A Soybean Sucrose Non-Fermenting Protein Kinase 1 Gene, GmSNF1, Positively Regulates Plant Response to Salt and Salt–Alkali Stress in Transgenic Plants
Abstract
1. Introduction
2. Results
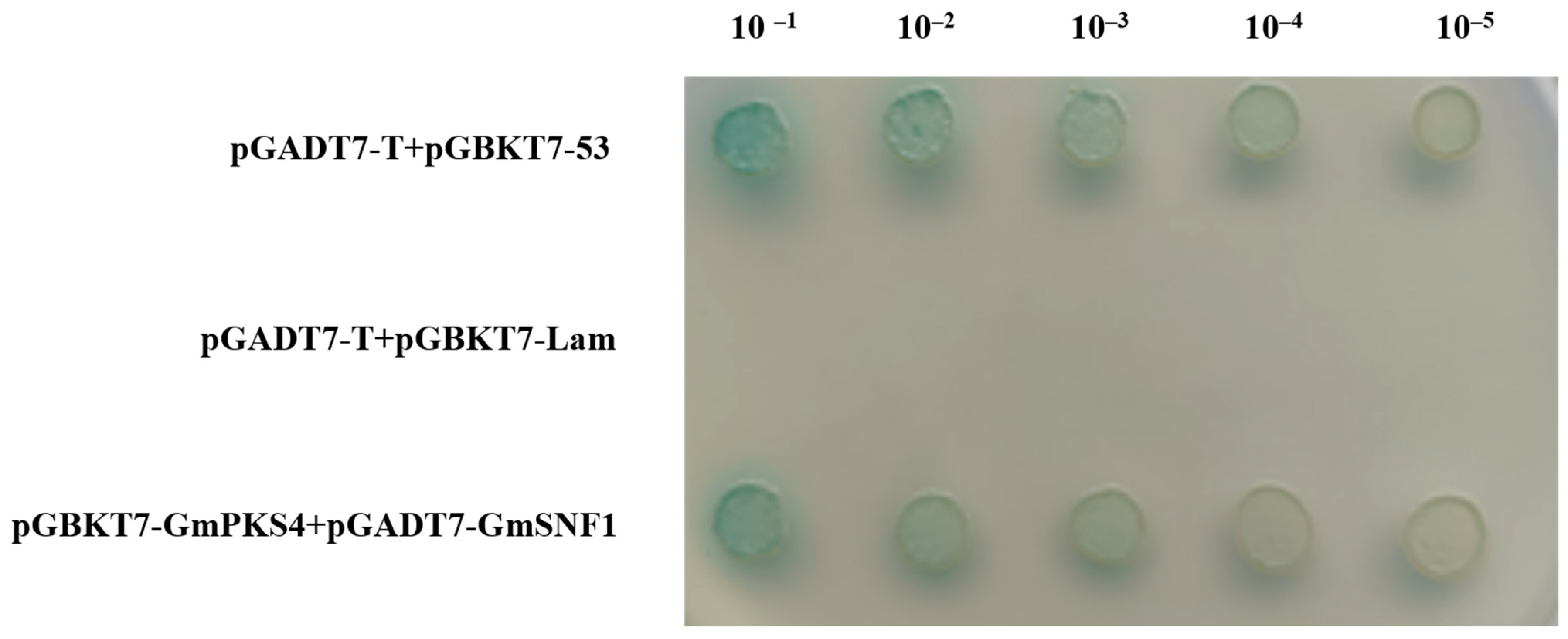
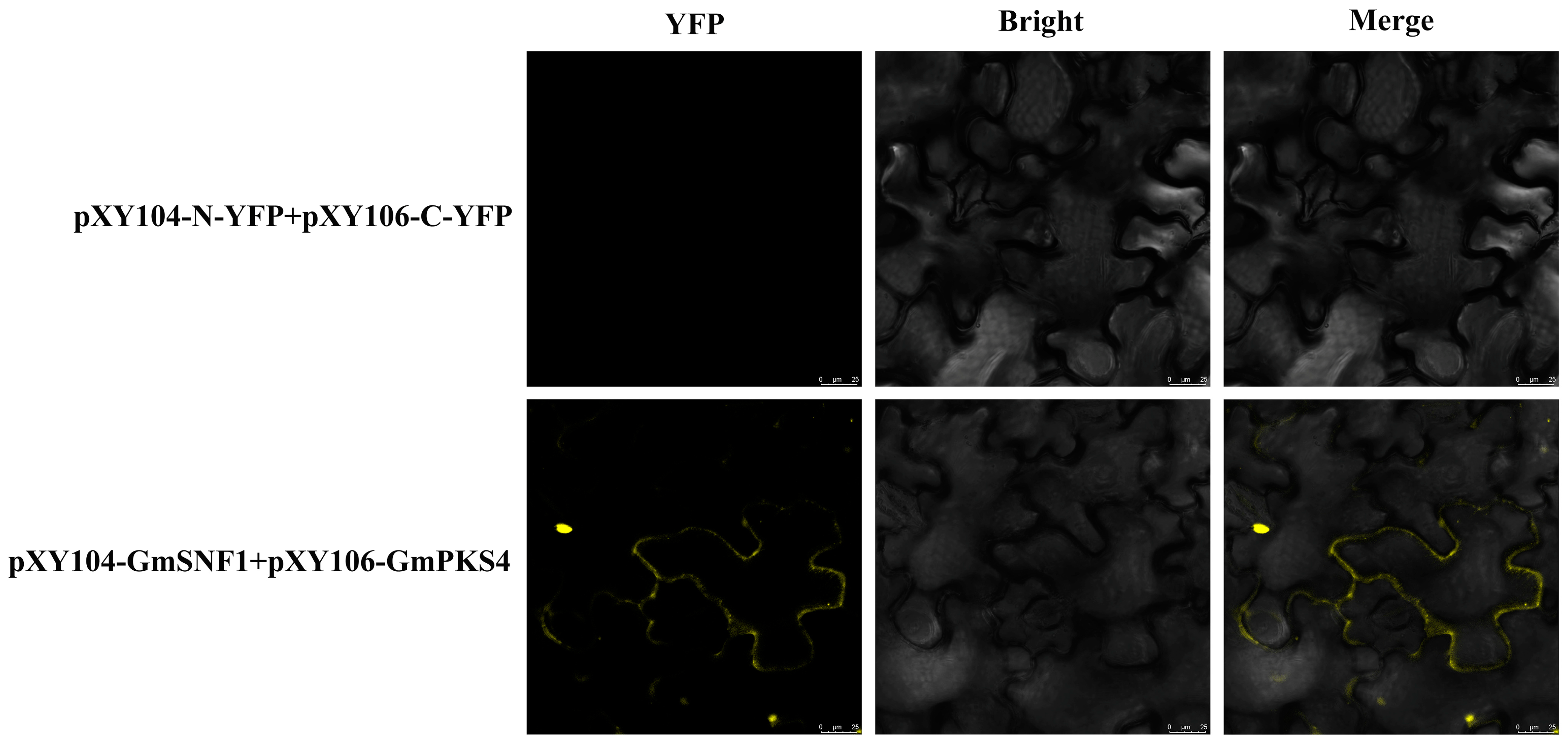
2.1. Analysis of the Interaction between GmSNF1 and GmPKS4 Proteins
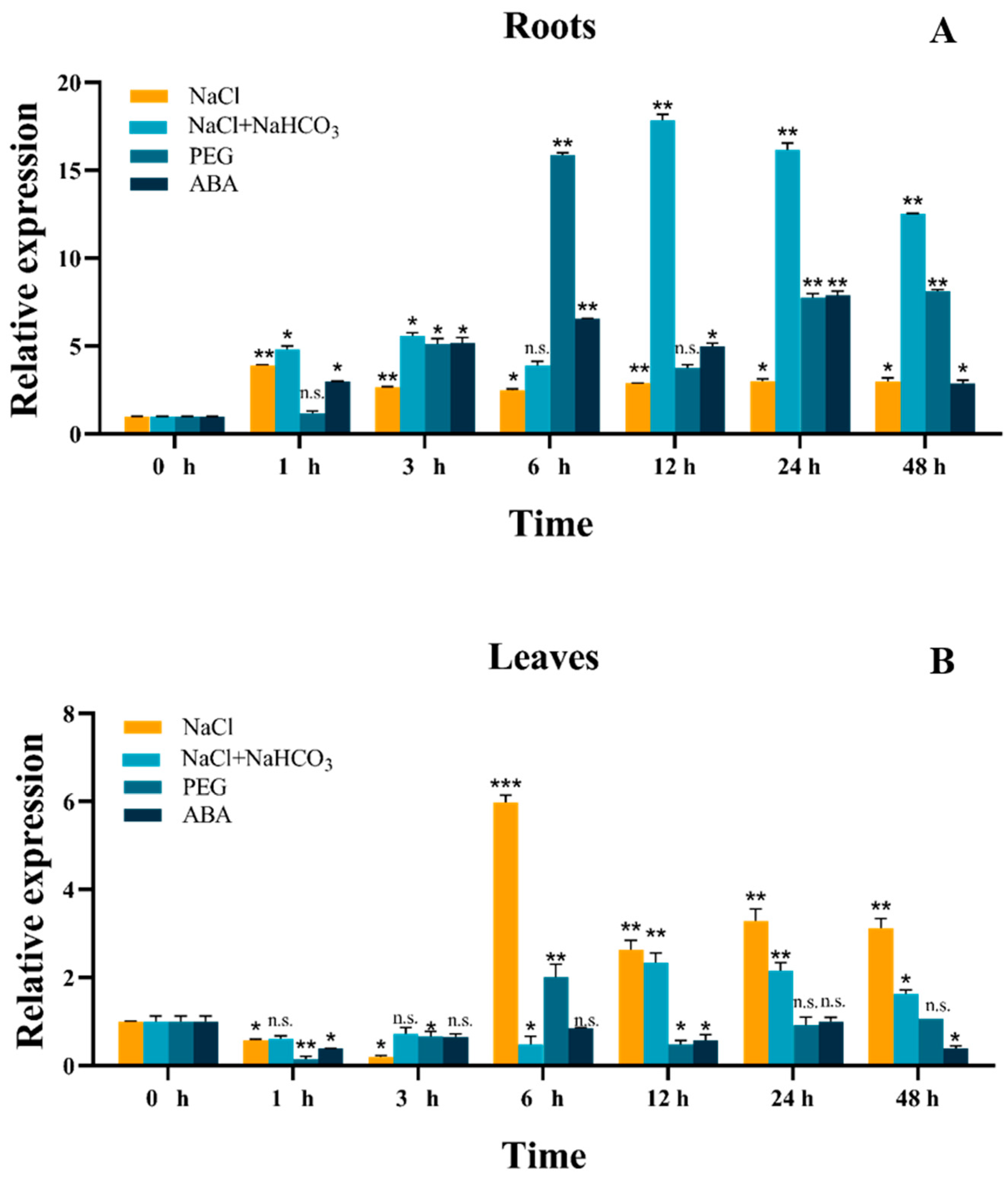
2.2. Expression Analysis of the GmSNF1 Gene under Abiotic Stresses
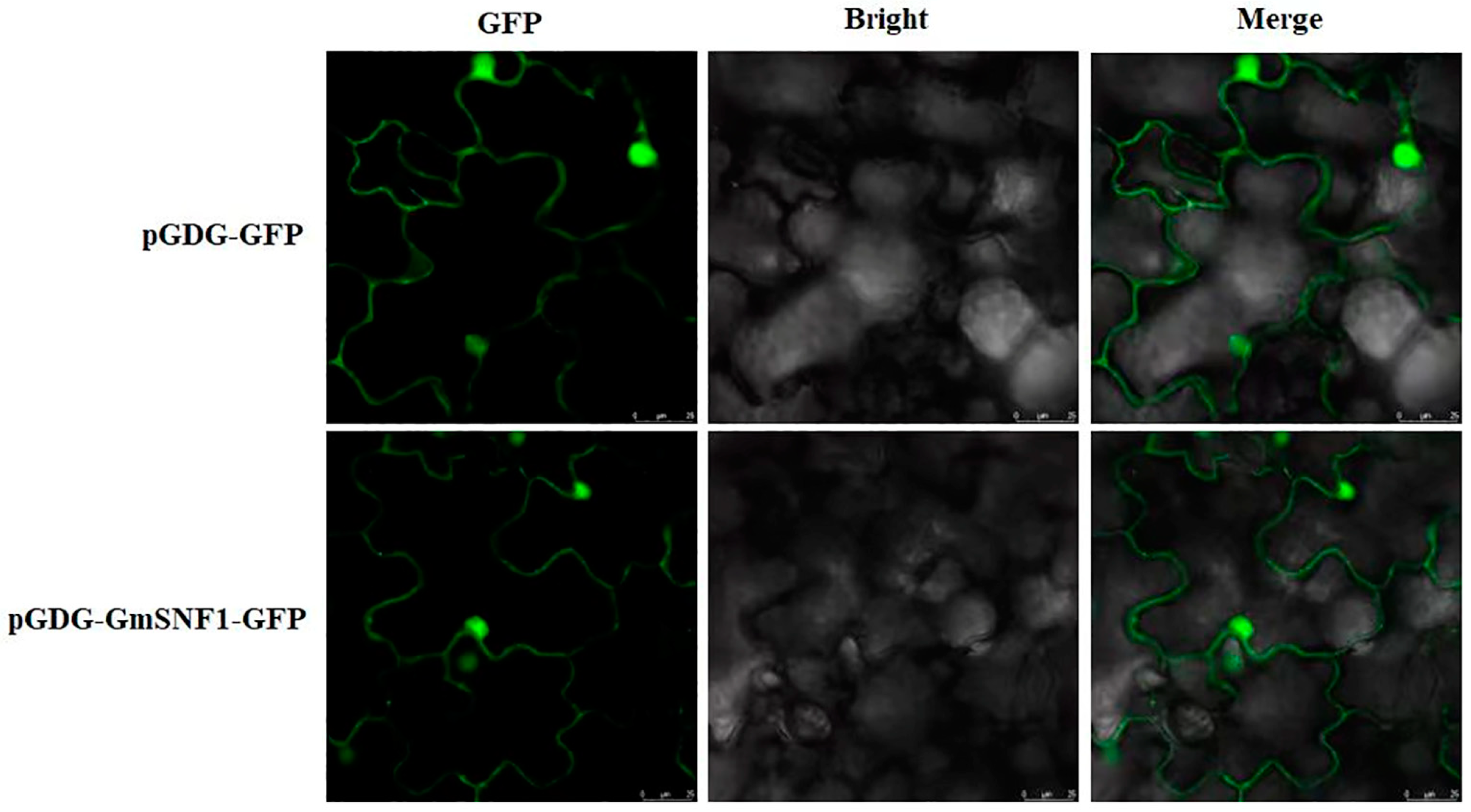
2.3. Subcellular Localization Analysis of the GmSNF1 Protein
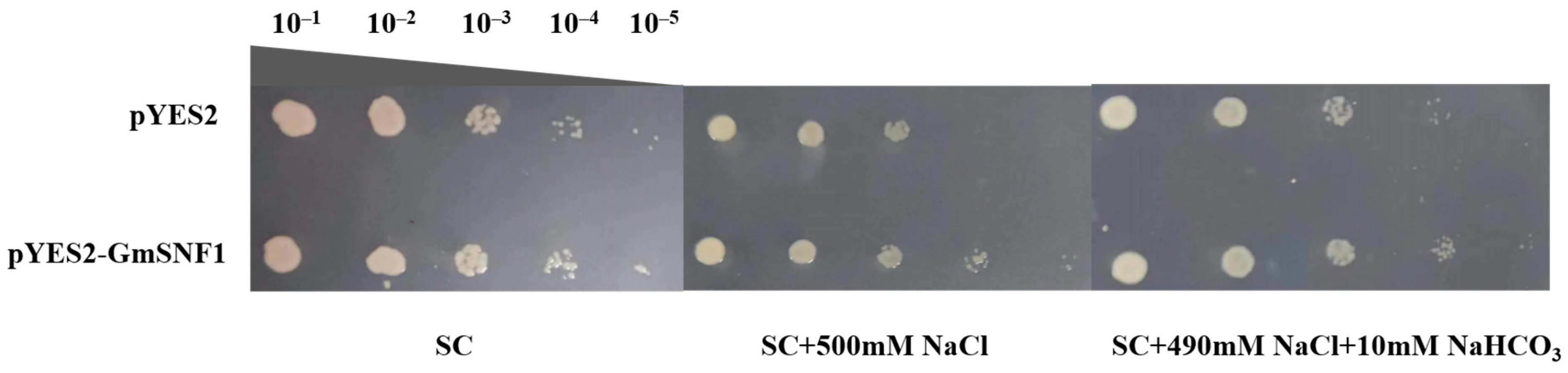
2.4. Heterologous Expression of the GmSNF1 Gene in a Yeast System Enhances Salt and Salt–Alkali Tolerance of Yeast Strains
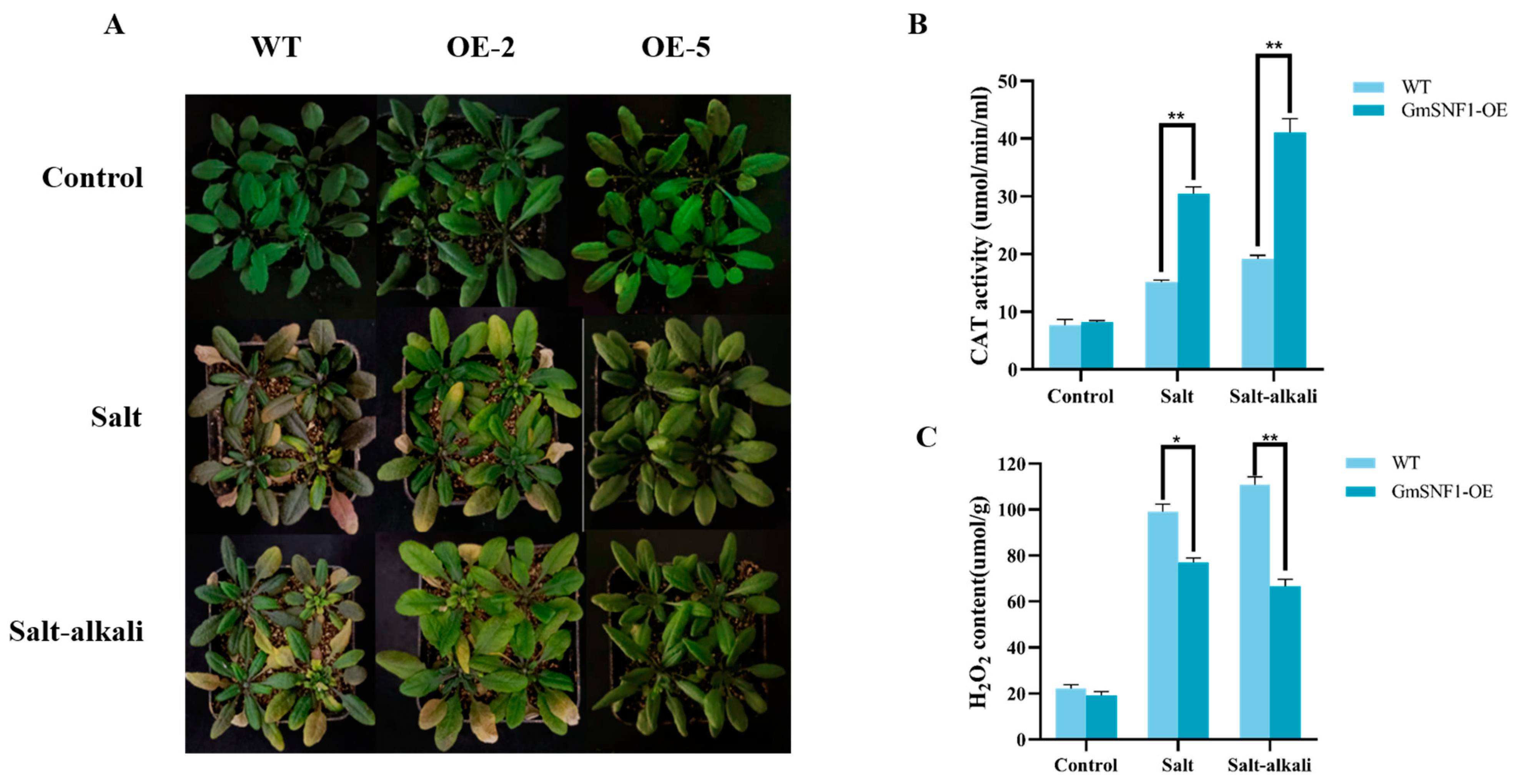
2.5. GmSNF1 Overexpression Enhances Salt and Salt–Alkali Stress Tolerance in Arabidopsis Plants
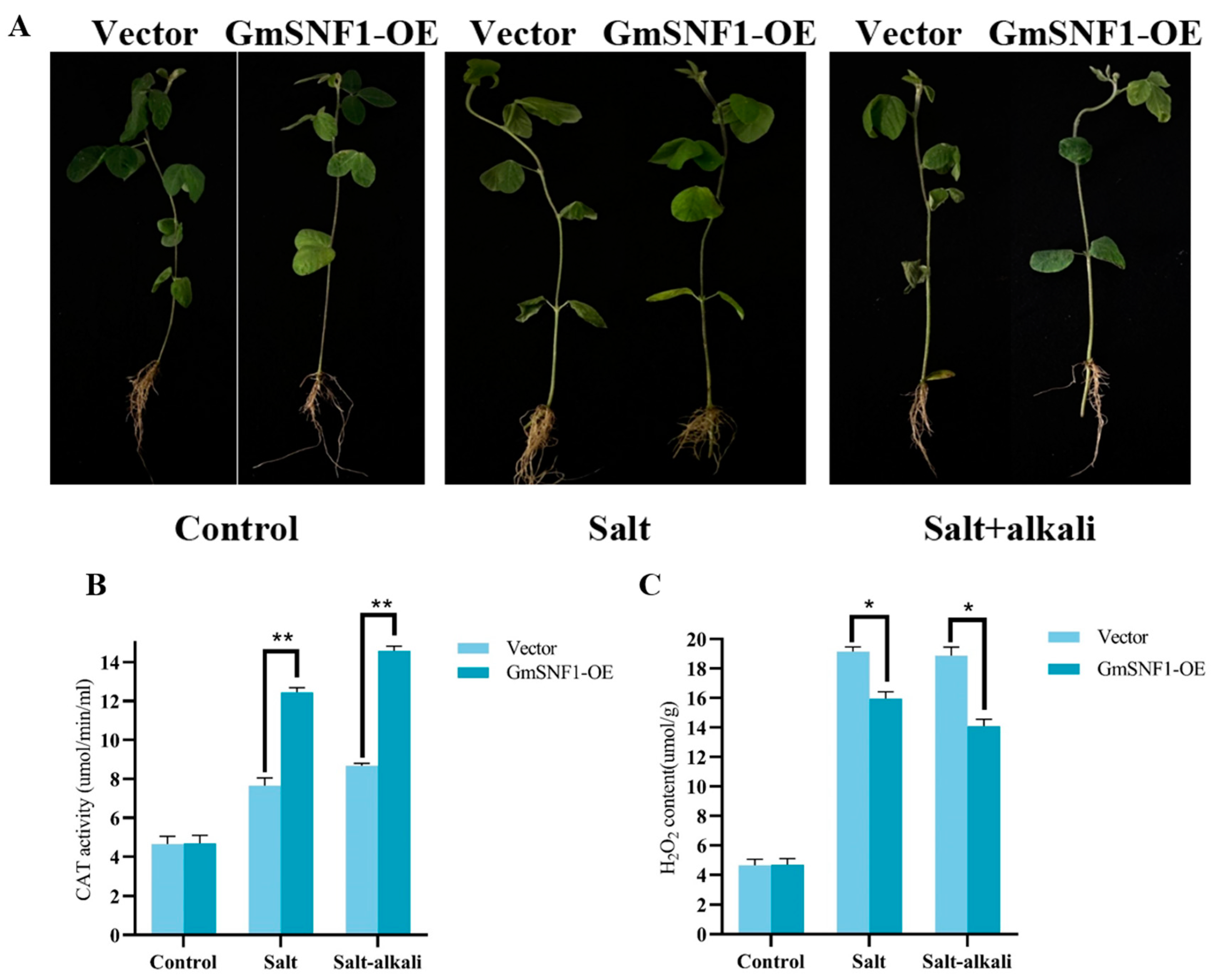
2.6. GmSNF1 Overexpression Enhances Salt and Salt-Alkali Stress Tolerance in Soybean Transgenic Hairy Root Complex Plants
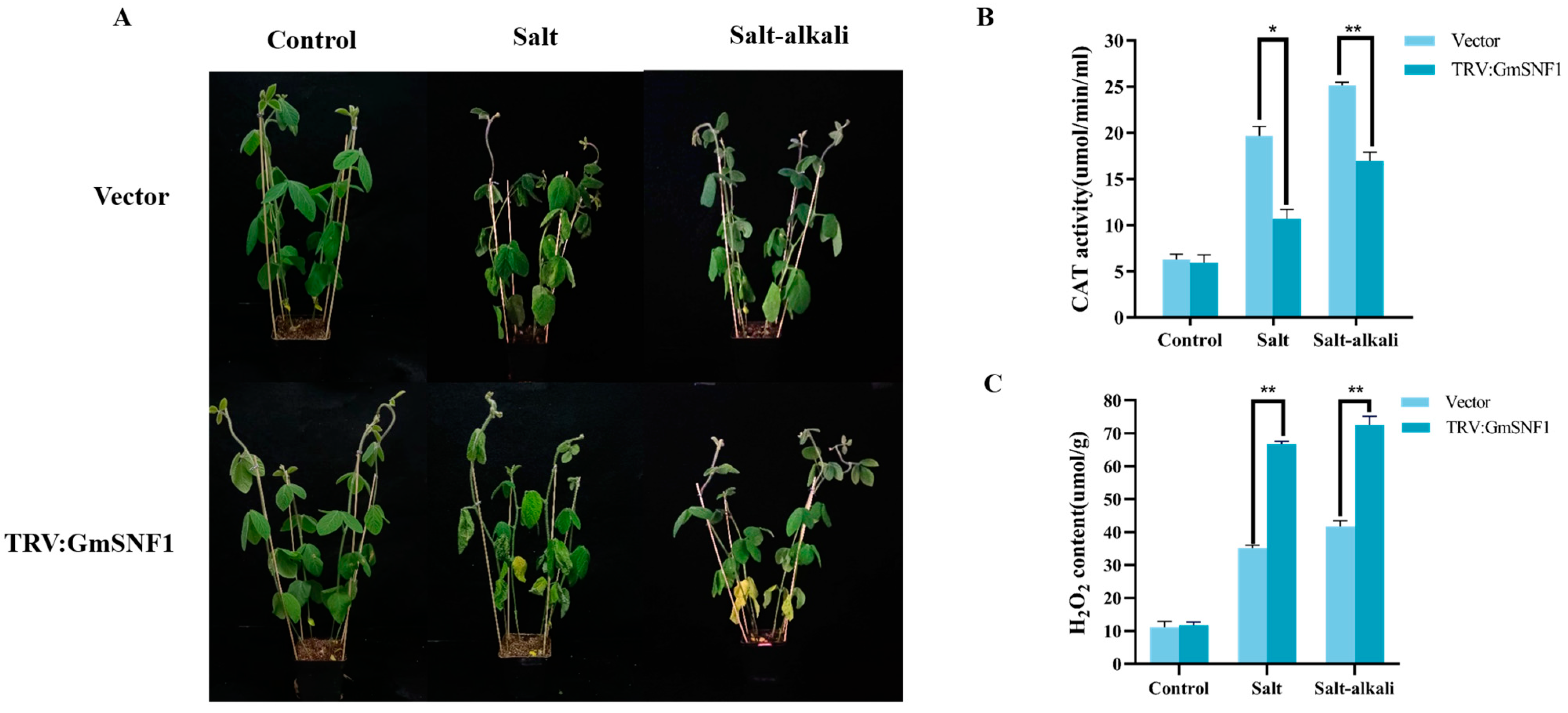
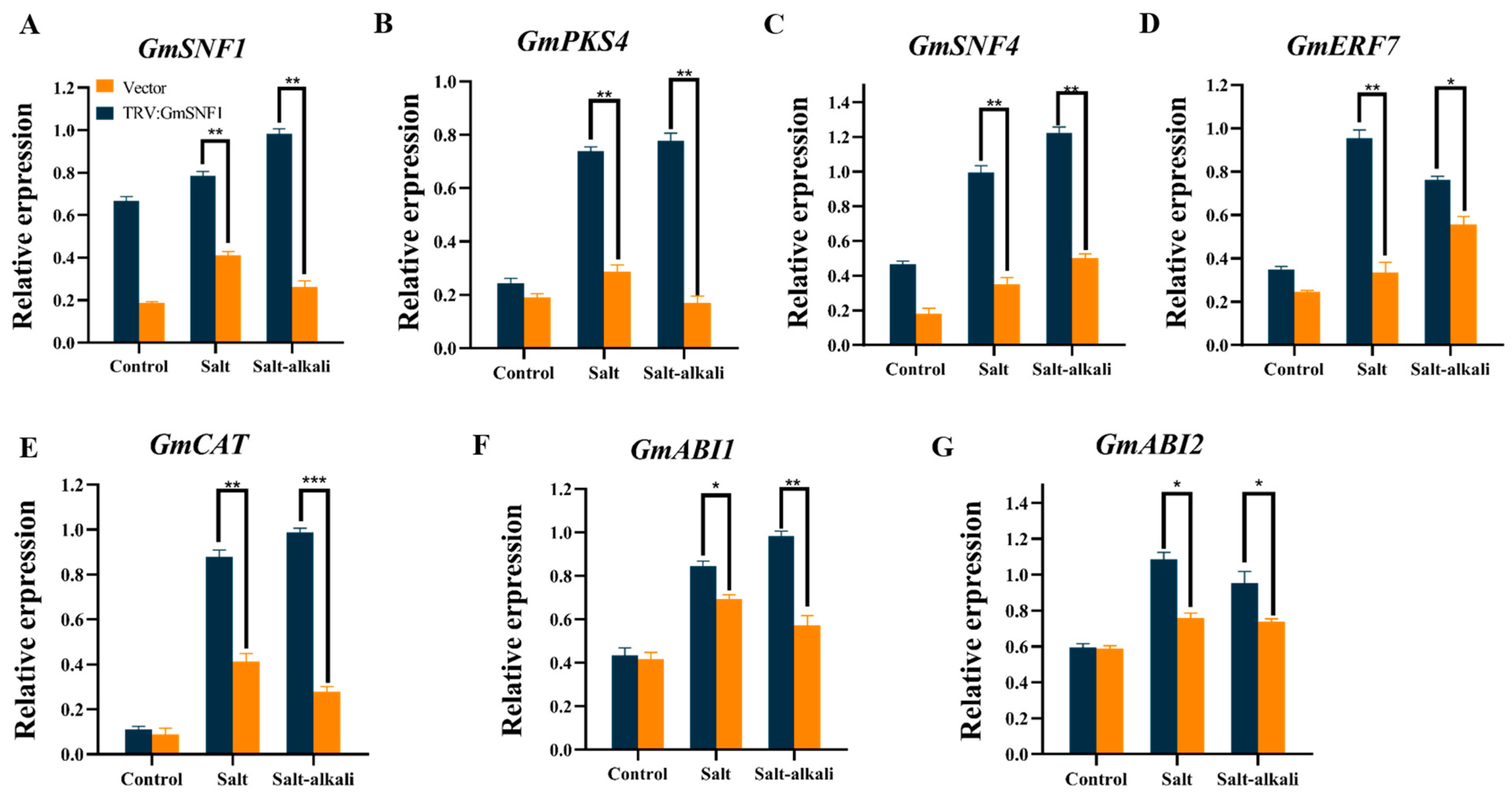
2.7. GmSNF1 Gene Silencing Enhances the Sensitivity of Soybean to Salt and Salt-Alkali Stress
3. Discussion
4. Materials and Methods
4.1. Yeast Two-Hybrid Validation
4.2. BiFC Verification
4.3. Plant Materials
4.4. qRT-PCR
4.5. Cloning and Subcellular Localization Analysis of the GmSNF1 Gene
4.6. Heterologous Expression of GmSNF1 in the Yeast System
4.7. Overexpression of the GmSNF1 Gene in A. thaliana and Analysis of Stress Tolerance
4.8. Stress Tolerance Analysis of Soybean Hairy Root Complex Plants Overexpressing the GmSNF1 Gene
