. Agents Chemother. 2016, 60, 6023–6033. [Google Scholar] [CrossRef]
- Hurley, J.H. The Sugar Kinase/Heat Shock Protein 70/Actin Superfamily: Implications of Conserved Structure for Mechanism. Annu. Rev. Biophys. Biomol. Struct. 1996, 25, 137–162. [Google Scholar] [CrossRef]
- Nishimasu, H.; Fushinobu, S.; Shoun, H.; Wakagi, T. Identification and Characterization of an ATP-Dependent Hexokinase with Broad Substrate Specificity from the Hyperthermophilic Archaeon Sulfolobus Tokodaii. J. Bacteriol. 2006, 188, 2014–2019. [Google Scholar] [CrossRef][Green Version]
- Roy, S.; Vivoli Vega, M.; Harmer, N.J. Carbohydrate Kinases: A Conserved Mechanism across Differing Folds. Catalysts 2019, 9, 29. [Google Scholar] [CrossRef]
- Roth, E.F. Malarial Parasite Hexokinase and Hexokinase-Dependent Glutathione Reduction in the Plasmodium Falciparum-Infected Human Erythrocyte. J. Biol. Chem. 1987, 262, 15678–15682. [Google Scholar] [CrossRef] [PubMed]
- Haeussler, K.; Berneburg, I.; Jortzik, E.; Hahn, J.; Rahbari, M.; Schulz, N.; Preuss, J.; Zapol’skii, V.A.; Bode, L.; Pinkerton, A.B.; et al. Glucose 6-Phosphate Dehydrogenase 6-Phosphogluconolactonase: Characterization of the Plasmodium Vivax Enzyme and Inhibitor Studies. Malar. J. 2019, 18, 22. [Google Scholar] [CrossRef] [PubMed]
- Gardner, M.J.; Hall, N.; Fung, E.; White, O.; Berriman, M.; Hyman, R.W.; Carlton, J.M.; Pain, A.; Nelson, K.E.; Bowman, S.; et al. Genome Sequence of the Human Malaria Parasite Plasmodium Falciparum. Nature 2002, 419, 498–511. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.S.; Darling, J.E.; Suryadi, J.; Morris, J.C.; Drew, M.E.; Subramaniam, S. Plasmodium Vivax and Human Hexokinases Share Similar Active Sites but Display Distinct Quaternary Architectures. IUCrJ 2020, 7, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Miseta, A.; Csutora, P. Relationship between the Occurrence of Cysteine in Proteins and the Complexity of Organisms. Mol. Biol. Evol. 2000, 17, 1232–1239. [Google Scholar] [CrossRef]
- Kehr, S.; Jortzik, E.; Delahunty, C.; Yates, J.R.; Rahlfs, S.; Becker, K. Protein S-Glutathionylation in Malaria Parasites. Antioxid. Redox Signal. 2011, 15, 2855–2865. [Google Scholar] [CrossRef]
- Wang, L.; Delahunty, C.; Prieto, J.H.; Rahlfs, S.; Jortzik, E.; Yates, J.R.; Becker, K. Protein S-Nitrosylation in Plasmodium Falciparum. Antioxid. Redox Signal. 2014, 20, 2923–2935. [Google Scholar] [CrossRef]
- Bennett, W.S.; Steitz, T.A. Glucose-Induced Conformational Change in Yeast Hexokinase. Proc. Natl. Acad. Sci. USA 1978, 75, 4848–4852. [Google Scholar] [CrossRef]
- Kuser, P.; Cupri, F.; Bleicher, L.; Polikarpov, I. Crystal Structure of Yeast Hexokinase PI in Complex with Glucose: A Classical “Induced Fit” Example Revised. Proteins 2008, 72, 731–740. [Google Scholar] [CrossRef]
- Liu, S.; Ammirati, M.J.; Song, X.; Knafels, J.D.; Zhang, J.; Greasley, S.E.; Pfefferkorn, J.A.; Qiu, X. Insights into Mechanism of Glucokinase Activation: Observation of Multiple Distinct Protein Conformations. J. Biol. Chem. 2012, 287, 13598–13610. [Google Scholar] [CrossRef]
- Dalle-Donne, I.; Rossi, R.; Giustarini, D.; Colombo, R.; Milzani, A. S-Glutathionylation in Protein Redox Regulation. Free. Radic. Biol. Med. 2007, 43, 883–898. [Google Scholar] [CrossRef] [PubMed]
- Jortzik, E.; Wang, L.; Becker, K. Thiol-Based Posttranslational Modifications in Parasites. Antioxid. Redox Signal. 2012, 17, 657–673. [Google Scholar] [CrossRef] [PubMed]
