Abiotic Stress-Induced Leaf Senescence: Regulatory Mechanisms and Application
Abstract
1. Introduction
2. Abiotic Stress-Induced Leaf Senescence
2.1. Nitrogen Deficiency-Induced Leaf Senescence
2.2. Drought Stress-Induced Leaf Senescence
2.3. Salt Stress-Induced Leaf Senescence
2.4. Darkness-Induced Leaf Senescence
2.5. Low Oxygen-Induced Leaf Senescence
2.6. Extreme Temperatures Stress-Induced Leaf Senescence
2.7. Other Abiotic Stresses-Induced Leaf Senescence
3. Improvement of Stress Tolerance through Regulation of Leaf Senescence
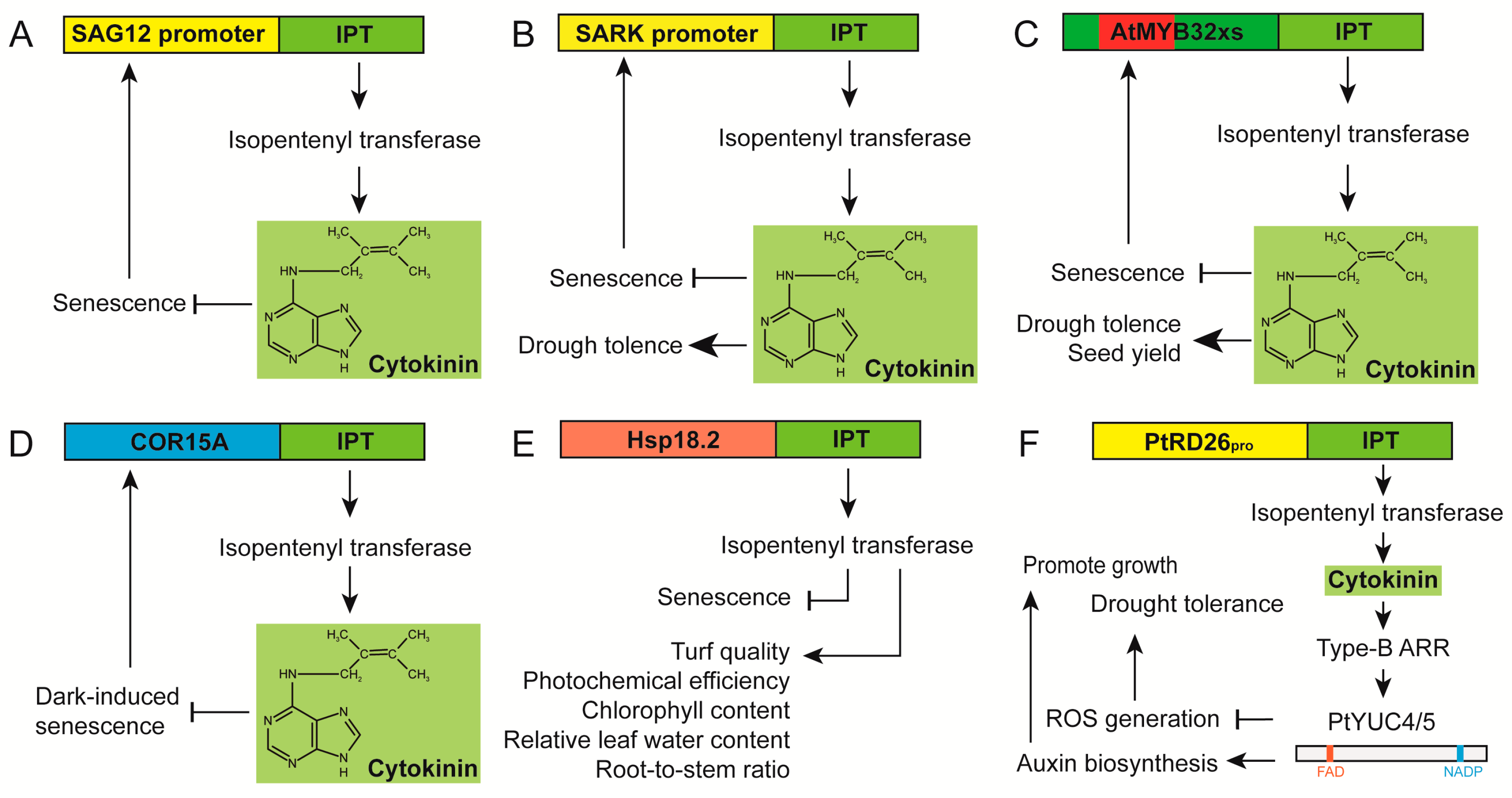
3.1. Utilization of Senescence-Specific or Stress-Associated Promoters
3.2. Modulation of Expression of Senescence Associated Genes
4. Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ROS | Reactive Oxygen Species |
| SAG | Senescence-Associated Gene |
| NLA | Nitrogen Limitation Adaptation |
| PHO2 | PHOSPHATE2 |
| UBC | Ubiquitin-conjugating enzyme |
| UBP12 | Ubiquitin-specific protease 12 |
| GDS1 | GROWTH, DEVELOPMENT AND SPLICING 1 |
| ORE1 | ORESARA1 |
| JUB1 | JUNGBRUNNEN1 |
| ERF | ETHYLENE RESPONSE FACTOR |
| COR15A | COLD-REGULATED15A |
| ERD10 | EARLY RESPONSIVE TO DEHYDRATION10 |
| RD29A | RESPONSIVE TO DESICCATION29A |
| PIF | PHYTOCHROME-INTERACTING FACTOR |
| bZIP | basic LEUCINE ZIPPER |
| IDL | Individually Darkened Leaf |
| ABI5 | ABSCISIC ACID INSENSITIVE 5 |
| EEL | ENHANCED EM LEVELS |
| SGR1 | STAYGREEN 1 |
| NYC1 | NON-YELLOW COLORING 1 |
| GDL1 | GOLDEN2-LIKE 1 |
| LOX2 | LIPOXYGENASE 2 |
| AOS | ALLENE OXIDE SYNTHASE |
| SUB1A | SUBMERGENCE1A |
| HXK1 | Hexokinase-1 |
| PAP1 | PRODUCTION OF ANTHOCYANIN PIGMENT 1 |
| IPT | Isopentenyltransferase |
| SARK | Senescence-associated Receptor Kinase |
| Kn1 | Knotted1 |
| QTL | Quantitative Trait Loci |
| PILSP | Plants with Ideal Leaf Senescence Phenotype |
| ROS | Reactive Oxygen Species |
| SAG | Senescence-Associated Gene |
| NLA | Nitrogen Limitation Adaptation |
| PHO2 | PHOSPHATE2 |
| UBC | Ubiquitin-conjugating enzyme |
| UBP12 | Ubiquitin-specific protease 12 |
| GDS1 | GROWTH, DEVELOPMENT AND SPLICING 1 |
| ORE1 | ORESARA1 |
| JUB1 | JUNGBRUNNEN1 |
| ERF | ETHYLENE RESPONSE FACTOR |
| COR15A | COLD-REGULATED15A |
| ERD10 | EARLY RESPONSIVE TO DEHYDRATION10 |
References
- Guo, Y.; Gan, S. Leaf senescence: Signals, execution, and regulation. Curr. Top. Dev. Biol. 2005, 71, 83–112. [Google Scholar]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef]
- Wang, H.L.; Zhang, Y.; Wang, T.; Yang, Q.; Yang, Y.; Li, Z.; Li, B.; Wen, X.; Li, W.; Yin, W.; et al. An alternative splicing variant of PtRD26 delays leaf senescence by regulating multiple NAC transcription factors in Populus. Plant Cell 2021, 33, 1594–1614. [Google Scholar] [CrossRef]
- Keskitalo, J.; Bergquist, G.; Gardestrom, P.; Jansson, S. A cellular timetable of autumn senescence. Plant Physiol. 2005, 139, 1635–1648. [Google Scholar] [CrossRef] [PubMed]
- Cooke, J.E.; Weih, M. Nitrogen storage and seasonal nitrogen cycling in Populus: Bridging molecular physiology and ecophysiology. New Phytol. 2005, 167, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Andleeb, T.; Knight, E.; Borrill, P. Wheat NAM genes regulate the majority of early monocarpic senescence transcriptional changes including nitrogen remobilization genes. G3 2023, 13, jkac275. [Google Scholar] [CrossRef]
- Wei, S.; Li, X.; Lu, Z.; Zhang, H.; Ye, X.; Zhou, Y.; Li, J.; Yan, Y.; Pei, H.; Duan, F.; et al. A transcriptional regulator that boosts grain yields and shortens the growth duration of rice. Science 2022, 377, eabi8455. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ren, G.; Zhang, K.; Li, Z.; Miao, Y.; Guo, H. Leaf senescence: Progression, regulation, and application. Mol. Hortic. 2021, 1, 5. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, Y.; Tan, S.; Yang, Q.; Wang, H.-L.; Xia, X.; Luo, J.; Guo, H.; Zhang, Z.; Li, Z. LSD 4.0: An improved database for comparative studies of leaf senescence. Mol. Hortic. 2022, 2, 24. [Google Scholar] [CrossRef]
