Choosing Variant Interpretation Tools for Clinical Applications: Context Matters
Abstract
1. Introduction
2. Results
2.1. Estimating the Sensitivity, Specificity, and Coverage/Reject Rate of the Pathogenicity Predictors
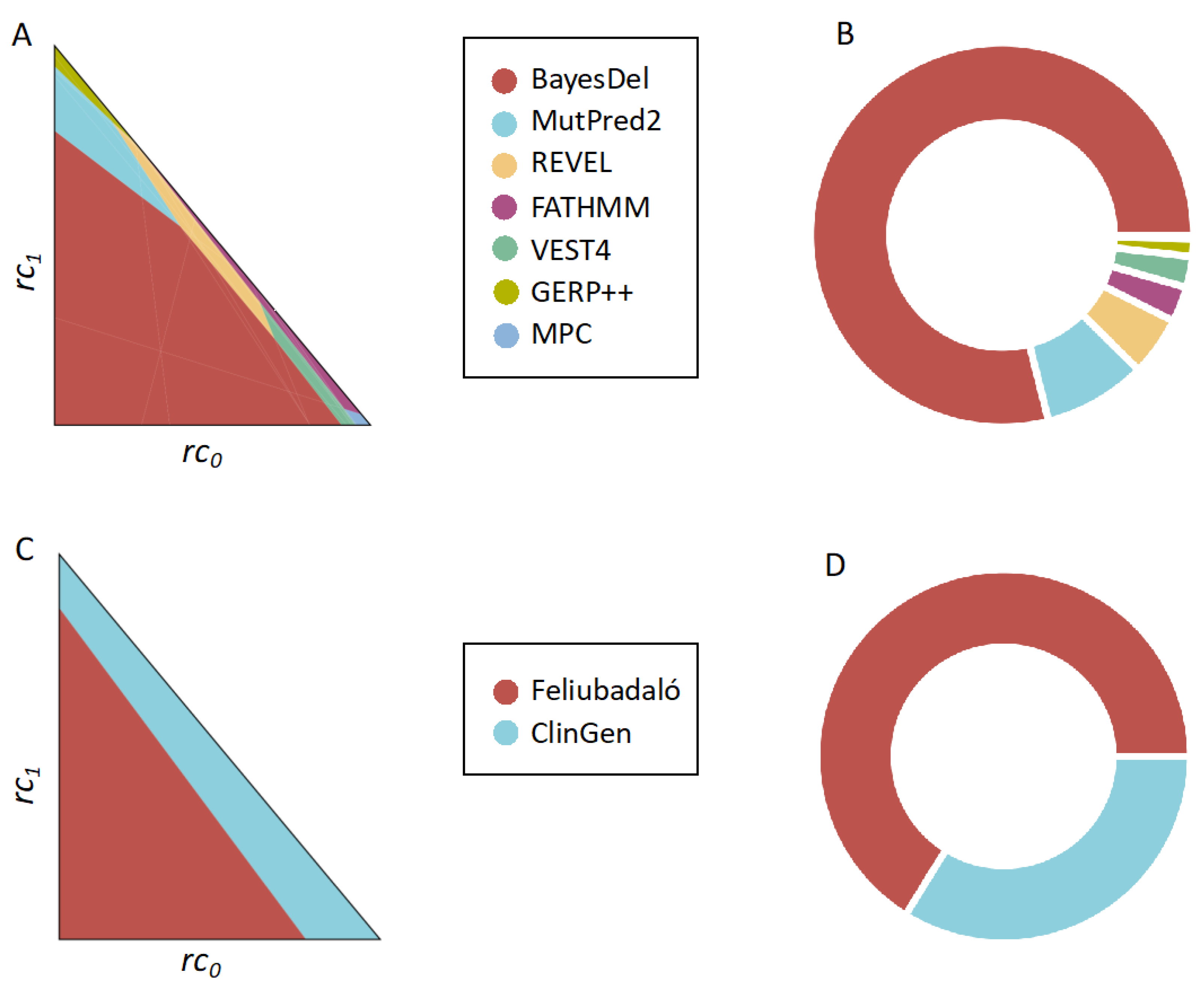
