Paradox: Curcumin, a Natural Antioxidant, Suppresses Osteosarcoma Cells via Excessive Reactive Oxygen Species
Abstract
1. Introduction
2. Results
2.1. CUR Decreased the Viability of MG-63, Which Was Reversed by NAC
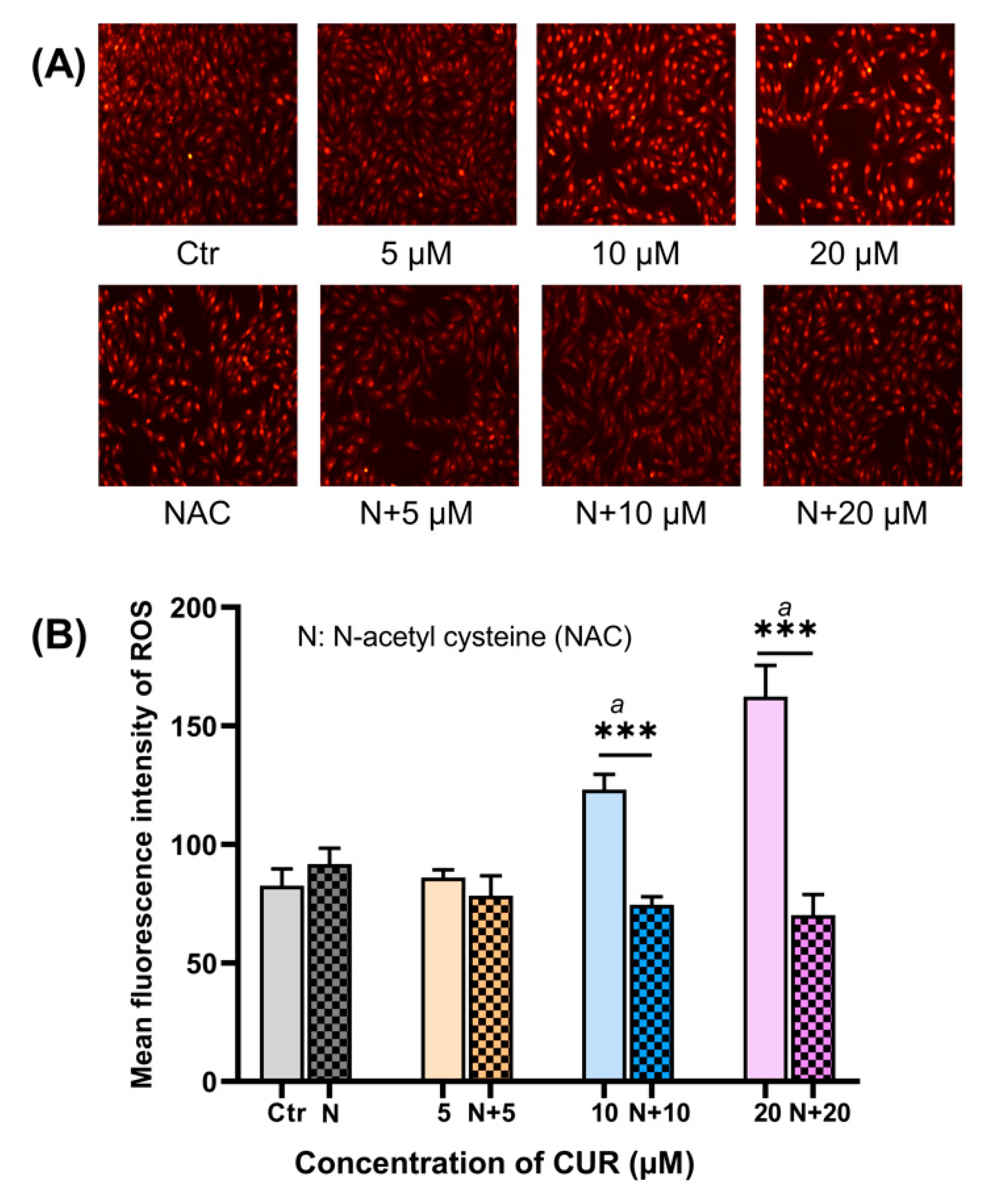
2.2. CUR Elevated Intracellular ROS in MG-63 Cells
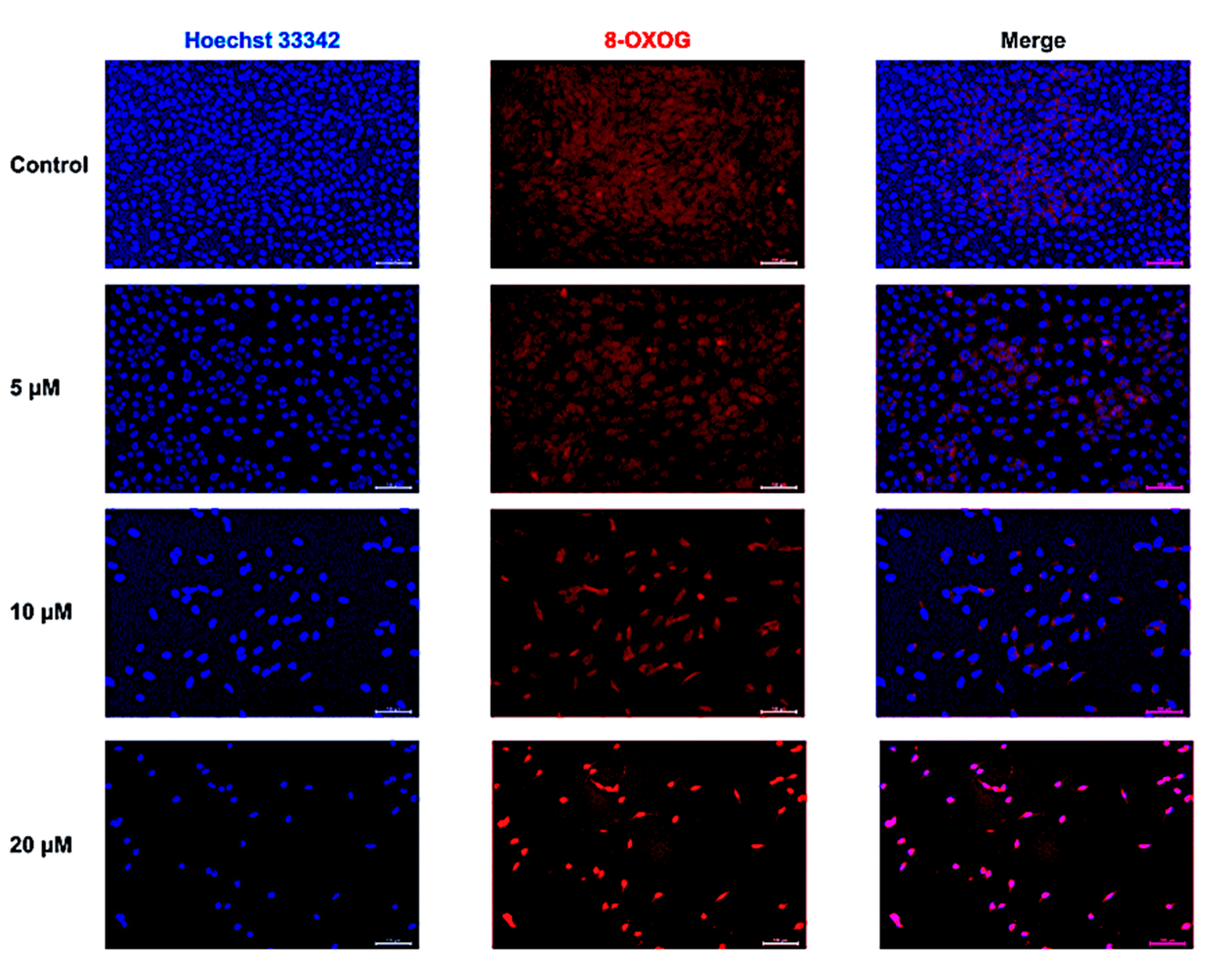
2.3. CUR-Induced Oxidative DNA Damage
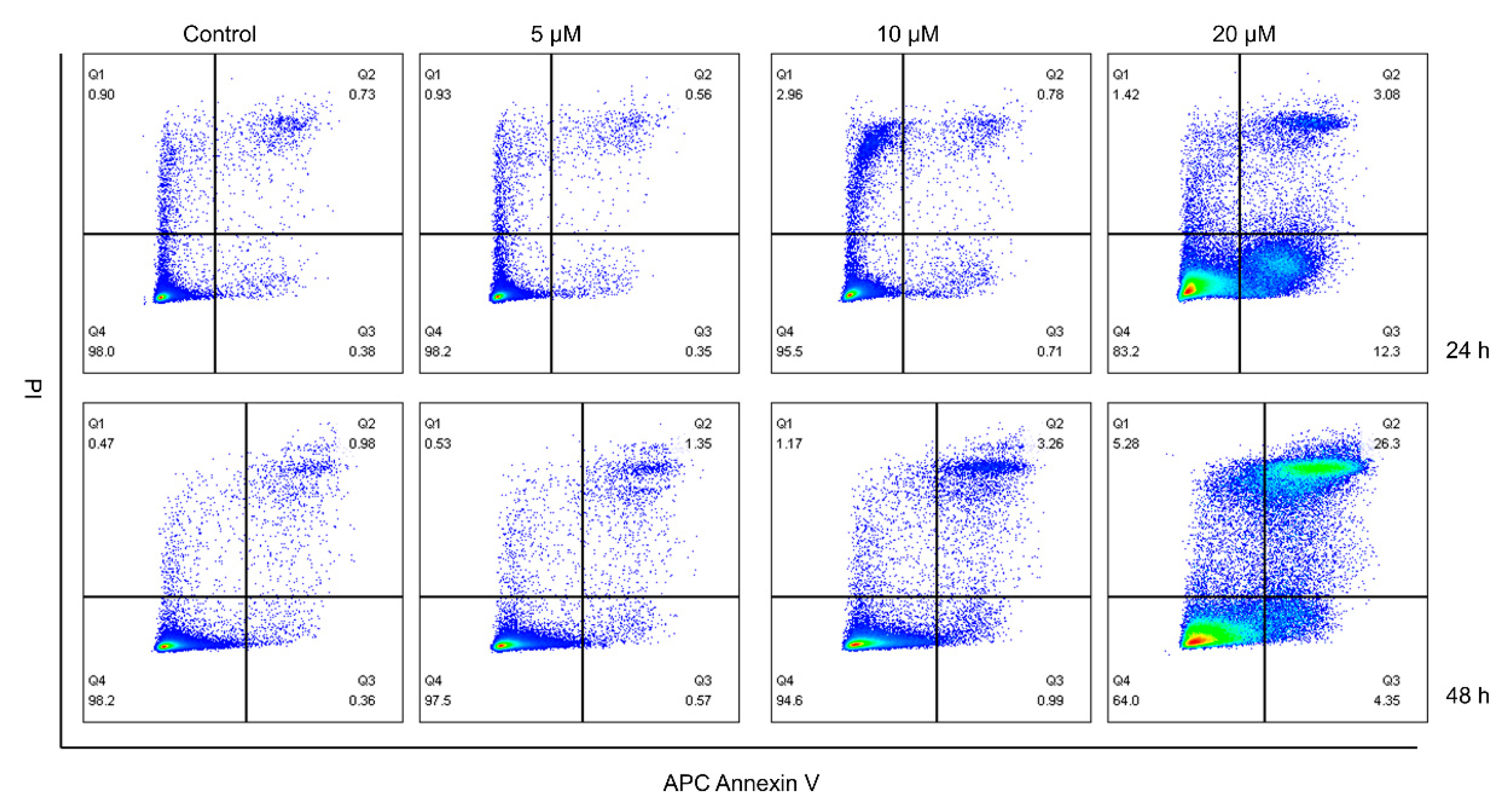
2.4. CUR Promoted the Apoptosis of MG-63 Cells
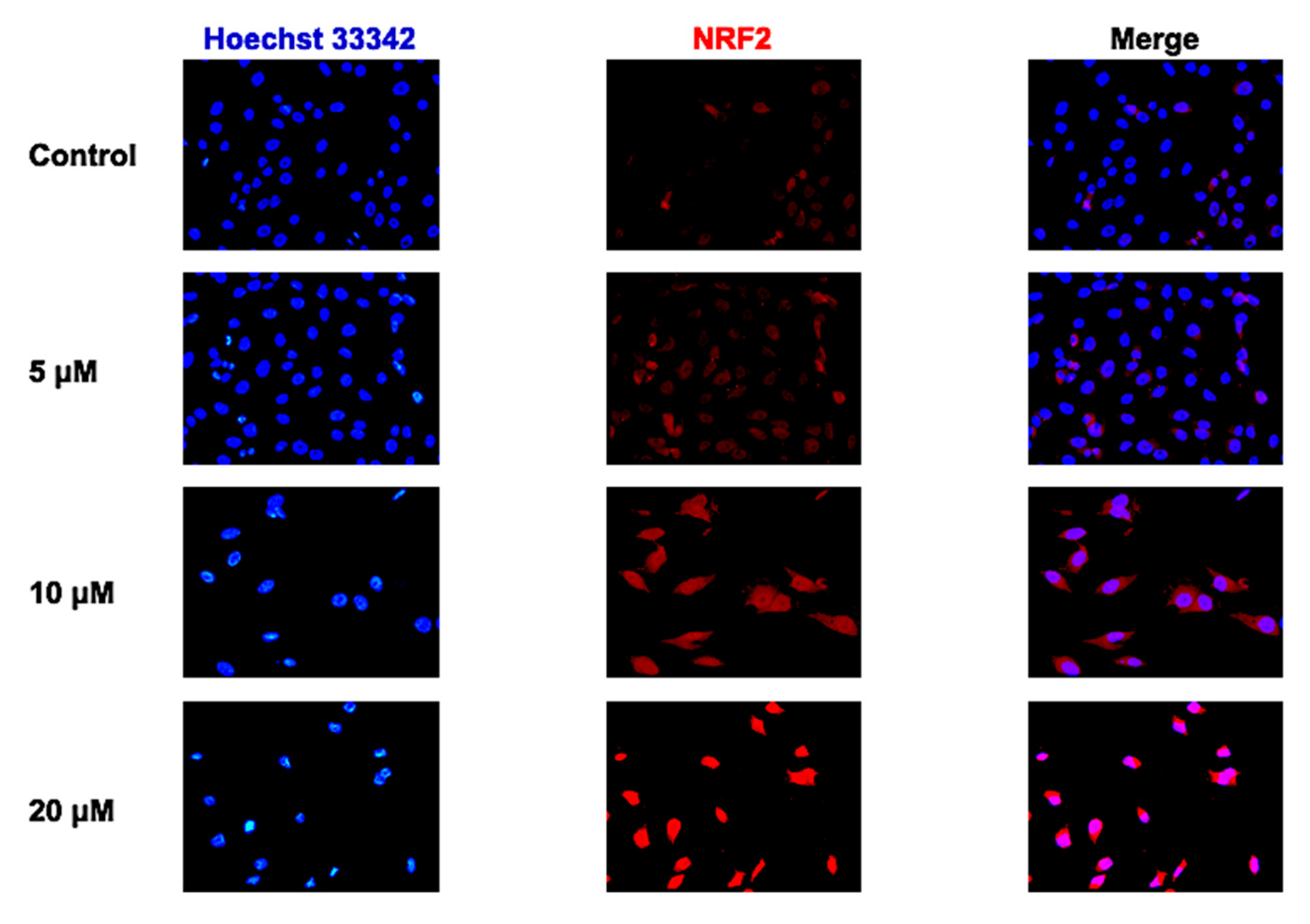
2.5. Effect of CUR on NRF2 Nuclear Translocation in MG-63 Cells
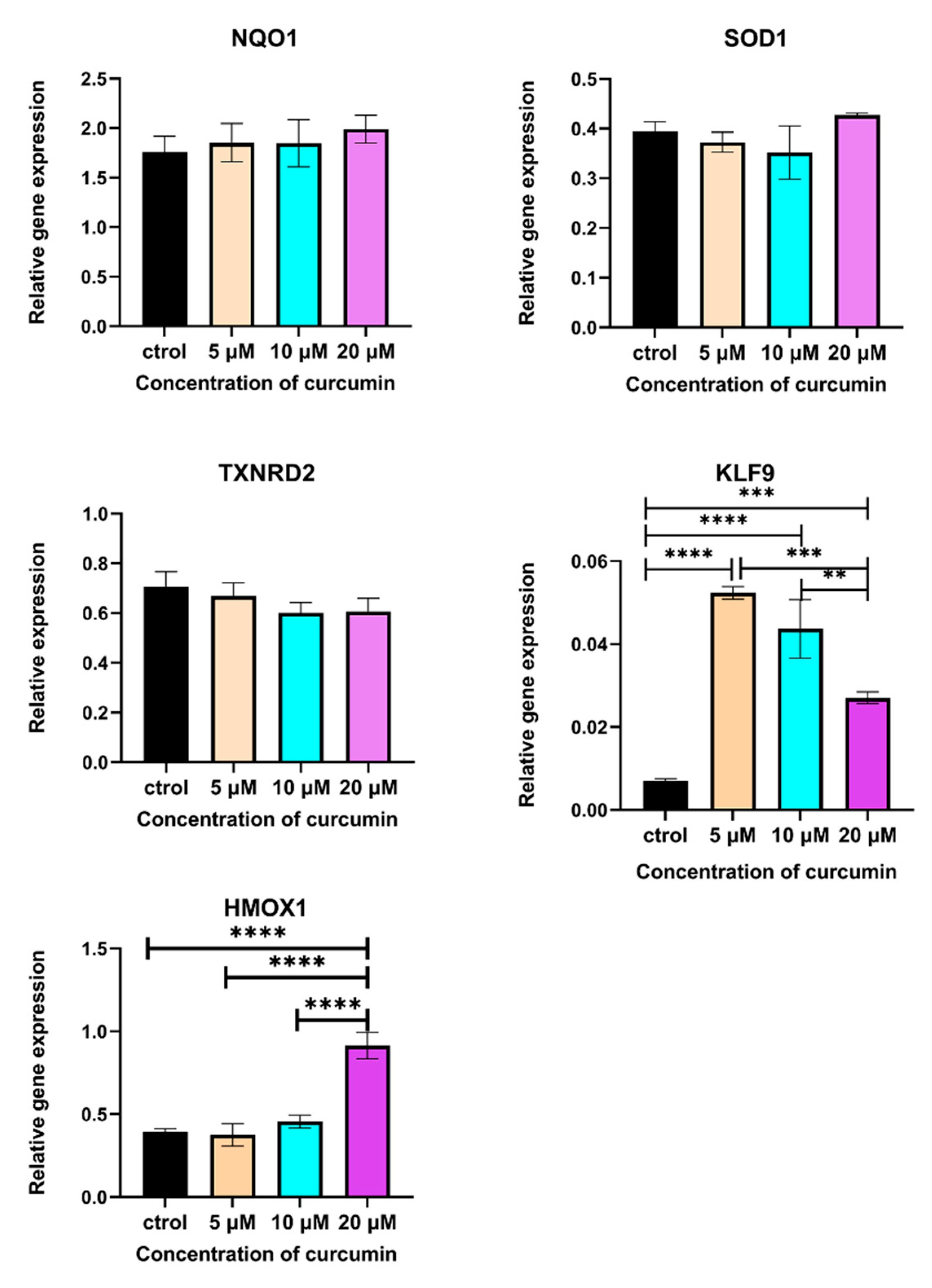
2.6. CUR Modulated the Expression of Some NRF2-Related Genes
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture
4.2. Cell Viability Assay
4.3. Quantitation of Introcellular ROS Level
4.4. Immunofluorescence Staining of 8-OXOG
4.5. Quantitation of Cell Apoptosis
4.6. Immunofluorescence Staining of NRF2
4.7. Reverse Transcription and Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kansara, M.; Teng, M.W.; Smyth, M.J.; Thomas, D.M. Translational Biology of Osteosarcoma. Nat. Rev. Cancer 2014, 14, 722–735. [Google Scholar] [CrossRef]
- Mirabello, L.; Troisi, R.J.; Savage, S.A. Osteosarcoma Incidence and Survival Rates from 1973 to 2004. Cancer 2009, 115, 1531–1543. [Google Scholar] [CrossRef] [PubMed]
- Dorfman, H.D.; Czerniak, B. Bone Cancers. Cancer 1995, 75, 203–210. [Google Scholar] [CrossRef]
- Ritter, J.; Bielack, S.S. Osteosarcoma. Ann. Oncol. 2010, 21 (Suppl. S7), vii320–vii325. [Google Scholar] [CrossRef] [PubMed]
- Hayyan, M.; Hashim, M.A.; AlNashef, I.M. Superoxide Ion: Generation and Chemical Implications. Chem. Rev. 2016, 116, 3029–3085. [Google Scholar] [CrossRef]