- Cao, J.; Liu, H.; Tan, S.; Li, Z. Transcription Factors-Regulated Leaf Senescence: Current Knowledge, Challenges and Approaches. Int. J. Mol. Sci. 2023, 24, 9245. [Google Scholar] [CrossRef]
- Moschen, S.; Di Rienzo, J.A.; Higgins, J.; Tohge, T.; Watanabe, M.; Gonzalez, S.; Rivarola, M.; Garcia-Garcia, F.; Dopazo, J.; Hopp, H.E.; et al. Integration of transcriptomic and metabolic data reveals hub transcription factors involved in drought stress response in sunflower (Helianthus annuus L.). Plant Mol. Biol. 2017, 94, 549–564. [Google Scholar] [CrossRef] [PubMed]
- Moschen, S.; Bengoa Luoni, S.; Paniego, N.B.; Hopp, H.E.; Dosio, G.A.; Fernandez, P.; Heinz, R.A. Identification of candidate genes associated with leaf senescence in cultivated sunflower (Helianthus annuus L.). PLoS ONE 2014, 9, e104379. [Google Scholar] [CrossRef] [PubMed]
- Trupkin, S.A.; Astigueta, F.H.; Baigorria, A.H.; Garcia, M.N.; Delfosse, V.C.; Gonzalez, S.A.; Perez de la Torre, M.C.; Moschen, S.; Lia, V.V.; Fernandez, P.; et al. Identification and expression analysis of NAC transcription factors potentially involved in leaf and petal senescence in Petunia hybrida. Plant Sci. 2019, 287, 110195. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Peng, J.; Wen, X.; Guo, H. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 2013, 25, 3311–3328. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, X.; Guo, C.; Wen, L.; Deng, Z.; Zhang, Z.; Li, W.; Liu, T.; Guo, Y. Senescence-Related Receptor Kinase 1 (SENRK1) functions downstream of WRKY53 in regulating leaf senescence in Arabidopsis. J. Exp. Bot. 2023. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Kim, D.; Paek, N.C. Salt Treatments and Induction of Senescence. Methods Mol. Biol. 2018, 1744, 141–149. [Google Scholar]
- Sade, N.; Del Mar Rubio-Wilhelmi, M.; Umnajkitikorn, K.; Blumwald, E. Stress-induced senescence and plant tolerance to abiotic stress. J. Exp. Bot. 2018, 69, 845–853. [Google Scholar] [CrossRef]
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf Senescence: Systems and Dynamics Aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef]
- Li, Z.; Kim, J.H.; Kim, J.; Lyu, J.I.; Zhang, Y.; Guo, H.; Nam, H.G.; Woo, H.R. ATM suppresses leaf senescence triggered by DNA double-strand break through epigenetic control of senescence-associated genes in Arabidopsis. New Phytol. 2020, 227, 473–484. [Google Scholar] [CrossRef]
- Han, D.; Du, M.; Zhou, Z.; Wang, S.; Li, T.; Han, J.; Xu, T.; Yang, G. Overexpression of a Malus baccata NAC Transcription Factor Gene MbNAC25 Increases Cold and Salinity Tolerance in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 1198. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Sun, C.H.; Zhang, Q.Y.; Gu, K.D.; Hao, Y.J. The basic helix-loop-helix transcription factor MdbHLH3 modulates leaf senescence in apple via the regulation of dehydratase-enolase-phosphatase complex 1. Hortic. Res. 2020, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Lira, B.S.; Gramegna, G.; Trench, B.A.; Alves, F.R.R.; Silva, E.M.; Silva, G.F.F.; Thirumalaikumar, V.P.; Lupi, A.C.D.; Demarco, D.; Purgatto, E.; et al. Manipulation of a Senescence-Associated Gene Improves Fleshy Fruit Yield. Plant Physiol. 2017, 175, 77–91. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Y.; Tureckova, V.; Xue, G.P.; Fernie, A.R.; Mueller-Roeber, B.; Balazadeh, S. The NAC Transcription Factor SlNAP2 Regulates Leaf Senescence and Fruit Yield in Tomato. Plant Physiol. 2018, 177, 1286–1302. [Google Scholar] [CrossRef]