4.9. VIGS Technology Silences GmSNF1 Gene
4.10. Stress-Related Gene Expression Analysis
4.11. Measurement of Physiological Indicators
4.12. Statistics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ding, X.; Liu, B.; Liu, H.; Sun, X.; Sun, X.; Wang, W.; Zheng, C. A new CIPK gene CmCIPK8 enhances salt tolerance in transgenic chrysanthemum. Sci. Hortic. 2023, 308, 111562. [Google Scholar] [CrossRef]
- Jiao, Y.; Bai, Z.; Xu, J.; Zhao, M.; Khan, Y.; Hu, Y.; Shi, L. Metabolomics and its physiological regulation process reveal the salt tolerant mechanism in Glycine soja seedling roots. Plant Physiol. Biochem. 2018, 126, 187–196. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, H.; Qiu, Z.; Hu, B.; Zeng, B.; Zhong, C.; Fan, C. Comprehensive Analysis of SnRK Gene Family and their Responses to Salt Stress in Eucalyptus grandis. Int. J. Mol. Sci. 2019, 20, 2786. [Google Scholar] [CrossRef]
- Wang, W.R.; Liang, J.H.; Wang, G.F.; Sun, M.X.; Xiao, Y.S. Overexpression of PpSnRK1α in tomato enhanced salt tolerance by regulating ABA signaling pathway and reactive oxygen metabolism. BMC Plant Biol. 2020, 20, 128. [Google Scholar] [CrossRef]
- de Dios Barajas-Lopez, J.; Moreno, J.R.; Gamez-Arjona, F.M.; Pardo, J.M.; Punkkinen, M.; Zhu, J.-K.; Quintero, F.J.; Fujii, H. Upstream kinases of plant Sn RKRK s are involved in salt stress tolerance. Plant J. 2018, 93, 107–118. [Google Scholar] [CrossRef]
- Ketehouli, T.; Zhou, Y.; Dai, S.; Carther, K.F.I.; Sun, D.; Li, Y.; Nguyen, Q.V.H.; Xu, H.; Wang, F.; Liu, W.; et al. A soybean calcineurin B-like protein-interacting protein kinase, GmPKS4, regulates plant responses to salt and alkali stresses. J. Plant Physiol. 2021, 256, 153331. [Google Scholar] [CrossRef]