- Han, D.; Williams, E.; Cadenas, E. Mitochondrial Respiratory Chain-Dependent Generation of Superoxide Anion and Its Release into the Intermembrane Space. Biochem. J. 2001, 353, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Juan, C.A.; Pérez de la Lastra, J.M.; Plou, F.J.; Pérez-Lebeña, E. The Chemistry of Reactive Oxygen Species (ROS) Revisited: Outlining Their Role in Biological Macromolecules (DNA, Lipids and Proteins) and Induced Pathologies. Int. J. Mol. Sci. 2021, 22, 4642. [Google Scholar] [CrossRef]
- Schieber, M.; Chandel, N.S. ROS Function in Redox Signaling and Oxidative Stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of Cancer Metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef]
- Edderkaoui, M.; Hong, P.; Vaquero, E.C.; Lee, J.K.; Fischer, L.; Friess, H.; Buchler, M.W.; Lerch, M.M.; Pandol, S.J.; Gukovskaya, A.S. Extracellular Matrix Stimulates Reactive Oxygen Species Production and Increases Pancreatic Cancer Cell Survival through 5-Lipoxygenase and NADPH Oxidase. Am. J. Physiol. Gastrointest. Liver Physiol. 2005, 289, G1137–G1147. [Google Scholar] [CrossRef]
- Moloney, J.N.; Cotter, T.G. ROS Signalling in the Biology of Cancer. Semin. Cell Dev. Biol. 2018, 80, 50–64. [Google Scholar] [CrossRef]
- Bukke, V.N.; Moola, A.; Serviddio, G.; Vendemiale, G.; Bellanti, F. Nuclear Factor Erythroid 2-Related Factor 2-Mediated Signaling and Metabolic Associated Fatty Liver Disease. World J. Gastroenterol. 2022, 28, 6909–6921. [Google Scholar] [CrossRef] [PubMed]
- Ghareghomi, S.; Habibi-Rezaei, M.; Arese, M.; Saso, L.; Moosavi-Movahedi, A.A. Nrf2 Modulation in Breast Cancer. Biomedicines 2022, 10, 2668. [Google Scholar] [CrossRef] [PubMed]