- Groitl, B.; Jakob, U. Thiol-Based Redox Switches. Biochim. Biophys. Acta 2014, 1844, 1335–1343. [Google Scholar] [CrossRef] [PubMed]
- Schipper, S.; Wu, H.; Furdui, C.M.; Poole, L.B.; Delahunty, C.M.; Park, R.; Yates, J.R.; Becker, K.; Przyborski, J.M. Identification of Sulfenylation Patterns in Trophozoite Stage Plasmodium Falciparum Using a Non-Dimedone Based Probe. Mol. Biochem. Parasitol. 2021, 242, 111362. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.E. Isozymes of Mammalian Hexokinase: Structure, Subcellular Localization and Metabolic Function. J. Exp. Biol. 2003, 206, 2049–2057. [Google Scholar] [CrossRef]
- Aleshin, A.E.; Zeng, C.; Bartunik, H.D.; Fromm, H.J.; Honzatko, R.B. Regulation of Hexokinase I: Crystal Structure of Recombinant Human Brain Hexokinase Complexed with Glucose and Phosphate. J. Mol. Biol. 1998, 282, 345–357. [Google Scholar] [CrossRef]
- He, C.; Chen, J.; Wang, H.; Wan, Y.; Zhou, J.; Dan, Z.; Zeng, Y.; Xu, W.; Zhu, Y.; Huang, W.; et al. Crystal Structures of Rice Hexokinase 6 with a Series of Substrates Shed Light on Its Enzymatic Mechanism. Biochem. Biophys. Res. Commun. 2019, 515, 614–620. [Google Scholar] [CrossRef]
- Kosow, D.P.; Rose, I.A. Product Inhibition of the Hexokinases. J. Biol. Chem. 1970, 245, 198–204. [Google Scholar] [CrossRef]
- Kuettner, E.B.; Kettner, K.; Keim, A.; Svergun, D.I.; Volke, D.; Singer, D.; Hoffmann, R.; Müller, E.-C.; Otto, A.; Kriegel, T.M.; et al. Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces Lactis: A Molecular Basis for Understanding the Control of Yeast Hexokinase Functions via Covalent Modification and Oligomerization. J. Biol. Chem. 2010, 285, 41019–41033. [Google Scholar] [CrossRef]
- Aleshin, A.E.; Kirby, C.; Liu, X.; Bourenkov, G.P.; Bartunik, H.D.; Fromm, H.J.; Honzatko, R.B. Crystal Structures of Mutant Monomeric Hexokinase I Reveal Multiple ADP Binding Sites and Conformational Changes Relevant to Allosteric Regulation. J. Mol. Biol. 2000, 296, 1001–1015. [Google Scholar] [CrossRef]
- Mulichak, A.M.; Wilson, J.E.; Padmanabhan, K.; Garavito, R.M. The Structure of Mammalian Hexokinase-1. Nat. Struct. Biol. 1998, 5, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Kosow, D.P.; Oski, F.A.; Warms, J.V.; Rose, I.A. Regulation of Mammalian Hexokinase: Regulatory Differences between Isoenzyme I and II. Arch. Biochem. Biophys. 1973, 157, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Colowick, S.P. The Hexokinases. In The Enzymes; Boyer, P.D., Ed.; Academic Press: Cambridge, MA, USA, 1973; Volume 9, pp. 1–48. [Google Scholar]
- Kosow, D.P.; Rose, I.A. Activators of Yeast Hexokinase. J. Biol. Chem. 1971, 246, 2618–2625. [Google Scholar] [CrossRef]
- Steinböck, F.; Choojun, S.; Held, I.; Roehr, M.; Kubicek, C.P. Characterization and Regulatory Properties of a Single Hexokinase from the Citric Acid Accumulating Fungus Aspergillus Niger. Biochim. Biophys. Acta 1994, 1200, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.Y.; Alechina, O.; Aleshin, A.E.; Fromm, H.J.; Honzatko, R.B. Identification of a Phosphate Regulatory Site and a Low Affinity Binding Site for Glucose 6-Phosphate in the N-Terminal Half of Human Brain Hexokinase. J. Biol. Chem. 1998, 273, 19548–19553. [Google Scholar] [CrossRef] [PubMed]
- Hoggett, J.G.; Kellett, G.L. Kinetics of the Monomer-Dimer Reaction of Yeast Hexokinase PI. Biochem. J. 1992, 287 Pt 2, 567–572. [Google Scholar] [CrossRef]