- Coccetti, P.; Nicastro, R.; Tripodi, F. Saccharomyces cerevisiaeConventional and emerging roles of the energy sensor Snf1/AMPK in Saccharomyces cerevisiae. Microb. Cell 2018, 5, 482–494. [Google Scholar] [CrossRef]
- Hardie, D.G.; Schaffer, B.E.; Brunet, A. AMPK: An Energy-Sensing Pathway with Multiple Inputs and Outputs. Trends Cell Biol. 2016, 26, 190–201. [Google Scholar] [CrossRef]
- Emanuelle, S.; Doblin, M.S.; Stapleton, D.I.; Bacic, A.; Gooley, P.R. Molecular Insights into the Enigmatic Metabolic Regulator, SnRK1. Trends Plant Sci. 2016, 21, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Huang, T.; Zhou, C.; Chen, W.; Cha, J.; Wei, X.; Xing, F.; Qian, M.; Ma, Q.; Duan, H.; et al. Characterization of subunits encoded by Sn RK1 and dissection of combinations among these subunits in sorghum(Sorghum bicolor L.). J. Agric. Sci.-Sri Lanka 2023, 22, 8. [Google Scholar] [CrossRef]
- Han, C.; Liu, Y.; Shi, W.; Qiao, Y.; Bai, M.Y. KIN10 promotes stomatal development through stabilization of the SPEECHLESS transcription factor. Nat. Commun. 2020, 11, 4214. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Su, Z.-Z.; Huang, L.; Xia, F.-N.; Qi, H.; Xie, L.-J.; Xiao, S.; Chen, Q.-F. The AMP-Activated Protein Kinase KIN10 Is Involved in the Regulation of Autophagy in Arabidopsis. Front. Plant Sci. 2017, 8, 1201. [Google Scholar] [CrossRef]
- Hardie, D.G.; Carling, D.; Carlson, M. The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 1998, 67, 821–855. [Google Scholar] [CrossRef] [PubMed]