- Tossetta, G.; Marzioni, D. Targeting the NRF2/KEAP1 Pathway in Cervical and Endometrial Cancers. Eur. J. Pharmacol. 2023, 941, 175503. [Google Scholar] [CrossRef]
- Tossetta, G.; Fantone, S.; Marzioni, D.; Mazzucchelli, R. Role of Natural and Synthetic Compounds in Modulating NRF2/KEAP1 Signaling Pathway in Prostate Cancer. Cancers 2023, 15, 3037. [Google Scholar] [CrossRef]
- Tossetta, G.; Fantone, S.; Piani, F.; Crescimanno, C.; Ciavattini, A.; Giannubilo, S.R.; Marzioni, D. Modulation of NRF2/KEAP1 Signaling in Preeclampsia. Cells 2023, 12, 1545. [Google Scholar] [CrossRef]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell Survival Responses to Environmental Stresses Via the Keap1-Nrf2-ARE Pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Kensler, T.W.; Motohashi, H. The KEAP1-NRF2 System: A Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol. Rev. 2018, 98, 1169–1203. [Google Scholar] [CrossRef]
- Robledinos-Antón, N.; Fernández-Ginés, R.; Manda, G.; Cuadrado, A. Activators and Inhibitors of NRF2: A Review of Their Potential for Clinical Development. Oxid. Med. Cell. Longev. 2019, 2019, 9372182. [Google Scholar] [CrossRef]
- Baird, L.; Yamamoto, M. The Molecular Mechanisms Regulating the KEAP1-NRF2 Pathway. Mol. Cell. Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef]
- Zucker, S.N.; Fink, E.E.; Bagati, A.; Mannava, S.; Bianchi-Smiraglia, A.; Bogner, P.N.; Wawrzyniak, J.A.; Foley, C.; Leonova, K.I.; Grimm, M.J.; et al. Nrf2 Amplifies Oxidative Stress via Induction of Klf9. Mol. Cell 2014, 53, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Parga, J.A.; Rodriguez-Perez, A.I.; Garcia-Garrote, M.; Rodriguez-Pallares, J.; Labandeira-Garcia, J.L. Angiotensin II Induces Oxidative Stress