- Nishimasu, H.; Fushinobu, S.; Shoun, H.; Wakagi, T. Crystal Structures of an ATP-Dependent Hexokinase with Broad Substrate Specificity from the Hyperthermophilic Archaeon Sulfolobus Tokodaii. J. Biol. Chem. 2007, 282, 9923–9931. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Wilson, J.E. Kinetic and Regulatory Properties of HK I(+), a Modified Form of the Type I Isozyme of Mammalian Hexokinase in Which Interactions between the N- and C-Terminal Halves Have Been Disrupted. Arch. Biochem. Biophys. 2002, 399, 109–115. [Google Scholar] [CrossRef]
- Sebastian, S.; Wilson, J.E.; Mulichak, A.; Garavito, R.M. Allosteric Regulation of Type I Hexokinase: A Site-Directed Mutational Study Indicating Location of the Functional Glucose 6-Phosphate Binding Site in the N-Terminal Half of the Enzyme. Arch. Biochem. Biophys. 1999, 362, 203–210. [Google Scholar] [CrossRef]
- Saito, T.; Maeda, T.; Nakazawa, M.; Takeuchi, T.; Nozaki, T.; Asai, T. Characterisation of Hexokinase in Toxoplasma Gondii Tachyzoites. Int. J. Parasitol. 2002, 32, 961–967. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, H.; Guo, F.; Sun, M.; Zhu, G. A Unique Hexokinase in Cryptosporidium Parvum, an Apicomplexan Pathogen Lacking the Krebs Cycle and Oxidative Phosphorylation. Protist 2014, 165, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.; Neyaz, M.K.; Ikramul Hasan, S. Exploiting Unique Structural and Functional Properties of Malarial Glycolytic Enzymes for Antimalarial Drug Development. Malar. Res. Treat. 2014, 2014, 451065. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Tilley, L.; Vennerstrom, J.L.; Roberts, D.; Rogerson, S.; Ginsburg, H. Oxidative Stress in Malaria Parasite-Infected Erythrocytes: Host-Parasite Interactions. Int. J. Parasitol. 2004, 34, 163–189. [Google Scholar] [CrossRef] [PubMed]
- Pereira, D.M.S.; Carvalho Júnior, A.R.; da Costa Brito Lacerda, E.M.; da Silva, L.C.N.; Marinho, C.R.F.; André, E.; Fernandes, E.S. Oxidative and Nitrosative Stresses in Cerebral Malaria: Can We Target Them to Avoid a Bad Prognosis? J. Antimicrob. Chemother. 2020, 75, 1363–1373. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Sharma, N.; Sharma, G.P.; Mishra, N. Redox Interactome in Malaria Parasite Plasmodium Falciparum. Parasitol. Res. 2021, 120, 423–434. [Google Scholar] [CrossRef]
- Sturm, N.; Jortzik, E.; Mailu, B.M.; Koncarevic, S.; Deponte, M.; Forchhammer, K.; Rahlfs, S.; Becker, K. Identification of Proteins Targeted by the Thioredoxin Superfamily in Plasmodium Falciparum. PLoS Pathog. 2009, 5, e1000383. [Google Scholar] [CrossRef]
- Heneberg, P. Redox Regulation of Hexokinases. Antioxid. Redox Signal. 2019, 30, 415–442. [Google Scholar] [CrossRef]
- Rodríguez-Saavedra, C.; Morgado-Martínez, L.E.; Burgos-Palacios, A.; King-Díaz, B.; López-Coria, M.; Sánchez-Nieto, S. Moonlighting Proteins: The Case of the Hexokinases. Front. Mol. Biosci. 2021, 8, 701975. [Google Scholar] [CrossRef]
- Miller, S.; Ross-Inta, C.; Giulivi, C. Kinetic and Proteomic Analyses of S-Nitrosoglutathione-Treated Hexokinase A: Consequences for Cancer Energy Metabolism. Amino Acids 2007, 32, 593–602. [Google Scholar] [CrossRef]
- Mongin, A.A.; Dohare, P.; Jourd’heuil, D. Selective Vulnerability of Synaptic Signaling and Metabolism to Nitrosative Stress. Antioxid. Redox Signal. 2012, 17, 992–1012. [Google Scholar] [CrossRef]
