Transcriptome Analysis Reveals the Molecular Basis of Overfeeding-Induced Diabetes in Zebrafish
Abstract
1. Introduction
2. Results
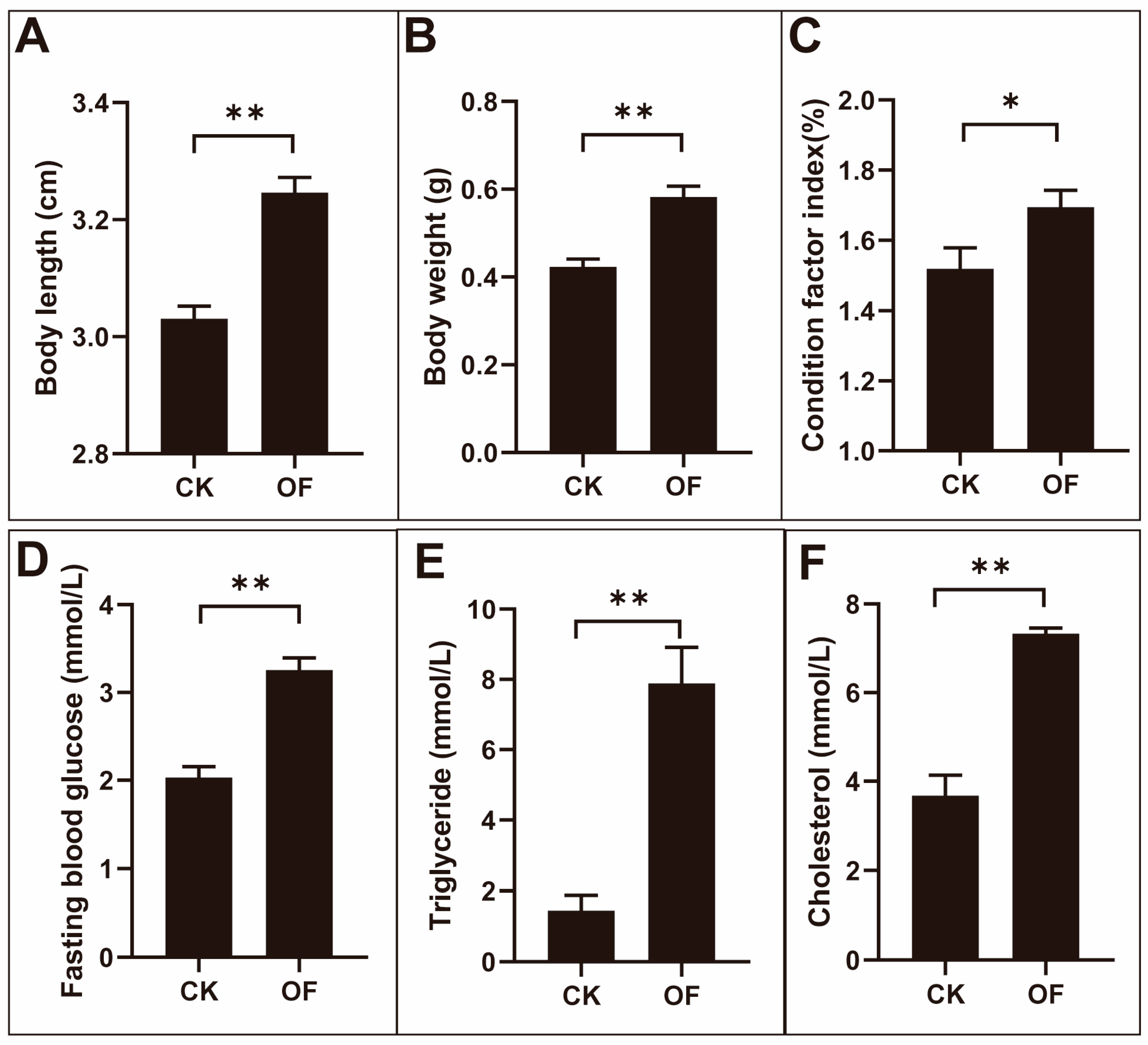
2.1. The Elevation of Fasting Blood Glucose Levels in Male Zebrafish after Four Weeks of Overfeeding
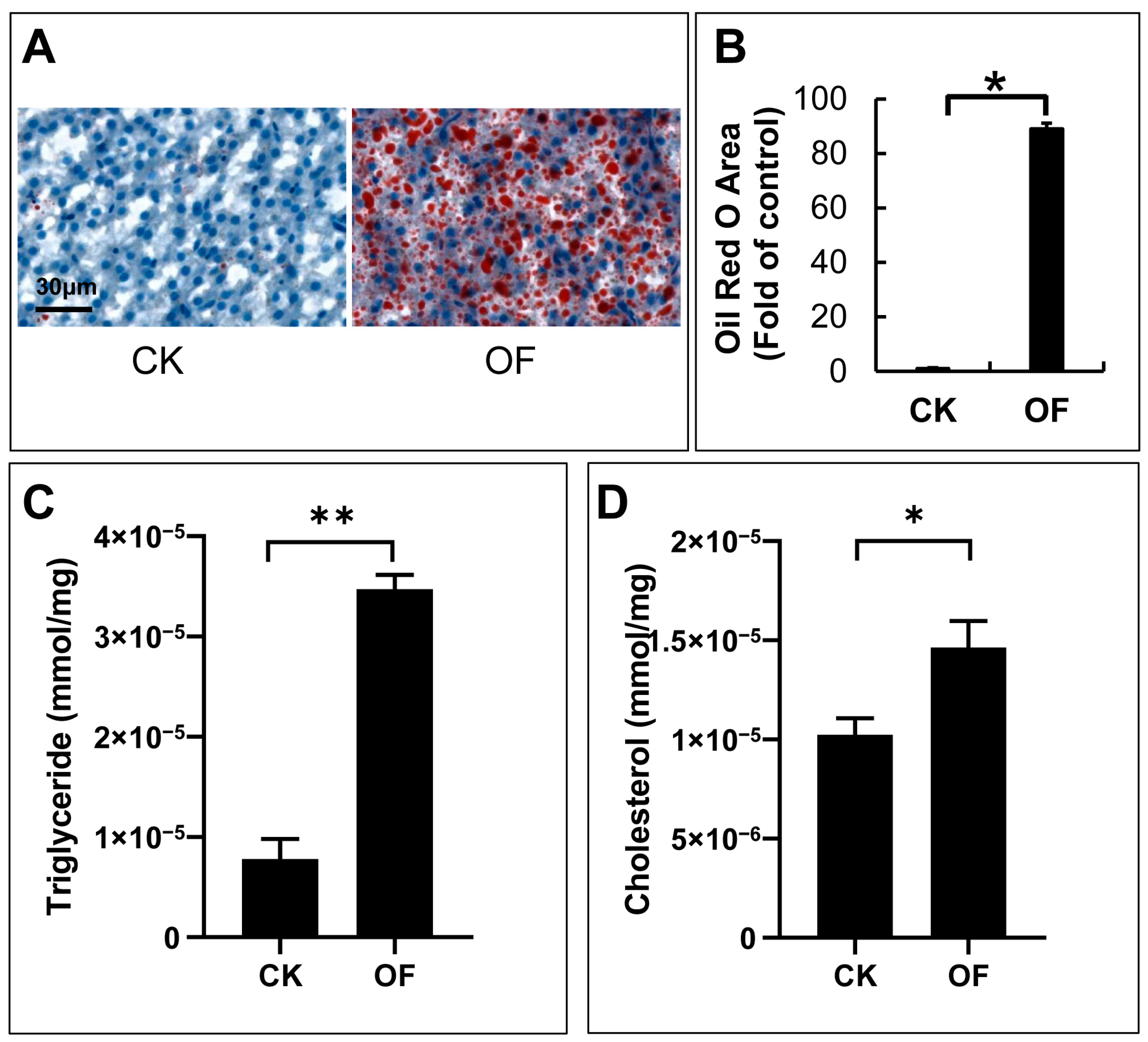
2.2. The Hepatic Accumulation of Lipid Droplets in Male Zebrafish after Four Weeks of Overfeeding
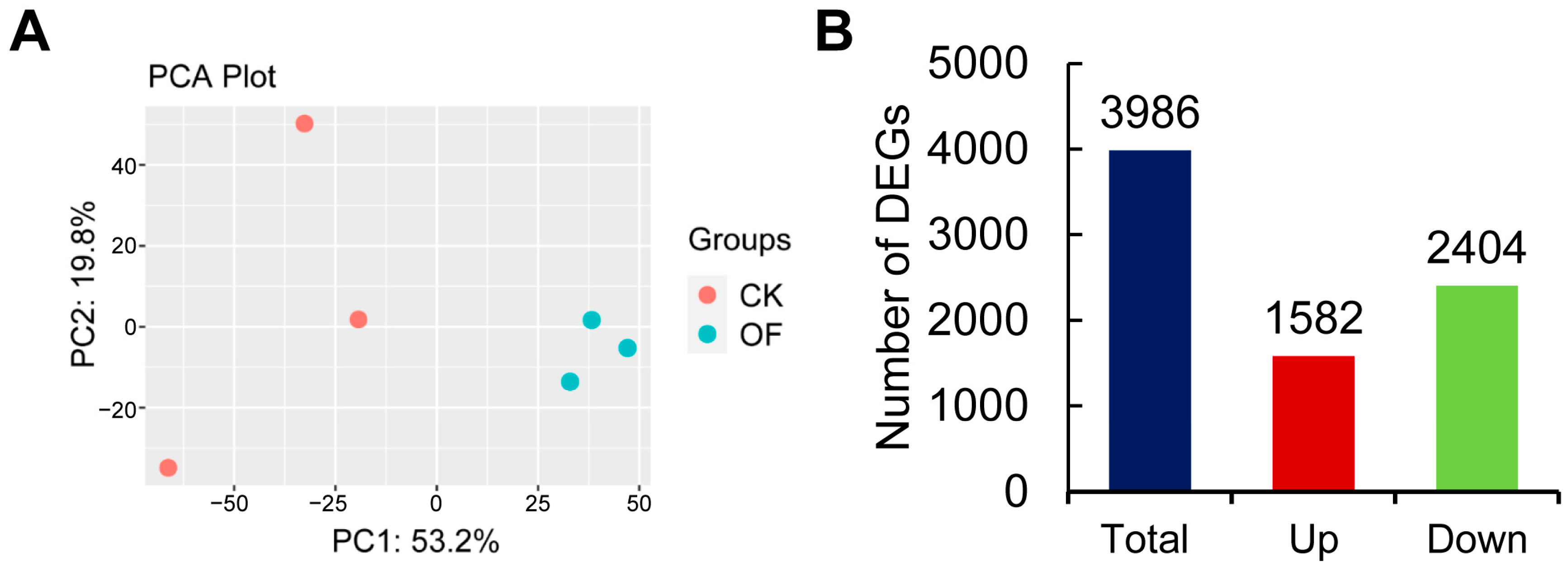
2.3. RNA-Seq Analysis to Identify Differentially Expressed Genes
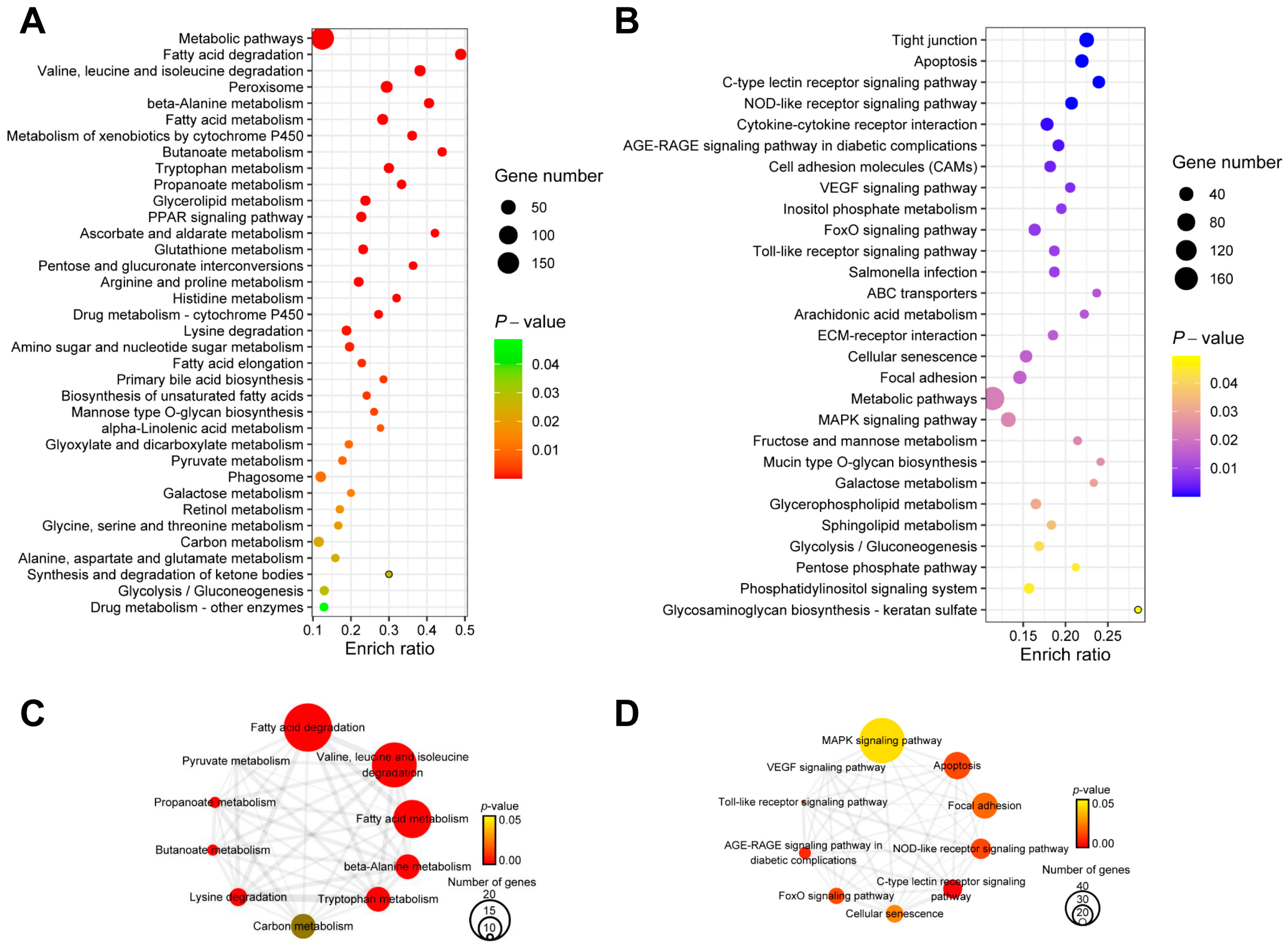
2.4. KEGG Enrichment Analysis for DEGs
2.5. GO Enrichment Analysis of DEGs
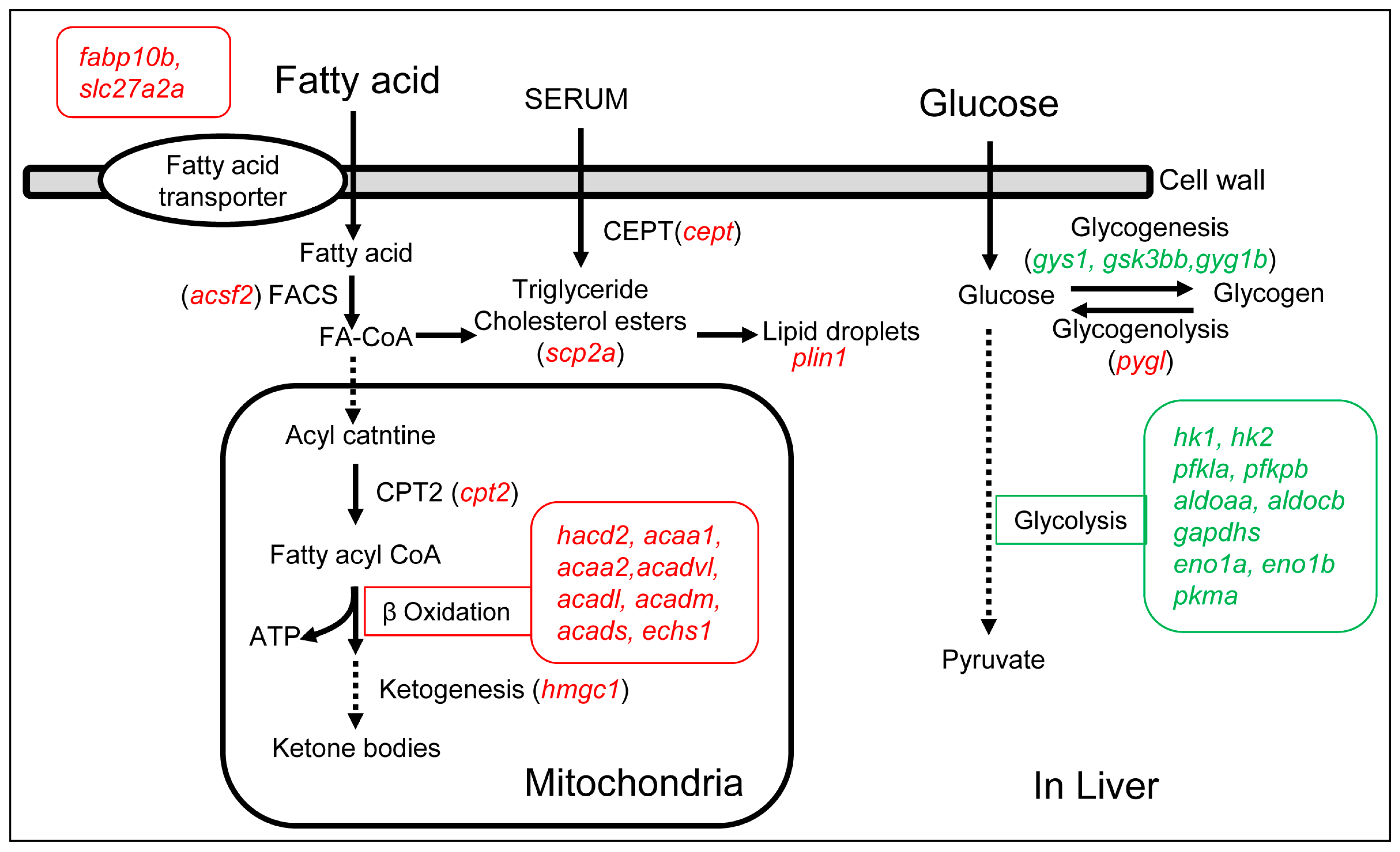
2.6. Overfeeding Up-Regulated Fatty Acid Metabolism Genes and Down-Regulated Glucose Metabolism Genes in Zebrafish Liver
3. Discussion
4. Materials and Methods
4.1. Methods of Overfeeding Experiments
4.2. Blood Collection and Measurement
4.3. Staining with Oil Red O and Histology
4.4. Measurement of Total Cholesterol and Triglyceride
4.5. Sample Collection and Analysis for RNA-Sequencing
4.6. Bioinformatics Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.N.; Mbanya, J.C.; et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef] [PubMed]
- DeFronzo, R.A.; Ferrannini, E.; Groop, L.; Henry, R.R.; Herman, W.H.; Holst, J.J.; Hu, F.B.; Kahn, C.R.; Raz, I.; Shulman, G.I.; et al. Type 2 diabetes mellitus. Nat. Rev. Dis. Primers 2015, 1, 15019. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2009, 32 (Suppl. S1), S62–S67. [Google Scholar] [CrossRef] [PubMed]
- Otero, Y.F.; Stafford, J.M.; McGuinness, O.P. Pathway-selective Insulin Resistance and Metabolic Disease: The Importance of Nutrient Flux*. J. Biol. Chem. 2014, 289, 20462–20469. [Google Scholar] [CrossRef]
- Wu, X.; Williams, K.J. NOX4 pathway as a source of selective insulin resistance and responsiveness. Arter. Thromb. Vasc. Biol. 2012, 32, 1236–1245. [Google Scholar] [CrossRef]
- American Diabetes Association. 2. Classification and Diagnosis of Diabetes. Diabetes Care 2017, 40 (Suppl. S1), S11–S24. [Google Scholar]
- Perego, C.; Da Dalt, L.; Pirillo, A.; Galli, A.; Catapano, A.L.; Norata, G.D. Cholesterol metabolism, pancreatic β-cell function and diabetes. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2019, 1865, 2149–2156. [Google Scholar] [CrossRef]
- Hiukka, A.; Fruchart-Najib, J.; Leinonen, E.; Hilden, H.; Fruchart, J.-C.; Taskinen, M.-R. Alterations of lipids and apolipoprotein CIII in very low density lipoprotein subspecies in type 2 diabetes. Diabetologia 2005, 48, 1207–1215. [Google Scholar] [CrossRef]