- Emanuelle, S.; Doblin, M.S.; Gooley, P.R.; Gentry, M.S. The UBA domain of SnRK1 promotes activation and maintains catalytic activity. Biochem. Biophys. Res. Commun. 2018, 497, 127–132. [Google Scholar] [CrossRef]
- Martínez-Barajas, E.; Coello, P. Review: How do SnRK1 protein kinases truly work? Plant Sci. 2020, 291, 110330. [Google Scholar] [CrossRef] [PubMed]
- Cordero, M.D.; Viollet, B. AMP-activated Protein Kinase Volume 107||Plant SnRK1 Kinases: Structure, Regulation, and Function. Exp. Suppl. 2016, 107, 403–438. [Google Scholar] [CrossRef]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef]
- Sun, D.; Fang, X.; Xiao, C.; Ma, Z.; He, K. Kinase SnRK1.1 Regulates nitrate channel SLAH3 Engaged in Nitrate-Dependent Alleviation of Ammonium Toxicity. Plant Physiol. 2021, 186, 731–749. [Google Scholar] [CrossRef]
- Pan, L. Role of the Phyllostachys edulis SnRK1a gene in plant growth and stress tolerance. S. Afr. J. Bot. 2020, 130, 414–421. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, L.; Wu, X.; Wang, S.; Zhang, P.; Li, M.; Jiang, J.; Ding, X.; Cao, X. Functional characterization of wild soybean (Glycine soja) GsSnRK1.1 protein kinase in plant resistance to abiotic stresses. J. Plant Physiol. 2023, 280, 153881. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Qiang, L.; Song, H.X.; Rong, X.M.; Ismail, A.M. Responses of Contrasting Rice (Oryza sativa L.) Genotypes to Salt Stress as Affected by Nutrient Concentrations. Chin. Agric. Sci. 2011, 10, 12. [Google Scholar] [CrossRef]
- Ye, Y.; Jia, X.; Xue, M.; Gao, Y.; Yue, H.; Ma, F.; Gong, X. MpSnRK2.10 confers salt stress tolerance in apple via the ABA signaling pathway. Sci. Hortic. 2022, 298, 110998. [Google Scholar] [CrossRef]