and Upregulates Neuroprotective Signaling from the NRF2 and KLF9 Pathway in Dopaminergic Cells. Free Radic. Biol. Med. 2018, 129, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Chhunchha, B.; Kubo, E.; Singh, D.P. Sulforaphane-Induced Klf9/Prdx6 Axis Acts as a Molecular Switch to Control Redox Signaling and Determines Fate of Cells. Cells 2019, 8, 1159. [Google Scholar] [CrossRef] [PubMed]
- Ak, T.; Gülçin, İ. Antioxidant and Radical Scavenging Properties of Curcumin. Chem. Biol. Interact. 2008, 174, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Kunnumakkara, A.B.; Aggarwal, B.B. Curcumin as “Curecumin”: From Kitchen to Clinic. Biochem. Pharmacol. 2008, 75, 787–809. [Google Scholar] [CrossRef]
- Shin, J.W.; Chun, K.-S.; Kim, D.-H.; Kim, S.-J.; Kim, S.H.; Cho, N.-C.; Na, H.-K.; Surh, Y.-J. Curcumin Induces Stabilization of Nrf2 Protein through Keap1 Cysteine Modification. Biochem. Pharmacol. 2020, 173, 113820. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Gupta, S.C.; Sung, B. Curcumin: An Orally Bioavailable Blocker of TNF and Other pro-Inflammatory Biomarkers. Br. J. Pharmacol. 2013, 169, 1672–1692. [Google Scholar] [CrossRef]
- Giordano, A.; Tommonaro, G. Curcumin and Cancer. Nutrients 2019, 11, 2376. [Google Scholar] [CrossRef]
- Tossetta, G.; Fantone, S.; Giannubilo, S.R.; Marzioni, D. The Multifaced Actions of Curcumin in Pregnancy Outcome. Antioxidants 2021, 10, 126. [Google Scholar] [CrossRef]
- Li, Z.; Li, D.; Chen, R.; Gao, S.; Xu, Z.; Li, N. Cell Death Regulation: A New Way for Natural Products to Treat Osteoporosis. Pharmacol. Res. 2023, 187, 106635. [Google Scholar] [CrossRef]