- Bhowmik, B.; Siddiquee, T.; Mujumder, A.; Afsana, F.; Ahmed, T.; Mdala, I.A.; Moreira, N.C.D.V.; Khan, A.K.A.; Hussain, A.; Holmboe-Ottesen, G.; et al. Serum Lipid Profile and Its Association with Diabetes and Prediabetes in a Rural Bangladeshi Population. Int. J. Environ. Res. Public Health 2018, 15, 1944. [Google Scholar] [CrossRef]
- Wang, L.; Yan, N.; Zhang, M.; Pan, R.; Dang, Y.; Niu, Y. The association between blood glucose levels and lipids or lipid ratios in type 2 diabetes patients: A cross-sectional study. Front. Endocrinol. 2022, 13, 969080. [Google Scholar] [CrossRef]
- Taskinen, M.-R.; Borén, J. New insights into the pathophysiology of dyslipidemia in type 2 diabetes. Atherosclerosis 2015, 239, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Vergès, B. Abnormal hepatic apolipoprotein B metabolism in type 2 diabetes. Atherosclerosis 2010, 211, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Lazar, M.A. Dissociating fatty liver and diabetes. Trends Endocrinol. Metab. 2012, 24, 4–12. [Google Scholar] [CrossRef]
- Lee, I.-M.; Shiroma, E.J.; Lobelo, F.; Puska, P.; Blair, S.N.; Katzmarzyk, P.T.; Kahlmeier, S.; the Lancet Physical Activity Series Working Group. Effect of physical inactivity on major non-communicable diseases worldwide: An analysis of burden of disease and life expectancy. Lancet 2012, 380, 219–229. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Su, Y.; You, Y.; Ma, Y.; Yang, G.; Song, Y.; Liu, X.; Wang, M.; Zhang, L.; et al. The metabolic side effects of 12 antipsychotic drugs used for the treatment of schizophrenia on glucose: A network meta-analysis. BMC Psychiatry 2017, 17, 373. [Google Scholar] [CrossRef]
- Malik, V.S.; Popkin, B.M.; Bray, G.A.; Despres, J.P.; Hu, F.B. Sugar-sweetened beverages, obesity, type 2 diabetes mellitus, and cardiovascular disease risk. Circulation 2010, 121, 1356–1364. [Google Scholar] [CrossRef] [PubMed]
- Malik, V.S.; Popkin, B.M.; Bray, G.A.; Despres, J.P.; Willett, W.C.; Hu, F.B. Sugar-sweetened beverages and risk of metabolic syndrome and type 2 diabetes: A meta-analysis. Diabetes Care 2010, 33, 2477–2483. [Google Scholar] [CrossRef] [PubMed]
- Wild, S.H.; Roglic, G.; Green, A.; Sicree, R.; King, H. Global Prevalence of Diabetes: Estimates for the Year 2000 and Projections for 2030. Diabetes Care 2004, 27, 2569. [Google Scholar] [CrossRef]
- Risérus, U.; Willett, W.C.; Hu, F.B. Dietary fats and prevention of type 2 diabetes. Prog. Lipid Res. 2009, 48, 44–51. [Google Scholar] [CrossRef]
- Hu, E.A.; Pan, A.; Malik, V.; Sun, Q. White rice consumption and risk of type 2 diabetes: Meta-analysis and systematic review. BMJ 2012, 344, e1454. [Google Scholar] [CrossRef]
- Carris, N.W.; Magness, R.R.; Labovitz, A.J. Prevention of Diabetes Mellitus in Patients with Prediabetes. Am. J. Cardiol. 2019, 123, 507–512. [Google Scholar] [CrossRef]