- Gan, S.; Amasino, R.M. Inhibition of leaf senescence by autoregulated production of cytokinin. Science 1995, 270, 1986–1988. [Google Scholar] [CrossRef]
- Rivero, R.M.; Kojima, M.; Gepstein, A.; Sakakibara, H.; Mittler, R.; Gepstein, S.; Blumwald, E. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl. Acad. Sci. USA 2007, 104, 19631–19636. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, W.Q.; Zhang, G.L.; Kaminek, M.; Dobrev, P.; Xu, J.; Gruissem, W. Senescence-inducible expression of isopentenyl transferase extends leaf life, increases drought stress resistance and alters cytokinin metabolism in cassava. J. Integr. Plant Biol. 2010, 52, 653–669. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S.S. Translational researches on leaf senescence for enhancing plant productivity and quality. J. Exp. Bot. 2014, 65, 3901–3913. [Google Scholar] [CrossRef]
- Park, B.S.; Yao, T.; Seo, J.S.; Wong, E.C.C.; Mitsuda, N.; Huang, C.H.; Chua, N.H. Arabidopsis NITROGEN LIMITATION ADAPTATION regulates ORE1 homeostasis during senescence induced by nitrogen deficiency. Nat. Plants 2018, 4, 898–903. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Jeong, J.S.; Seo, J.S.; Park, B.S.; Chua, N.H. Arabidopsis ubiquitin-specific proteases UBP12 and UBP13 shape ORE1 levels during leaf senescence induced by nitrogen deficiency. New Phytol. 2019, 223, 1447–1460. [Google Scholar] [CrossRef]
- Fan, H.; Quan, S.; Ye, Q.; Zhang, L.; Liu, W.; Zhu, N.; Zhang, X.; Ruan, W.; Yi, K.; Crawford, N.M.; et al. A molecular framework underlying low-nitrogen-induced early leaf senescence in Arabidopsis thaliana. Mol. Plant 2023, 16, 756–774. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Hannam, C.; Gu, H.; Bi, Y.M.; Rothstein, S.J. A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J. 2007, 50, 320–337. [Google Scholar] [CrossRef]
- Liu, W.; Sun, Q.; Wang, K.; Du, Q.; Li, W.X. Nitrogen Limitation Adaptation (NLA) is involved in source-to-sink remobilization of nitrate by mediating the degradation of NRT1.7 in Arabidopsis. New Phytol. 2017, 214, 734–744. [Google Scholar] [CrossRef] [PubMed]
- Delhaize, E.; Randall, P.J. Characterization of a Phosphate-Accumulator Mutant of Arabidopsis thaliana. Plant Physiol. 1995, 107, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Rengel, Z.; Delhaize, E. Uptake and translocation of phosphate by pho2 mutant and wild-type seedlings of Arabidopsis thaliana. Planta 1998, 205, 251–256. [Google Scholar] [CrossRef]
- Kim, D.W.; Jeon, S.J.; Hwang, S.M.; Hong, J.C.; Bahk, J.D. The C3H-type zinc finger protein GDS1/C3H42 is a nuclear-speckle-localized protein that is essential for normal growth and development in Arabidopsis. Plant Sci. 2016, 250, 141–153. [Google Scholar] [CrossRef]
- Liebsch, D.; Keech, O. Dark-induced leaf senescence: New insights into a complex light-dependent regulatory pathway. New Phytol. 2016, 212, 563–570. [Google Scholar] [CrossRef]
- Song, Y.; Yang, C.; Gao, S.; Zhang, W.; Li, L.; Kuai, B. Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol. Plant 2014, 7, 1776–1787. [Google Scholar] [CrossRef]
- Li, N.; Bo, C.; Zhang, Y.; Wang, L. Phytochrome Interacting Factors PIF4 and PIF5 promote heat stress induced leaf senescence in Arabidopsis. J. Exp. Bot. 2021, 72, 4577–4589. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Jeong, J.; Kang, M.Y.; Kim, J.; Paek, N.C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 2014, 5, 4636. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Munne-Bosch, S.; Alegre, L. Die and let live: Leaf senescence contributes to plant survival under drought stress. Funct. Plant Biol. 2004, 31, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Seo, P.J.; Lee, H.J.; Park, C.M. A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 2012, 70, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Hickman, R.; Hill, C.; Penfold, C.A.; Breeze, E.; Bowden, L.; Moore, J.D.; Zhang, P.; Jackson, A.; Cooke, E.; Bewicke-Copley, F.; et al. A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 2013, 75, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef]