- Xu, C.; Wang, M.; Guo, W.; Sun, W.; Liu, Y. Curcumin in Osteosarcoma Therapy: Combining with Immunotherapy, Chemotherapeutics, Bone Tissue Engineering Materials and Potential Synergism with Photodynamic Therapy. Front. Oncol. 2021, 11, 672490. [Google Scholar] [CrossRef]
- Angulo, P.; Kaushik, G.; Subramaniam, D.; Dandawate, P.; Neville, K.; Chastain, K.; Anant, S. Natural Compounds Targeting Major Cell Signaling Pathways: A Novel Paradigm for Osteosarcoma Therapy. J. Hematol. Oncol. 2017, 10, 10. [Google Scholar] [CrossRef]
- Sarkar, N.; Bose, S. Liposome-Encapsulated Curcumin-Loaded 3D Printed Scaffold for Bone Tissue Engineering. ACS Appl. Mater. Interfaces 2019, 11, 17184–17192. [Google Scholar] [CrossRef]
- Devassy, J.G.; Nwachukwu, I.D.; Jones, P.J.H. Curcumin and Cancer: Barriers to Obtaining a Health Claim. Nutr. Rev. 2015, 73, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.-F.; Gong, Y.-X.; Li, H.-F.; Sun, F.-L.; Li, W.-L.; Chen, D.-Q.; Xie, D.-P.; Ren, C.-X.; Guo, X.-Y.; Wang, Z.-Y.; et al. Curcumin Activates ROS Signaling to Promote Pyroptosis in Hepatocellular Carcinoma HepG2 Cells. In Vivo 2021, 35, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Kim, H.S.; Jung, E.-J.; Lee, J.Y.; Tsang, B.K.; Lim, J.M.; Song, Y.S. Curcumin Induces ER Stress-Mediated Apoptosis through Selective Generation of Reactive Oxygen Species in Cervical Cancer Cells. Mol. Carcinog. 2016, 55, 918–928. [Google Scholar] [CrossRef] [PubMed]
- Papiez, M.; Krzyściak, W.; Szade, K.; Bukowska-Straková, K.; Bystrowska, B.; Jozkowicz, A.; Dulak, J.; Kozakowska, M.; Hajduk, K. Curcumin Enhances the Cytogenotoxic Effect of Etoposide in Leukemia Cells through Induction of Reactive Oxygen Species. Drug Des. Devel. Ther. 2016, 10, 557–570. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kunwar, A.; Barik, A.; Mishra, B.; Rathinasamy, K.; Pandey, R.; Priyadarsini, K.I. Quantitative Cellular Uptake, Localization and Cytotoxicity of Curcumin in Normal and Tumor Cells. Biochim. Biophys. Acta Gen. Subj. 2008, 1780, 673–679. [Google Scholar] [CrossRef]
- Feng, T.; Wei, Y.; Lee, R.; Zhao, L. Liposomal Curcumin and Its Application in Cancer. Int. J. Nanomed. 2017, 12, 6027–6044. [Google Scholar] [CrossRef]
- Kumari, S.; Sharma, N.; Sahi, S.V. Advances in Cancer Therapeutics: Conventional Thermal Therapy to Nanotechnology-Based Photothermal Therapy. Pharmaceutics 2021, 13, 1174. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, X.; Wang, X.; Guan, X.; Zhang, W.; Ma, J. Recent Advances in Selective Photothermal Therapy of Tumor. J. Nanobiotechnology 2021, 19, 335. [Google Scholar] [CrossRef] [PubMed]
- Kadkhoda, J.; Tarighatnia, A.; Tohidkia, M.R.; Nader, N.D.; Aghanejad, A. Photothermal Therapy-Mediated Autophagy in Breast Cancer Treatment: Progress and Trends. Life Sci. 2022, 298, 120499. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, A.; Jameei, A.; Karande, A.A.; Chakravarty, A.R. BODIPY-Attached Zinc(II) Complexes of Curcumin Drug for Visible Light Assisted Photo-Sensitization, Cellular Imaging and Targeted PDT. Eur. J. Med. Chem. 2021, 220, 113438. [Google Scholar] [CrossRef]
- Zhong, Y.; Zhang, X.; Yang, L.; Liang, F.; Zhang, J.; Jiang, Y.; Chen, X.; Ren, F. Hierarchical Dual-Responsive Cleavable Nanosystem for Synergetic Photodynamic/Photothermal Therapy against Melanoma. Mater. Sci. Eng. C 2021, 131, 112524. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Chen, X.; Zhang, C.; Yang, T.; Deng, Z.; Song, Y.; Huang, L.; Li, F.; Li, Q.; Lin, S.; et al. EF24 Induces Ferroptosis in Osteosarcoma Cells through HMOX1. Biomed. Pharmacother. 