- Elo, B.; Villano, C.M.; Govorko, D.; White, L.A. Larval zebrafish as a model for glucose metabolism: Expression of phos-phoenolpyruvate carboxykinase as a marker for exposure to anti-diabetic compounds. J. Mol. Endocrinol. 2007, 38, 433–440. [Google Scholar] [CrossRef]
- Zang, L.; Shimada, Y.; Nishimura, N. Development of a Novel Zebrafish Model for Type 2 Diabetes Mellitus. Sci. Rep. 2017, 7, 1461. [Google Scholar] [CrossRef]
- Wang, G.; Rajpurohit, S.K.; Delaspre, F.; Walker, S.L.; White, D.T.; Ceasrine, A.; Kuruvilla, R.; Li, R.J.; Shim, J.S.; Liu, J.O.; et al. First quantitative high-throughput screen in zebrafish identifies novel pathways for increasing pancreatic β-cell mass. Elife 2015, 4, e08261. [Google Scholar] [CrossRef] [PubMed]
- Choi, T.-Y.; Choi, T.-I.; Lee, Y.-R.; Choe, S.-K.; Kim, C.-H. Zebrafish as an animal model for biomedical research. Exp. Mol. Med. 2021, 53, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Baek, J.S.; Fang, L.; Li, A.C.; Miller, Y.I. Ezetimibe and Simvastatin Reduce Cholesterol Levels in Zebrafish Larvae Fed a High-Cholesterol Diet. Cholesterol 2012, 2012, 564705. [Google Scholar] [CrossRef] [PubMed]
- Pedroso, G.L.; Hammes, T.O.; Escobar, T.D.; Fracasso, L.B.; Forgiarini, L.F.; da Silveira, T.R. Blood collection for biochemical analysis in adult zebrafish. J. Vis. Exp. JoVE 2012, 63, e3865. [Google Scholar]
- Zang, L.; Maddison, L.A.; Chen, W. Zebrafish as a Model for Obesity and Diabetes. Front. Cell Dev. Biol. 2018, 6, 91. [Google Scholar] [CrossRef]
- Salehpour, A.; Rezaei, M.; Khoradmehr, A.; Tahamtani, Y.; Tamadon, A. Which Hyperglycemic Model of Zebrafish (Danio rerio) Suites My Type 2 Diabetes Mellitus Research? A Scoring System for Available Methods. Front. Cell Dev. Biol. 2021, 9, 652061. [Google Scholar] [CrossRef]
- Arjmand, B.; Alavi-Moghadam, S.; Kokabi-Hamidpour, S.; Arjmand, R.; Rezaei-Tavirani, M.; Larijani, B.; Goodarzi, P.; Mehrdad, N.; Rajaeinejad, M. Development and Validation of Type 2 Diabetic Zebrafish Model for Cell-Based Treatments. In Methods in Molecular Biology; Springer: New York, NY, USA, 2023; pp. 1–11. [Google Scholar] [CrossRef]
- Kim, I.; Seok, S.H.; Lee, H.-Y. Development of a Zebrafish Larvae Model for Diabetic Heart Failure with Reduced Ejection Fraction. Korean Circ. J. 2023, 53, 34–46. [Google Scholar] [CrossRef]
- Zang, L.; Shimada, Y.; Nishimura, Y.; Tanaka, T.; Nishimura, N.; Michel, M.; Page-McCaw, P.S.; Chen, W.; Cone, R.D.; Kuninaga, S.; et al. A Novel, Reliable Method for Repeated Blood Collection from Aquarium Fish. Zebrafish 2013, 10, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.C.; Harhay, M.O.; Harhay, M.N. The Value of Anthropometric Measures in Nutrition and Metabolism: Comment on Anthropometrically Predicted Visceral Adipose Tissue and Blood-Based Biomarkers: A Cross-Sectional Analysis. Nutr. Metab. Insights 2019, 12, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Kleinert, M.; Clemmensen, C.; Hofmann, S.M.; Moore, M.C.; Renner, S.; Woods, S.C.; Huypens, P.; Beckers, J.; de Angelis, M.H.; Schürmann, A.; et al. Animal models of obesity and diabetes mellitus. Nat. Rev. Endocrinol. 