- Claeys, H.; Inze, D. The agony of choice: How plants balance growth and survival under water-limiting conditions. Plant Physiol. 2013, 162, 1768–1779. [Google Scholar] [CrossRef]
- Athar, H.U.; Zulfiqar, F.; Moosa, A.; Ashraf, M.; Zafar, Z.U.; Zhang, L.; Ahmed, N.; Kalaji, H.M.; Nafees, M.; Hossain, M.A.; et al. Salt stress proteins in plants: An overview. Front. Plant Sci. 2022, 13, 999058. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Ashraf, M. Nanoparticles potentially mediate salt stress tolerance in plants. Plant Physiol. Biochem. 2021, 160, 257–268. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef]
- Fricke, W. Energy costs of salinity tolerance in crop plants: Night-time transpiration and growth. New Phytol. 2020, 225, 1152–1165. [Google Scholar] [CrossRef]
- Munns, R.; Day, D.A.; Fricke, W.; Watt, M.; Arsova, B.; Barkla, B.J.; Bose, J.; Byrt, C.S.; Chen, Z.H.; Foster, K.J.; et al. Energy costs of salt tolerance in crop plants. New Phytol. 2020, 225, 1072–1090. [Google Scholar] [CrossRef]
- Tyerman, S.D.; Munns, R.; Fricke, W.; Arsova, B.; Barkla, B.J.; Bose, J.; Bramley, H.; Byrt, C.; Chen, Z.; Colmer, T.D.; et al. Energy costs of salinity tolerance in crop plants. New Phytol. 2019, 221, 25–29. [Google Scholar] [CrossRef]
- Kim, H.J.; Nam, H.G.; Lim, P.O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef]
- Park, S.J.; Park, S.; Kim, Y.; Hyeon, D.Y.; Park, H.; Jeong, J.; Jeong, U.; Yoon, Y.S.; You, D.; Kwak, J.; et al. Ethylene responsive factor34 mediates stress-induced leaf senescence by regulating salt stress-responsive genes. Plant Cell Environ. 2022, 45, 1719–1733. [Google Scholar] [CrossRef]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Kohler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef]
- Peng, J.; Li, Z.; Wen, X.; Li, W.; Shi, H.; Yang, L.; Zhu, H.; Guo, H. Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet. 2014, 10, e1004664. [Google Scholar] [CrossRef]
- Kim, Y.S.; Sakuraba, Y.; Han, S.H.; Yoo, S.C.; Paek, N.C. Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol. 2013, 54, 1660–1672. [Google Scholar] [CrossRef]
- Mahmood, K.; Xu, Z.; El-Kereamy, A.; Casaretto, J.A.; Rothstein, S.J. The Arabidopsis Transcription Factor ANAC032 Represses Anthocyanin Biosynthesis in Response to High Sucrose and Oxidative and Abiotic Stresses. Front. Plant Sci. 2016, 7, 1548. [Google Scholar] [CrossRef]
- Mahmood, K.; El-Kereamy, A.; Kim, S.H.; Nambara, E.; Rothstein, S.J. ANAC032 Positively Regulates Age-Dependent and Stress-Induced Senescence in Arabidopsis thaliana. Plant Cell Physiol. 2016, 57, 2029–2046. [Google Scholar] [CrossRef]
- Mito, T.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M.; Matsui, K. Generation of chimeric repressors that confer salt tolerance in Arabidopsis and rice. Plant Biotechnol. J. 2011, 9, 736–746. [Google Scholar] [CrossRef]
- Yang, S.D.; Seo, P.J.; Yoon, H.K.; Park, C.M. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 2011, 23, 2155–2168. [Google Scholar] [CrossRef]
- Wu, A.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.I.; Asensi-Fabado, M.A.; Munne-Bosch, S.; Antonio, C.; Tohge, T.; et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 24, 482–506. [Google Scholar] [CrossRef]
- Podzimska-Sroka, D.; O’Shea, C.; Gregersen, P.L.; Skriver, K. NAC Transcription Factors in Senescence: From Molecular Structure to Function in Crops. Plants 2015, 4, 412–448. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, W.; Liu, L.; Chen, T.; Zhou, F.; Lin, Y. Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol. 2013, 13, 132. [Google Scholar] [CrossRef]