- Chen, N.; Pan, L.; Yang, Z.; Su, M.; Xu, J.; Jiang, X.; Yin, X.; Wang, T.; Wan, F.; Chi, X. ArabidopsisA MYB-related transcription factor from peanut, AhMYB30, improves freezing and salt stress tolerance in transgenic through both DREB/CBF and ABA-signaling pathways. Front. Plant Sci. 2023, 14, 1136626. [Google Scholar] [CrossRef]
- Quintero, F.J.; Martinez-Atienza, J.; Villalta, I.; Jiang, X.; Kim, W.; Ali, Z.; Fujii, H.; Mendoza, I.; Yun, D.; Zhu, J.; et al. Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. USA 2011, 108, 2611–2616. [Google Scholar] [CrossRef]
- Liu, X.; An, X.; Liu, X.; Hu, D.; Wang, X.; You, C.; Hao, Y. MdSnRK1.1 interacts with MdJAZ18 to regulate sucrose-induced anthocyanin and proanthocyanidin accumulation in apple. J. Exp. Bot. 2017, 68, 2977–2990. [Google Scholar] [CrossRef]
- Tsai, A.Y.; Gazzarrini, S. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 2012, 69, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef] [PubMed]
- Polge, C.; Thomas, M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007, 12, 20–28. [Google Scholar] [CrossRef]
- Zhao, Y.-F.; Han, Y.-Q.; Hou, S.-P.; Liu, Z.-W.; Han, Y.-H.; Li, Y.-Q. Genome-wide characterization of SNF1-related kinase1 (SnRK1) gene and its expression under sugar signaling and abiotic stress in cereal grains. J. Plant Physiol. 2022, 58, 18. [Google Scholar] [CrossRef]
- Nietzsche, M.; Schießl, I.; Börnke, F. The complex becomes more complex: Protein-protein interactions of SnRK1 with DUF581 family proteins provide a framework for cell- and stimulus type-specific SnRK1 signaling in plants. Front. Plant Sci. 2014, 5, 54. [Google Scholar] [CrossRef]