2018, 14, 140–162. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Liu, H.; Yu, J.; He, S.; Li, P.; Ma, C.; Zhang, H.; Xu, L.; Ping, F.; Li, W.; et al. Triglyceride is independently correlated with insulin resistance and islet beta cell function: A study in population with different glucose and lipid metabolism states. Lipids Health Dis. 2020, 19, 121. [Google Scholar] [CrossRef]
- Gleeson, M.; Connaughton, V.; Arneson, L.S. Induction of hyperglycaemia in zebrafish (Danio rerio) leads to morphological changes in the retina. Acta Diabetol. 2007, 44, 157–163. [Google Scholar] [CrossRef]
- Alvarez, Y.; Chen, K.; Reynolds, A.L.; Waghorne, N.; O’Connor, J.J.; Kennedy, B.N. Predominant cone photoreceptor dys-function in a hyperglycaemic model of non-proliferative diabetic retinopathy. Dis. Models Mech. 2010, 3, 236–245. [Google Scholar] [CrossRef]
- Carnovali, M.; Luzi, L.; Banfi, G.; Mariotti, M. Chronic hyperglycemia affects bone metabolism in adult zebrafish scale model. Endocrine 2016, 54, 808–817. [Google Scholar] [CrossRef]
- Singh, A.; Castillo, H.A.; Brown, J.; Kaslin, J.; Dwyer, K.M.; Gibert, Y. High glucose levels affect retinal patterning during zebrafish embryogenesis. Sci. Rep. 2019, 9, 4121. [Google Scholar] [CrossRef]
- Mohammadi, H.; Manouchehri, H.; Changizi, R.; Bootorabi, F.; Khorramizadeh, M.R. Correction to: Concurrent metformin and silibinin therapy in diabetes: Assessments in zebrafish (Danio rerio) animal model. J. Diabetes Metab. Disord. 2020, 20, 1099. [Google Scholar] [CrossRef]
- Capiotti, K.M.; Antonioli, R., Jr.; Kist, L.W.; Bogo, M.R.; Bonan, C.D.; Da Silva, R.S. Persistent impaired glucose metabolism in a zebrafish hyperglycemia model. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2014, 171, 58–65. [Google Scholar] [CrossRef]
- Hu, F.B.; Manson, J.E.; Stampfer, M.J.; Colditz, G.; Liu, S.; Solomon, C.G.; Willett, W.C. Diet, Lifestyle, and the Risk of Type 2 Diabetes Mellitus in Women. N. Engl. J. Med. 2001, 345, 790–797. [Google Scholar] [CrossRef]