- Zhao, D.; Derkx, A.P.; Liu, D.C.; Buchner, P.; Hawkesford, M.J. Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol. 2015, 17, 904–913. [Google Scholar] [CrossRef]
- Liu, G.; Li, X.; Jin, S.; Liu, X.; Zhu, L.; Nie, Y.; Zhang, X. Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS ONE 2014, 9, e86895. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Piao, W.; Lim, J.H.; Han, S.H.; Kim, Y.S.; An, G.; Paek, N.C. Rice ONAC106 Inhibits Leaf Senescence and Increases Salt Tolerance and Tiller Angle. Plant Cell Physiol. 2015, 56, 2325–2339. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, Y.; Li, H.; Wang, T.; Zhang, J.; Ouyang, B.; Ye, Z. Molecular and functional characterization of ShNAC1, an NAC transcription factor from Solanum habrochaites. Plant Sci. 2018, 271, 9–19. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, X.; Duan, H.; Lian, C.; Liu, C.; Yin, W.; Xia, X. Three stress-responsive NAC transcription factors from Populus euphratica differentially regulate salt and drought tolerance in transgenic plants. Physiol. Plant. 2018, 162, 73–97. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Y.; Zou, D.; Zhao, Y.; Wang, H.L.; Zhang, Y.; Xia, X.; Luo, J.; Guo, H.; Zhang, Z. LSD 3.0: A comprehensive resource for the leaf senescence research community. Nucleic Acids Res. 2020, 48, D1069–D1075. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.D.; Yang, C.C.; Qin, R.; Alamin, M.; Yue, E.K.; Jin, X.L.; Shi, C.H. A guanine insert in OsBBS1 leads to early leaf senescence and salt stress sensitivity in rice (Oryza sativa L.). Plant Cell Rep. 2018, 37, 933–946. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Tao, H.; Xu, S.; Li, K.; Zafar, S.; Cao, W.; Yang, Y. Overexpression of salt-induced protein (salT) delays leaf senescence in rice. Genet. Mol. Biol. 2019, 42, 80–86. [Google Scholar] [CrossRef]
- Allu, A.D.; Soja, A.M.; Wu, A.; Szymanski, J.; Balazadeh, S. Salt stress and senescence: Identification of cross-talk regulatory components. J. Exp. Bot. 2014, 65, 3993–4008. [Google Scholar] [CrossRef]
- de Wit, M.; Galvao, V.C.; Fankhauser, C. Light-Mediated Hormonal Regulation of Plant Growth and Development. Annu. Rev. Plant Biol. 2016, 67, 513–537. [Google Scholar] [CrossRef]
- Weaver, L.M.; Amasino, R.M. Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol. 2001, 127, 876–886. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, T.; Liu, J.; Li, H.; Liu, B. Shade-Induced Leaf Senescence in Plants. Plants 2023, 12, 1550. [Google Scholar] [CrossRef]
- Brouwer, B.; Ziolkowska, A.; Bagard, M.; Keech, O.; Gardestrom, P. The impact of light intensity on shade-induced leaf senescence. Plant Cell Environ. 2012, 35, 1084–1098. [Google Scholar] [CrossRef]
- Liebsch, D.; Juvany, M.; Li, Z.; Wang, H.L.; Ziolkowska, A.; Chrobok, D.; Boussardon, C.; Wen, X.; Law, S.R.; Janeckova, H.; et al. Metabolic control of arginine and ornithine levels paces the progression of leaf senescence. Plant Physiol. 2022, 189, 1943–1960. [Google Scholar] [CrossRef]
- Wu, H.Y.; Liu, L.A.; Shi, L.; Zhang, W.F.; Jiang, C.D. Photosynthetic acclimation during low-light-induced leaf senescence in post-anthesis maize plants. Photosynth. Res. 2021, 150, 313–326. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S.S. Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 2012, 35, 644–655. [Google Scholar] [CrossRef]
- Buchanan-Wollaston, V.; Page, T.; Harrison, E.; Breeze, E.; Lim, P.O.; Nam, H.G.; Lin, J.F.; Wu, S.H.; Swidzinski, J.; Ishizaki, K.; et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005, 42, 567–585. [Google Scholar] [CrossRef] [PubMed]