- Mair, A.; Pedrotti, L.; Wurzinger, B.; Anrather, D.; Simeunovic, A.; Weiste, C.; Valerio, C.; Dietrich, K.; Kirchler, T.; Nägele, T. SnRK1-triggered switch of bZIP63 dimerization mediates the low-energy response in plants. Elife 2015, 4, e5828. [Google Scholar] [CrossRef]
- Cho, H.Y.; Wen, T.N.; Wang, Y.T.; Shih, M.C. Quantitative phosphoproteomics of protein kinase SnRK1 regulated protein phosphorylation in Arabidopsis under submergence. J. Exp. Bot. 2016, 67, 2745–2760. [Google Scholar] [CrossRef]
- Nukarinen, E.; Nägele, T.; Pedrotti, L.; Wurzinger, B.; Mair, A.; Landgraf, R.; Börnke, F.; Hanson, J.; Teige, M.; Baena-Gonzalez, E.; et al. Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 2016, 6, 31697. [Google Scholar] [CrossRef]
- Zhao, T.T.; Wang, J.G.; Wang, W.Z.; Feng, C.L.; Feng, X.Y.; Shen, L.B.; Zhang, S.Z. Structure, Intracellular Localisation and Expression Analysis of Sucrose Nonfermenting-Related Kinase ShSnRK1α in Sugarcane. Sugar Tech. 2023, 25, 69–76. [Google Scholar] [CrossRef]
- Blanco, N.E.; Liebsch, D.; Díaz, M.G.; Strand, S.; Whelan, J. Dual and dynamic intracellular localization of Arabidopsis thaliana SnRK1.1. J. Exp. Bot. 2019, 70, 2325–2338. [Google Scholar] [CrossRef]
- Teymennet-Ramírez, K.V.; Martínez-Morales, F.; Trejo-Hernández, M.R. Yeast Surface Display System: Strategies for Improvement and Biotechnological Applications. Front. Bioeng. Biotechnol. 2021, 9, 794742. [Google Scholar] [CrossRef] [PubMed]