- Scherer, P.E. Adipose Tissue: From Lipid Storage Compartment to Endocrine Organ. Diabetes 2006, 55, 1537–1545. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Mao, Y.; Cui, T.; Tang, D.; Wang, X.L. Impact of a Combined High Cholesterol Diet and High Glucose Environment on Vasculature. PLoS ONE 2013, 8, e81485. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; Lai, K.; Zhong, Q.; Demin, K.A.; Kalueff, A.V.; Song, C. High-glucose/high-cholesterol diet in zebrafish evokes diabetic and affective pathogenesis: The role of peripheral and central inflammation, microglia and apoptosis. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2019, 96, 109752. [Google Scholar] [CrossRef] [PubMed]
- Kahn, S.E.; Hull, R.L.; Utzschneider, K.M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006, 444, 840–846. [Google Scholar] [CrossRef] [PubMed]
- James, D.E.; Stöckli, J.; Birnbaum, M.J. The aetiology and molecular landscape of insulin resistance. Nat. Rev. Mol. Cell Biol. 2021, 22, 751–771. [Google Scholar] [CrossRef]
- Chen, C.; Liu, D. Establishment of Zebrafish Models for Diabetes Mellitus and Its Microvascular Complications. J. Vasc. Res. 2022, 59, 251–260. [Google Scholar] [CrossRef]
- Samuel, V.T.; Shulman, G.I. Nonalcoholic Fatty Liver Disease, Insulin Resistance, and Ceramides. N. Engl. J. Med. 2019, 381, 1866–1869. [Google Scholar] [CrossRef]
- Kumar, A.; Sundaram, K.; Mu, J.; Dryden, G.W.; Sriwastva, M.K.; Lei, C.; Zhang, L.; Qiu, X.; Xu, F.; Yan, J.; et al. High-fat diet-induced upregulation of exosomal phosphatidylcholine contributes to insulin resistance. Nat. Commun. 2021, 12, 213–234. [Google Scholar] [CrossRef] [PubMed]
- Huebschmann, A.G.; Huxley, R.R.; Kohrt, W.M.; Zeitler, P.; Regensteiner, J.G.; Reusch, J.E.B. Sex differences in the burden of type 2 diabetes and cardiovascular risk across the life course. Diabetologia 2019, 62, 1761–1772. [Google Scholar] [CrossRef]
- Mauvais-Jarvis, F. Gender differences in glucose homeostasis and diabetes. Physiol. Behav. 2018, 187, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Huebbe, P.; Rimbach, G. Evolution of human apolipoprotein E (APOE) isoforms: Gene structure, protein function and interaction with dietary factors. Ageing Res. Rev. 2017, 37, 146–161. [Google Scholar] [CrossRef] [PubMed]
- Sahini, N.; Borlak, J. Recent insights into the molecular pathophysiology of lipid droplet formation in hepatocytes. Prog. Lipid Res. 2014, 54, 86–112. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-L.; Qian, Y.-C.; Limbu, S.M.; Wang, J.; Chen, L.-Q.; Zhang, M.-L.; Du, Z.-Y. Lipolysis and lipophagy play individual and interactive roles in regulating triacylglycerol and cholesterol homeostasis and mitochondrial form in zebrafish. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2021, 1866, 158988. [Google Scholar] [CrossRef] [PubMed]
- Verstraete, K.; Verschueren, K.H.G.; Dansercoer, A.; Savvides, S.N. Acetyl-CoA is produced by the citrate synthase homology module of ATP-citrate lyase. Nat. Struct. Mol. Biol. 2021, 28, 636–638. [Google Scholar] [CrossRef]