2021, 136, 111202. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.-C.; Chen, H.-H.; Qu, Y.-Y.; Xu, C.-W.; Yang, C.; Liu, Y. MicroRNA-221 Promotes Cisplatin Resistance in Osteosarcoma Cells by Targeting PPP2R2A. Biosci. Rep. 2019, 39, BSR20190198. [Google Scholar] [CrossRef]
- Hussain, Y.; Islam, L.; Khan, H.; Filosa, R.; Aschner, M.; Javed, S. Curcumin–Cisplatin Chemotherapy: A Novel Strategy in Promoting Chemotherapy Efficacy and Reducing Side Effects. Phytother. Res. 2021, 35, 6514–6529. [Google Scholar] [CrossRef]
- Howells, L.M.; Iwuji, C.O.O.; Irving, G.R.B.; Barber, S.; Walter, H.; Sidat, Z.; Griffin-Teall, N.; Singh, R.; Foreman, N.; Patel, S.R.; et al. Curcumin Combined with FOLFOX Chemotherapy Is Safe and Tolerable in Patients with Metastatic Colorectal Cancer in a Randomized Phase IIa Trial. J. Nutr. 2019, 149, 1133–1139. [Google Scholar] [CrossRef]
- Tan, B.L.; Norhaizan, M.E. Curcumin Combination Chemotherapy: The Implication and Efficacy in Cancer. Molecules 2019, 24, 2527. [Google Scholar] [CrossRef]
- Cooke, M.S.; Evans, M.D.; Dizdaroglu, M.; Lunec, J. Oxidative DNA Damage: Mechanisms, Mutation, and Disease. FASEB J. 2003, 17, 1195–1214. [Google Scholar] [CrossRef]
- Wang, J.; Nagy, N.; Masucci, M.G. The Epstein–Barr Virus Nuclear Antigen-1 Upregulates the Cellular Antioxidant Defense to Enable B-Cell Growth Transformation and Immortalization. Oncogene 2020, 39, 603–616. [Google Scholar] [CrossRef] [PubMed]

| Target Gene | Primer Nucleotide Sequences | Amplicon Size | Accession No. NCBI Genebank |
|---|---|---|---|
| HPRT | FW GCTGACCTGCTGGATTACAT REV CTTGCGACCTTGACCATCT | 260 | NM_000194 |