- Romero, J.M.; Bizzozero, O.A. Intracellular Glutathione Mediates the Denitrosylation of Protein Nitrosothiols in the Rat Spinal Cord. J. Neurosci. Res. 2009, 87, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Moriarty, N.W.; Tronrud, D.E.; Adams, P.D.; Karplus, P.A. A New Default Restraint Library for the Protein Backbone in Phenix: A Conformation-Dependent Geometry Goes Mainstream. Acta Crystallogr. D Biol. Crystallogr. 2016, 72, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Secondary-Structure Matching (SSM), a New Tool for Fast Protein Structure Alignment in Three Dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2256–2268. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]

| PDB Accession Code | 7ZZI (PfHK) |
|---|---|
| Space group | P3121 |
| Unit cell parameters | |
| a, b, c (Å) | 125.24, 125.24, 120.93 |
| α, β, γ (°) | 90, 90, 120 |
| Data collection | |
| Beamline | SLS beam line X10SA |
| Temperature (K) | 100 |
| Resolution range | 49.5–2.8 (2.9–2.8) |
| Wilson B-factor Å2 | 95.8 |
| Total reflections | 278,108 (26,474) |
| Unique reflections | 27,459 (2644) |
| Multiplicity | 10.1 (10.0) |
| Completeness (%) | 99.7 (97.1) |
| Mean I/σ (I) | 13.1 (0.8) |
| Wilson B-factor | 95.8 |
| R-merge (%) | 0.12 (3.41) |
| R-pim (%) | 0.04 (1.12) |
| CC1/2 (%) | 0.999 (0.305) |
| Molecules per ASU | 2 |
| Refinement | |
| Reflections used in refinement | 27,456 (2644) |
| Reflections used for R-free | 2746 (264) |
| R-work | 0.199 (0.428) |
| R-free | 0.268 (0.481) |
| Protein residues | 930 |
| Macromolecules average B-factor Å2 | 89.9 |
| Ligands average B-factor Å2 | 82.8 |
| Solvent average B-factor Å2 | 82.2 |
| RMS deviations | |
| Bonds (Å) | 0.01 |
| Angles (°) | 1.17 |
| Ramachandran favored (%) | 96.11 |
| KM (mM) | ||
|---|---|---|
| Glucose | ATP | |
| PfHK, present study | 0.13 ± 0.01 | 0.83 ± 0.08 |
| PfHK, Harris et al. 2013 [7] | 0.62 ± 0.06 | 0.66 ± 0.08 |
| PfHK, Roth et al. 1987 [12] * | 0.431 ± 0.021 | 3.1 ± 1.4 |
| CpHK, Yu et al. 2015 [43] | 0.138 | 0.673 |
| TgHK, Saito et al. 2002 [42] | 0.0080 ± 0.0008 | 1.05 ± 0.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dillenberger, M.; Werner, A.-D.; Velten, A.-S.; Rahlfs, S.; Becker, K.; Fritz-Wolf, K. Structural Analysis of Plasmodium falciparum Hexokinase Provides Novel Information about Catalysis Due to a Plasmodium-Specific Insertion. Int. J. Mol. Sci. 2023, 24, 12739. https://doi.org/10.3390/ijms241612739
Dillenberger M, Werner A-D, Velten A-S, Rahlfs S, Becker K, Fritz-Wolf K. Structural Analysis of Plasmodium falciparum Hexokinase Provides Novel Information about Catalysis Due to a Plasmodium-Specific Insertion. International Journal of Molecular Sciences. 2023; 24(16):12739. https://doi.org/10.3390/ijms241612739
Chicago/Turabian StyleDillenberger, Melissa, Anke-Dorothee Werner, Ann-Sophie Velten, Stefan Rahlfs, Katja Becker, and Karin Fritz-Wolf. 2023. "Structural Analysis of Plasmodium falciparum Hexokinase Provides Novel Information about Catalysis Due to a Plasmodium-Specific Insertion" International Journal of Molecular Sciences 24, no. 16: 12739. https://doi.org/10.3390/ijms241612739
APA StyleDillenberger, M., Werner, A.-D., Velten, A.-S., Rahlfs, S., Becker, K., & Fritz-Wolf, K. (2023). Structural Analysis of Plasmodium falciparum Hexokinase Provides Novel Information about Catalysis Due to a Plasmodium-Specific Insertion. International Journal of Molecular Sciences, 24(16), 12739. https://doi.org/10.3390/ijms241612739