- Basak, S.; Mallick, R.; Banerjee, A.; Pathak, S.; Duttaroy, A.K. Chapter Five—Cytoplasmic fatty acid-binding proteins in metabolic diseases and cancers. In Advances in Protein Chemistry and Structural Biology; Donev, R., Ed.; Academic Press: Cambridge, MA, USA, 2022; Volume 132, pp. 143–174. [Google Scholar]
- Veech, R.L. The therapeutic implications of ketone bodies: The effects of ketone bodies in pathological conditions: Ketosis, ketogenic diet, redox states, insulin resistance, and mitochondrial metabolism. Prostaglandins Leukot. Essent. Fatty Acids 2004, 70, 309–319. [Google Scholar] [CrossRef]
- Karagianni, P.; Talianidis, I. Transcription factor networks regulating hepatic fatty acid metabolism. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2015, 1851, 2–8. [Google Scholar] [CrossRef]
- Pinti, M.V.; Fink, G.K.; Hathaway, Q.A.; Durr, A.J.; Kunovac, A.; Hollander, J.M. Mitochondrial dysfunction in type 2 diabetes mellitus: An organ-based analysis. Am. J. Physiol. Metab. 2019, 316, E268–E285. [Google Scholar] [CrossRef]
- Bhatti, J.S.; Bhatti, G.K.; Reddy, P.H. Mitochondrial dysfunction and oxidative stress in metabolic disorders—A step towards mitochondria based therapeutic strategies. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2017, 1863, 1066–1077. [Google Scholar] [CrossRef]
- Guo, X.; Li, H.; Xu, H.; Woo, S.; Dong, H.; Lu, F.; Lange, A.J.; Wu, C. Glycolysis in the control of blood glucose homeostasis. Acta Pharm. Sin. B 2012, 2, 358–367. [Google Scholar] [CrossRef]
- Dall’Agnese, A.; Platt, J.M.; Zheng, M.M.; Friesen, M.; Dall’Agnese, G.; Blaise, A.M.; Spinelli, J.B.; Henninger, J.E.; Tevonian, E.N.; Hannett, N.M.; et al. The dynamic clustering of insulin receptor underlies its signaling and is disrupted in insulin resistance. Nat. Commun. 2022, 13, 7522. [Google Scholar] [CrossRef] [PubMed]
- Merry, T.L.; Kuhlow, D.; Laube, B.; Pöhlmann, D.; Pfeiffer, A.F.H.; Kahn, C.R.; Ristow, M.; Zarse, K. Impairment of insulin signalling in peripheral tissue fails to extend murine lifespan. Aging Cell 2017, 16, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Kawano, Y.; Nishiumi, S.; Saito, M.; Yano, Y.; Azuma, T.; Yoshida, M. Identification of Lipid Species Linked to the Progression of Non-Alcoholic Fatty Liver Disease. Curr. Drug Targets 2015, 16, 1293–1300. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, S.-O.; Andersson, L.; Håversen, L.; Olsson, C.; Myhre, S.; Rutberg, M.; Mobini, R.; Li, L.; Lu, E.; Borén, J.; et al. The formation of lipid droplets: Possible role in the development of insulin resistance/type 2 diabetes. Prostaglandins Leukot. Essent. Fat. Acids 2011, 85, 215–218. [Google Scholar] [CrossRef]