- van der Graaff, E.; Schwacke, R.; Schneider, A.; Desimone, M.; Flugge, U.I.; Kunze, R. Transcription analysis of arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 2006, 141, 776–792. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.F.; Wu, S.H. Molecular events in senescing Arabidopsis leaves. Plant J. 2004, 39, 612–628. [Google Scholar] [CrossRef]
- Khanna, R.; Shen, Y.; Marion, C.M.; Tsuchisaka, A.; Theologis, A.; Schafer, E.; Quail, P.H. The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 2007, 19, 3915–3929. [Google Scholar] [CrossRef] [PubMed]
- Qiu, K.; Li, Z.; Yang, Z.; Chen, J.; Wu, S.; Zhu, X.; Gao, S.; Gao, J.; Ren, G.; Kuai, B.; et al. EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399. [Google Scholar] [CrossRef] [PubMed]
- Rauf, M.; Arif, M.; Dortay, H.; Matallana-Ramirez, L.P.; Waters, M.T.; Gil Nam, H.; Lim, P.O.; Mueller-Roeber, B.; Balazadeh, S. ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Rep. 2013, 14, 382–388. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef]
- Fukao, T.; Xu, K.; Ronald, P.C.; Bailey-Serres, J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 2006, 18, 2021–2034. [Google Scholar] [CrossRef]
- Lee, S.C.; Mustroph, A.; Sasidharan, R.; Vashisht, D.; Pedersen, O.; Oosumi, T.; Voesenek, L.A.; Bailey-Serres, J. Molecular characterization of the submergence response of the Arabidopsis thaliana ecotype Columbia. New Phytol. 2011, 190, 457–471. [Google Scholar] [CrossRef]
- Vashisht, D.; Hesselink, A.; Pierik, R.; Ammerlaan, J.M.; Bailey-Serres, J.; Visser, E.J.; Pedersen, O.; van Zanten, M.; Vreugdenhil, D.; Jamar, D.C.; et al. Natural variation of submergence tolerance among Arabidopsis thaliana accessions. New Phytol. 2011, 190, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Gasch, P.; Fundinger, M.; Muller, J.T.; Lee, T.; Bailey-Serres, J.; Mustroph, A. Redundant ERF-VII Transcription Factors Bind to an Evolutionarily Conserved cis-Motif to Regulate Hypoxia-Responsive Gene Expression in Arabidopsis. Plant Cell 2016, 28, 160–180. [Google Scholar] [CrossRef]
- Loreti, E.; Valeri, M.C.; Novi, G.; Perata, P. Gene Regulation and Survival under Hypoxia Requires Starch Availability and Metabolism. Plant Physiol. 2018, 176, 1286–1298. [Google Scholar] [CrossRef] [PubMed]
- Giuntoli, B.; Perata, P. Group VII Ethylene Response Factors in Arabidopsis: Regulation and Physiological Roles. Plant Physiol. 2018, 176, 1143–1155. [Google Scholar] [CrossRef]
- Fukao, T.; Yeung, E.; Bailey-Serres, J. The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell 2011, 23, 412–427. [Google Scholar] [CrossRef]
- Fukao, T.; Yeung, E.; Bailey-Serres, J. The submergence tolerance gene SUB1A delays leaf senescence under prolonged darkness through hormonal regulation in rice. Plant Physiol. 2012, 160, 1795–1807. [Google Scholar] [CrossRef]
- Alpuerto, J.B.; Hussain, R.M.; Fukao, T. The key regulator of submergence tolerance, SUB1A, promotes photosynthetic and metabolic recovery from submergence damage in rice leaves. Plant Cell Environ. 2016, 39, 672–684. [Google Scholar] [CrossRef]
- Jespersen, D.; Yu, J.; Huang, B. Metabolite responses to exogenous application of nitrogen, cytokinin, and ethylene inhibitors in relation to heat-induced senescence in creeping bentgrass. PLoS ONE 2015, 10, e0123744. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, D.; Zhang, J.; Huang, B. Chlorophyll loss associated with heat-induced senescence in bentgrass. Plant Sci. 2016, 249, 1–12. [Google Scholar] [CrossRef]
- Kim, C.; Kim, S.J.; Jeong, J.; Park, E.; Oh, E.; Park, Y.I.; Lim, P.O.; Choi, G. High Ambient Temperature Accelerates Leaf Senescence via Phytochrome-Interacting Factor 4 and 5 in Arabidopsis. Mol. Cells 2020, 43, 645–661. [Google Scholar] [PubMed]