| GUSB | FW CGCACAAGAGTGGTGCTGAG REV GGAGGTGTCAGTCAGGTA TT | 234 | NM_000181 |
| PBGD | FW TCCAAGCGGAGCCATGTCTG REV CCTGTGGTGGACATAGCAAT | 192 | NM_000190 |
| NQO1 | FW CACTGATCGT ACTGGCTCACTC REV ACAGACTCGGCAGGATACTGAA | 202 | NM_000903 |
| SOD1 | FW GACTGACTGAAGGCCTGCAT REV TAGACACATCGGCCACACCA | 186 | NM_000454 |
| HMOX1 | FW TGCGTTCCTGCTCAACATCC REV CAGCAACTGTCGCCACCAG | 233 | NM_002133 |
| TXNRD2 | FW GGAGCATGTTGAGGTCT ATC REV ATCACCTGCGCATAGGAAG | 210 | NM_006440 |
| KLF9 | FW CTCCCATCTCAAAGCCCA TT REV AGGTGGTCACTCCTCATGAAG | 183 | NM_001206 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, C.; Wang, M.; Zandieh Doulabi, B.; Sun, Y.; Liu, Y. Paradox: Curcumin, a Natural Antioxidant, Suppresses Osteosarcoma Cells via Excessive Reactive Oxygen Species. Int. J. Mol. Sci. 2023, 24, 11975. https://doi.org/10.3390/ijms241511975
Xu C, Wang M, Zandieh Doulabi B, Sun Y, Liu Y. Paradox: Curcumin, a Natural Antioxidant, Suppresses Osteosarcoma Cells via Excessive Reactive Oxygen Species. International Journal of Molecular Sciences. 2023; 24(15):11975. https://doi.org/10.3390/ijms241511975
Chicago/Turabian StyleXu, Chunfeng, Mingjie Wang, Behrouz Zandieh Doulabi, Yuanyuan Sun, and Yuelian Liu. 2023. "Paradox: Curcumin, a Natural Antioxidant, Suppresses Osteosarcoma Cells via Excessive Reactive Oxygen Species" International Journal of Molecular Sciences 24, no. 15: 11975. https://doi.org/10.3390/ijms241511975
APA StyleXu, C., Wang, M., Zandieh Doulabi, B., Sun, Y., & Liu, Y. (2023). Paradox: Curcumin, a Natural Antioxidant, Suppresses Osteosarcoma Cells via Excessive Reactive Oxygen Species. International Journal of Molecular Sciences, 24(15), 11975. https://doi.org/10.3390/ijms241511975