- Wang, L.; Liu, J.; Miao, Z.; Pan, Q.; Cao, W. Lipid droplets and their interactions with other organelles in liver diseases. Int. J. Biochem. Cell Biol. 2021, 133, 105937. [Google Scholar] [CrossRef]
- Uehara, K.; Santoleri, D.; Whitlock, A.E.G.; Titchenell, P.M. Insulin Regulation of Hepatic Lipid Homeostasis. In Comprehensive Physiology; Wiley: Hoboken, NJ, USA, 2023; pp. 4785–4809. [Google Scholar]
- Zang, L.; Shimada, Y.; Nishimura, Y.; Tanaka, T.; Nishimura, N. Repeated Blood Collection for Blood Tests in Adult Zebrafish. J. Vis. Exp. JoVE 2015, 102, e53272. [Google Scholar]
- Li, X.; Ge, G.; Song, G.; Li, Q.; Cui, Z. Effects of Nutritionally Induced Obesity on Metabolic Pathways of Zebrafish. Int. J. Mol. Sci. 2023, 24, 1850. [Google Scholar] [CrossRef]
- Ge, G.; Long, Y.; Song, G.; Li, Q.; Cui, Z.; Yan, H. Transcriptomic Profiling Revealed Signaling Pathways Associated with the Spawning of Female Zebrafish under Cold Stress. Int. J. Mol. Sci. 2022, 23, 7494. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. feature Counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
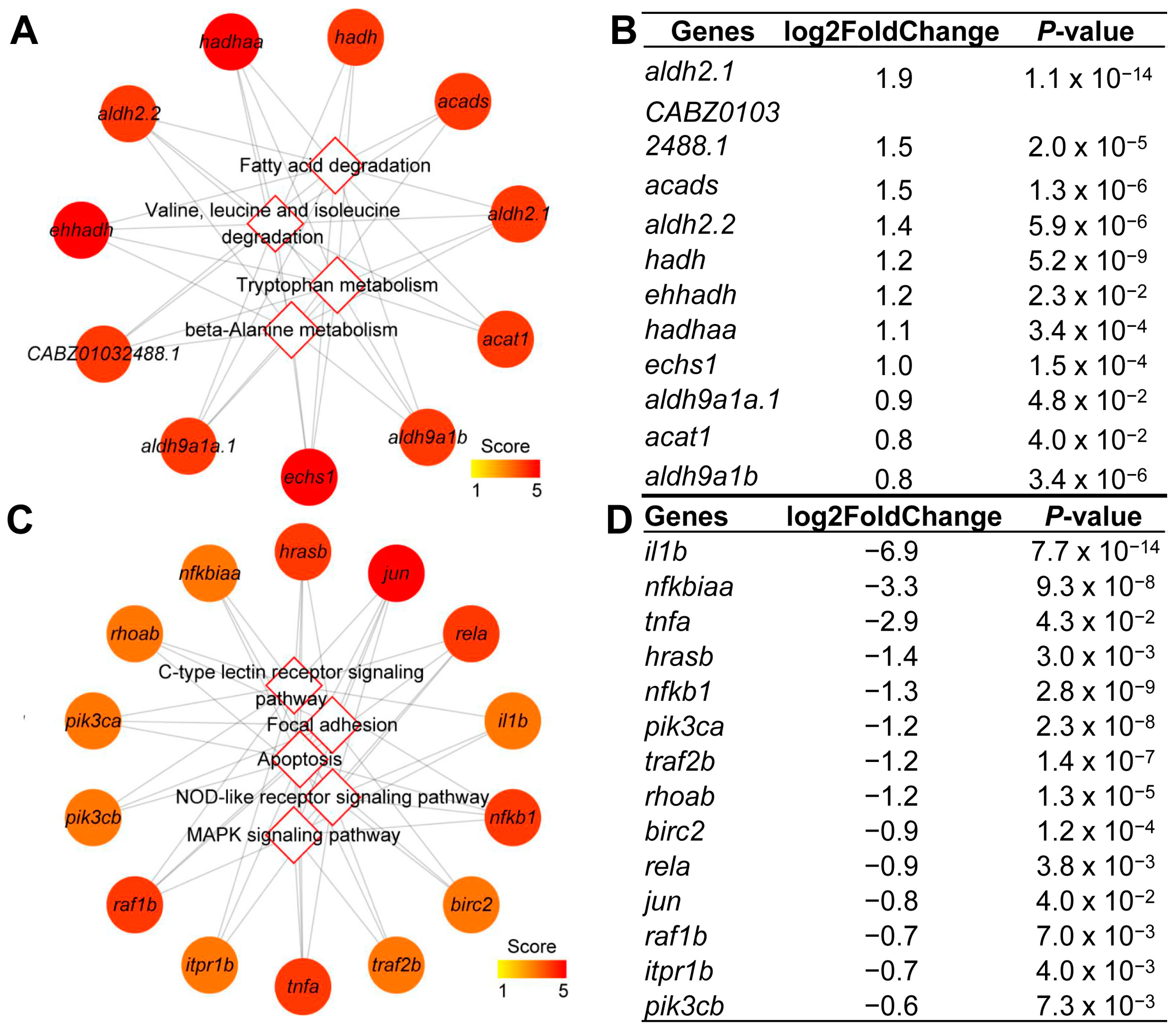
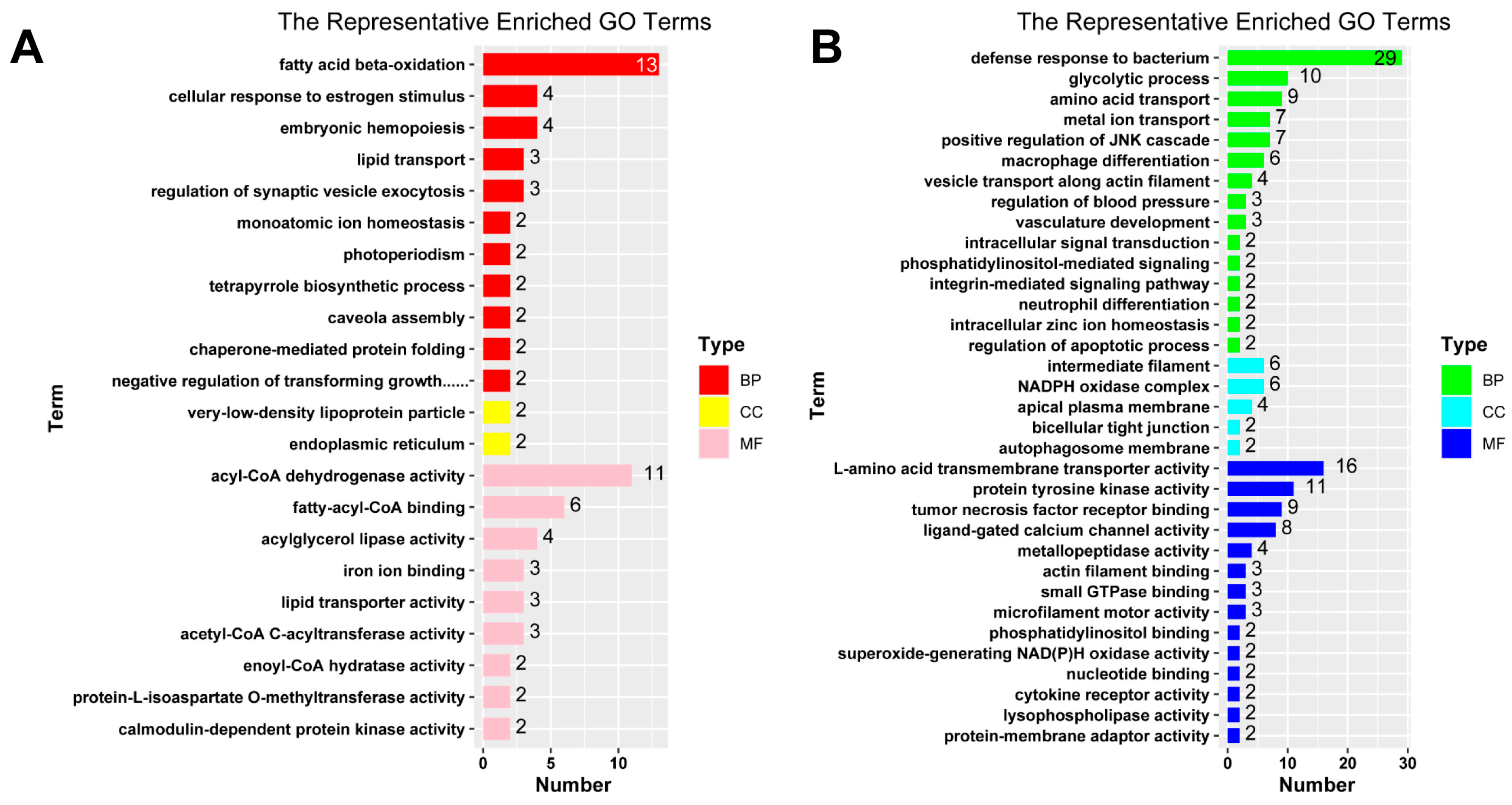
| Rank | Signaling Pathway | Score |
|---|---|---|
| 1 | Metabolic pathways | 2.11 × 108 |
| 2 | Valine, leucine and isoleucine degradation | 2.11 × 108 |
| 2 | Fatty acid degradation | 2.11 × 108 |
| 4 | Tryptophan metabolism | 2.07 × 108 |
| 5 | Lysine degradation | 2.07 × 108 |
| 6 | Carbon metabolism | 1.71 × 108 |
| 7 | Propanoate metabolism | 1.67 × 108 |
| 8 | Fatty acid metabolism | 1.67 × 108 |
| 9 | Butanoate metabolism | 1.63 × 108 |
| 10 | Beta-Alanine metabolism | 1.31 × 108 |
| Rank | Signaling Pathway | Score |
|---|---|---|
| 1 | C-type lectin receptor signaling pathway | 8.06 × 107 |
| 2 | AGE-RAGE signaling pathway in diabetic complications | 8.06 × 107 |
| 3 | NOD-like receptor signaling pathway | 8.06 × 107 |
| 3 | Apoptosis | 8.06 × 107 |
| 3 | Toll-like receptor signaling pathway | 8.06 × 107 |
| 3 | FoxO signaling pathway | 8.06 × 107 |
| 7 | Focal adhesion | 8.06 × 107 |
| 8 | Cellular senescence | 8.06 × 107 |
| 9 | Metabolic pathways | 7.98 × 107 |
| 10 | VEGF signaling pathway | 7.98 × 107 |
| Gene Name | Fold | Gene Description | Function |
| fabp10b | 2.8 | fatty acid binding protein 10b, liver basic | Fatty acid transporter |
| scp2a | 2.3 | sterol carrier protein 2a | Cholesterol transport |
| slc27a2a | 2.9 | solute carrier family 27 member 2a | Fatty acid transfer |
| acsf2 | 2.7 | acyl-CoA synthetase family member 2 | Fatty acyl-CoA synthase |
| cpt2 | 2.5 | carnitine palmitoyltransferase 2 | Carnitine palmitoyltransferase |
| hacd2 | 2.0 | 3-hydroxyacyl-CoA dehydratase 2 | Fatty acid β-oxidation |