- He, Y.; Zhang, X.; Shi, Y.; Xu, X.; Li, L.; Wu, J.L. Premature Senescence Leaf 50 Promotes Heat Stress Tolerance in Rice (Oryza sativa L.). Rice 2021, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Dai, J.; Ge, Q. Responses of Autumn Phenology to Climate Change and the Correlations of Plant Hormone Regulation. Sci. Rep. 2020, 10, 9039. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gao, C.; Ge, Q. Low temperature and short daylength interact to affect the leaf senescence of two temperate tree species. Tree Physiol. 2022, 42, 2252–2265. [Google Scholar] [CrossRef]
- Moore, B.; Zhou, L.; Rolland, F.; Hall, Q.; Cheng, W.H.; Liu, Y.X.; Hwang, I.; Jones, T.; Sheen, J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 2003, 300, 332–336. [Google Scholar] [CrossRef]
- Dai, N.; Schaffer, A.; Petreikov, M.; Shahak, Y.; Giller, Y.; Ratner, K.; Levine, A.; Granot, D. Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell 1999, 11, 1253–1266. [Google Scholar] [CrossRef]
- Wingler, A.; Purdy, S.; MacLean, J.A.; Pourtau, N. The role of sugars in integrating environmental signals during the regulation of leaf senescence. J. Exp. Bot. 2006, 57, 391–399. [Google Scholar] [CrossRef]
- Pourtau, N.; Jennings, R.; Pelzer, E.; Pallas, J.; Wingler, A. Effect of sugar-induced senescence on gene expression and implications for the regulation of senescence in Arabidopsis. Planta 2006, 224, 556–568. [Google Scholar] [CrossRef]
- Kim, J. Sugar metabolism as input signals and fuel for leaf senescence. Genes Genom. 2019, 41, 737–746. [Google Scholar] [CrossRef]
- Zhang, J.; Fei, L.; Dong, Q.; Zuo, S.; Li, Y.; Wang, Z. Cadmium binding during leaf senescence in Festuca arundinacea: Promotion phytoextraction efficiency by harvesting dead leaves. Chemosphere 2022, 289, 133253. [Google Scholar] [CrossRef]
- Piacentini, D.; Corpas, F.J.; D’Angeli, S.; Altamura, M.M.; Falasca, G. Cadmium and arsenic-induced-stress differentially modulates Arabidopsis root architecture, peroxisome distribution, enzymatic activities and their nitric oxide content. Plant Physiol. Biochem. 2020, 148, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Fei, L.; Zuo, S.; Zhang, J.; Wang, Z. Phytoextraction by harvesting dead leaves: Cadmium accumulation associated with the leaf senescence in Festuca arundinacea Schreb. Environ. Sci. Pollut. Res. Int. 2022, 29, 79214–79223. [Google Scholar] [CrossRef]
- Sykorova, B.; Kuresova, G.; Daskalova, S.; Trckova, M.; Hoyerova, K.; Raimanova, I.; Motyka, V.; Travnickova, A.; Elliott, M.C.; Kaminek, M. Senescence-induced ectopic expression of the A. tumefaciens ipt gene in wheat delays leaf senescence, increases cytokinin content, nitrate influx, and nitrate reductase activity, but does not affect grain yield. J. Exp. Bot. 2008, 59, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Calderini, O.; Bovone, T.; Scotti, C.; Pupilli, F.; Piano, E.; Arcioni, S. Delay of leaf senescence in Medicago sativa transformed with the ipt gene controlled by the senescence-specific promoter SAG12. Plant Cell Rep. 2007, 26, 611–615. [Google Scholar] [CrossRef] [PubMed]
- McCabe, M.S.; Garratt, L.C.; Schepers, F.; Jordi, W.J.; Stoopen, G.M.; Davelaar, E.; van Rhijn, J.H.; Power, J.B.; Davey, M.R. Effects of P(SAG12)-IPT gene expression on development and senescence in transgenic lettuce. Plant Physiol. 2001, 127, 505–516. [Google Scholar] [CrossRef]
- Xu, Y.; Burgess, P.; Zhang, X.; Huang, B. Enhancing cytokinin synthesis by overexpressing ipt alleviated drought inhibition of root growth through activating ROS-scavenging systems in Agrostis stolonifera. J. Exp. Bot. 2016, 67, 1979–1992. [Google Scholar] [CrossRef]
- Cowan, A.K.; Freeman, M.; Bjorkman, P.O.; Nicander, B.; Sitbon, F.; Tillberg, E. Effects of senescence-induced alteration in cytokinin metabolism on source-sink relationships and ontogenic and stress-induced transitions in tobacco. Planta 2005, 221, 801–814. [Google Scholar] [CrossRef]