- Qiao, K.; Wang, M.; Takano, T.; Liu, S. Overexpression of Acyl-CoA-Binding Protein 1 (ChACBP1) From Saline-Alkali-Tolerant Chlorella sp. Enhances Stress Tolerance in Arabidopsis. Front. Plant Sci. 2018, 9, 1772. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhu, Y.; Li, W.; Zhang, T.; Li, Y.; Kang, Y.; Wang, J.; Guo, J.; Jiang, X. Heterologous expression ofSesuvium portulacastrum SOS-related genes confer salt tolerance in yeast. Acta Physiol. Plant. 2023, 45, 58. [Google Scholar] [CrossRef]
- Meng, L.; Liu, H.L.; Lin, X.; Hu, X.P.; Teng, K.R.; Liu, S.X. Enhanced multi-stress tolerance and glucose utilization of Saccharomyces cerevisiae by overexpression of the SNF1 gene and varied beta isoform of Snf1 dominates in stresses. Microb. Cell Factories 2020, 19, 134. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Chan, Z. ROS Regulation During Abiotic Stress Responses in Crop Plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef]
- Jin, X.; Liu, Z.; Wu, W. POD, CAT and SOD enzyme activity of corn kernels as affected by low plasma pretreatment. Int. J. Food Prop. 2022, 26, 38–48. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Yan, X.; Guo, J. Physiological and transcriptomic analyses of yellow horn (Xanthoceras sorbifolia) provide important insights into salt and saline-alkali stress tolerance. PLoS ONE 2020, 15, e244365. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Sun, Y.; Shan, Z.; He, J.; Wang, N.; Gai, J.; Li, Y. Natural variation in the promoter of GsERD15B affects salt tolerance in soybean. Plant Biotechnol. J. 2021, 19, 1155–1169. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Wang, Y.; Li, Y.; Lei, T.; Yan, F.; Su, L.; Li, X.; Zhao, Y.; Sun, X.; Li, J. Isolation and molecular characterization of GmERF7, a soybean ethylene-response factor that increases salt stress tolerance in tobacco. Gene 2013, 513, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, P.; Dai, S.