| acaa1 | 2.3 | acetyl-CoA acyltransferase 1 | Fatty acid β-oxidation |
| acaa2 | 2.3 | acetyl-CoA acyltransferase 2 | Fatty acid β-oxidation |
| acadl | 2.4 | acyl-CoA dehydrogenase long chain | Fatty acid β-oxidation |
| acadm | 2.6 | acyl-CoA dehydrogenase medium chain | Fatty acid β-oxidation |
| acads | 2.8 | acyl-CoA dehydrogenase short chain | Fatty acid β-oxidation |
| acadvl | 1.9 | acyl-CoA dehydrogenase very long chain | Fatty acid β-oxidation |
| echs1 | 2.0 | enoyl CoA hydratase, short chain, 1, mitochondrial | Fatty acid β-oxidation |
| hmgcl | 1.6 | 3-hydroxy-3-methylglutaryl-CoA lyase | Ketogenesis |
| cetp | 2.1 | cholesteryl ester transfer protein, plasma | Cholesteryl ester transfer |
| plin1 | 14.1 | perilipin 1 | Lipid droplet-associated protein |
| Gene Name | Fold | Gene Description | Function |
|---|---|---|---|
| hk1 | 1.9 | hexokinase 1 | Glycolysis |
| hk2 | 6.0 | hexokinase 2 | Glycolysis |
| pfkla | 2.6 | phosphofructokinase, liver a | Glycolysis |
| pfkpb | 2.2 | phosphofructokinase, platelet b | Glycolysis |
| aldoaa | 2.0 | aldolase a, fructose-bisphosphate, a | Glycolysis |
| aldocb | 3.1 | aldolase C, fructose-bisphosphate, b | Glycolysis |
| gapdhs | 2.5 | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic | Glycolysis |
| eno1a | 4.3 | enolase 1a, (alpha) | Glycolysis |
| eno1b | 2.3 | enolase 1b, (alpha) | Glycolysis |
| pkma | 3.4 | pyruvate kinase M1/2a | Glycolysis |
| gys1 | 1.6 | glycogen synthase 1 (muscle) | Glycogenesis |
| gsk3bb | 2.6 | glycogen synthase kinase 3 beta, genome duplicate b | Glycogenesis |
| gyg1b | 8.1 | glycogenin 1b | Glycogenesis |
| pygl | (2.5) | phosphorylase, glycogen, liver | Glycogenolysis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, G.; Ren, J.; Song, G.; Li, Q.; Cui, Z. Transcriptome Analysis Reveals the Molecular Basis of Overfeeding-Induced Diabetes in Zebrafish. Int. J. Mol. Sci. 2023, 24, 11994. https://doi.org/10.3390/ijms241511994
Ge G, Ren J, Song G, Li Q, Cui Z. Transcriptome Analysis Reveals the Molecular Basis of Overfeeding-Induced Diabetes in Zebrafish. International Journal of Molecular Sciences. 2023; 24(15):11994. https://doi.org/10.3390/ijms241511994
Chicago/Turabian StyleGe, Guodong, Jing Ren, Guili Song, Qing Li, and Zongbin Cui. 2023. "Transcriptome Analysis Reveals the Molecular Basis of Overfeeding-Induced Diabetes in Zebrafish" International Journal of Molecular Sciences 24, no. 15: 11994. https://doi.org/10.3390/ijms241511994
APA StyleGe, G., Ren, J., Song, G., Li, Q., & Cui, Z. (2023). Transcriptome Analysis Reveals the Molecular Basis of Overfeeding-Induced Diabetes in Zebrafish. International Journal of Molecular Sciences, 24(15), 11994. https://doi.org/10.3390/ijms241511994