- Kant, S.; Burch, D.; Badenhorst, P.; Palanisamy, R.; Mason, J.; Spangenberg, G. Regulated expression of a cytokinin biosynthesis gene IPT delays leaf senescence and improves yield under rainfed and irrigated conditions in canola (Brassica napus L.). PLoS ONE 2015, 10, e0116349. [Google Scholar] [CrossRef]
- Khodakovskaya, M.; Li, Y.; Li, J.; Vankova, R.; Malbeck, J.; McAvoy, R. Effects of cor15a-IPT gene expression on leaf senescence in transgenic Petunia x hybrida and Dendranthema x grandiflorum. J. Exp. Bot. 2005, 56, 1165–1175. [Google Scholar] [CrossRef]
- Wang, H.L.; Yang, Q.; Tan, S.; Wang, T.; Zhang, Y.; Yang, Y.; Yin, W.; Xia, X.; Guo, H.; Li, Z. Regulation of cytokinin biosynthesis using PtRD26(pro) -IPT module improves drought tolerance through PtARR10-PtYUC4/5-mediated reactive oxygen species removal in Populus. J. Integr. Plant Biol. 2022, 64, 771–786. [Google Scholar] [CrossRef]
- Ori, N.; Juarez, M.T.; Jackson, D.; Yamaguchi, J.; Banowetz, G.M.; Hake, S. Leaf senescence is delayed in tobacco plants expressing the maize homeobox gene knotted1 under the control of a senescence-activated promoter. Plant Cell 1999, 11, 1073–1080. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef]
- Shin, D.; Lee, S.; Kim, T.H.; Lee, J.H.; Park, J.; Lee, J.; Lee, J.Y.; Cho, L.H.; Choi, J.Y.; Lee, W.; et al. Natural variations at the Stay-Green gene promoter control lifespan and yield in rice cultivars. Nat. Commun. 2020, 11, 2819. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Woo, H.R.; Nam, H.G. Toward Systems Understanding of Leaf Senescence: An Integrated Multi-Omics Perspective on Leaf Senescence Research. Mol. Plant 2016, 9, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Kim, H.J.; Nam, H.G.; Lim, P.O. Plant leaf senescence and death-regulation by multiple layers of control and implications for aging in general. J. Cell. Sci. 2013, 126 Pt 21, 4823–4833. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Guo, P.; Xia, X.; Guo, H.; Li, Z. Multiple Layers of Regulation on Leaf Senescence: New Advances and Perspectives. Front. Plant Sci. 2021, 12, 788996. [Google Scholar] [CrossRef]
- Tseng, Y.C.; Chu, S.W. High spatio-temporal-resolution detection of chlorophyll fluorescence dynamics from a single chloroplast with confocal imaging fluorometer. Plant Methods 2017, 13, 43. [Google Scholar] [CrossRef]
- Iwai, M.; Yokono, M.; Kurokawa, K.; Ichihara, A.; Nakano, A. Live-cell visualization of excitation energy dynamics in chloroplast thylakoid structures. Sci. Rep. 2016, 6, 29940. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, S.; Gao, Y.; Kan, C.; Wang, H.L.; Yang, Q.; Xia, X.; Ishida, T.; Sawa, S.; Guo, H.; et al. CLE42 delays leaf senescence by antagonizing ethylene pathway in Arabidopsis. New Phytol. 2022, 235, 550–562. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, S.; Sha, Y.; Sun, L.; Li, Z. Abiotic Stress-Induced Leaf Senescence: Regulatory Mechanisms and Application. Int. J. Mol. Sci. 2023, 24, 11996. https://doi.org/10.3390/ijms241511996
Tan S, Sha Y, Sun L, Li Z. Abiotic Stress-Induced Leaf Senescence: Regulatory Mechanisms and Application. International Journal of Molecular Sciences. 2023; 24(15):11996. https://doi.org/10.3390/ijms241511996
Chicago/Turabian StyleTan, Shuya, Yueqi Sha, Liwei Sun, and Zhonghai Li. 2023. "Abiotic Stress-Induced Leaf Senescence: Regulatory Mechanisms and Application" International Journal of Molecular Sciences 24, no. 15: 11996. https://doi.org/10.3390/ijms241511996
APA StyleTan, S., Sha, Y., Sun, L., & Li, Z. (2023). Abiotic Stress-Induced Leaf Senescence: Regulatory Mechanisms and Application. International Journal of Molecular Sciences, 24(15), 11996. https://doi.org/10.3390/ijms241511996