-Y.; Yong, L.-T.; Zhou, B.-H.; Wang, N.; Dong, Y.-Y.; Liu, W.-C.; Wang, F.-W.; Yang, H.-Y.; Li, X.-W. A Soybean Sucrose Non-Fermenting Protein Kinase 1 Gene, GmSNF1, Positively Regulates Plant Response to Salt and Salt–Alkali Stress in Transgenic Plants. Int. J. Mol. Sci. 2023, 24, 12482. https://doi.org/10.3390/ijms241512482
Lu P, Dai S-Y, Yong L-T, Zhou B-H, Wang N, Dong Y-Y, Liu W-C, Wang F-W, Yang H-Y, Li X-W. A Soybean Sucrose Non-Fermenting Protein Kinase 1 Gene, GmSNF1, Positively Regulates Plant Response to Salt and Salt–Alkali Stress in Transgenic Plants. International Journal of Molecular Sciences. 2023; 24(15):12482. https://doi.org/10.3390/ijms241512482
Chicago/Turabian StyleLu, Ping, Si-Yu Dai, Ling-Tao Yong, Bai-Hui Zhou, Nan Wang, Yuan-Yuan Dong, Wei-Can Liu, Fa-Wei Wang, Hao-Yu Yang, and Xiao-Wei Li. 2023. "A Soybean Sucrose Non-Fermenting Protein Kinase 1 Gene, GmSNF1, Positively Regulates Plant Response to Salt and Salt–Alkali Stress in Transgenic Plants" International Journal of Molecular Sciences 24, no. 15: 12482. https://doi.org/10.3390/ijms241512482
APA StyleLu, P., Dai, S.-Y., Yong, L.-T., Zhou, B.-H., Wang, N., Dong, Y.-Y., Liu, W.-C., Wang, F.-W., Yang, H.-Y., & Li, X.-W. (2023). A Soybean Sucrose Non-Fermenting Protein Kinase 1 Gene, GmSNF1, Positively Regulates Plant Response to Salt and Salt–Alkali Stress in Transgenic Plants. International Journal of Molecular Sciences, 24(15), 12482. https://doi.org/10.3390/ijms241512482