2.2. Application of the MISC Framework to the Comparison of Pathogenicity Predictors across Clinical Scenarios
2.3. Application of the MISC+REJ Framework to the Comparison of Pathogenicity Predictors across Clinical Scenarios
2.4. Cost Analysis for TP53 Gene Computational Evidence Criteria
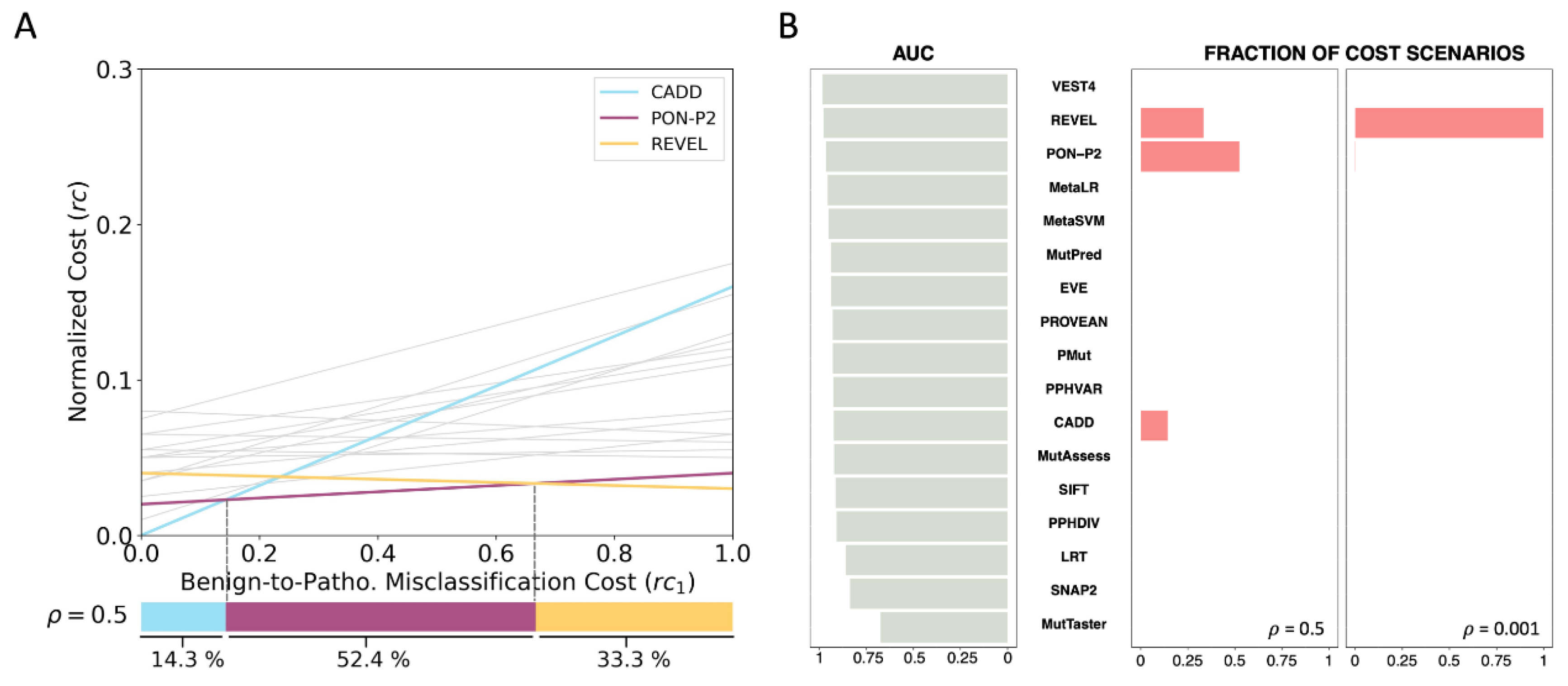
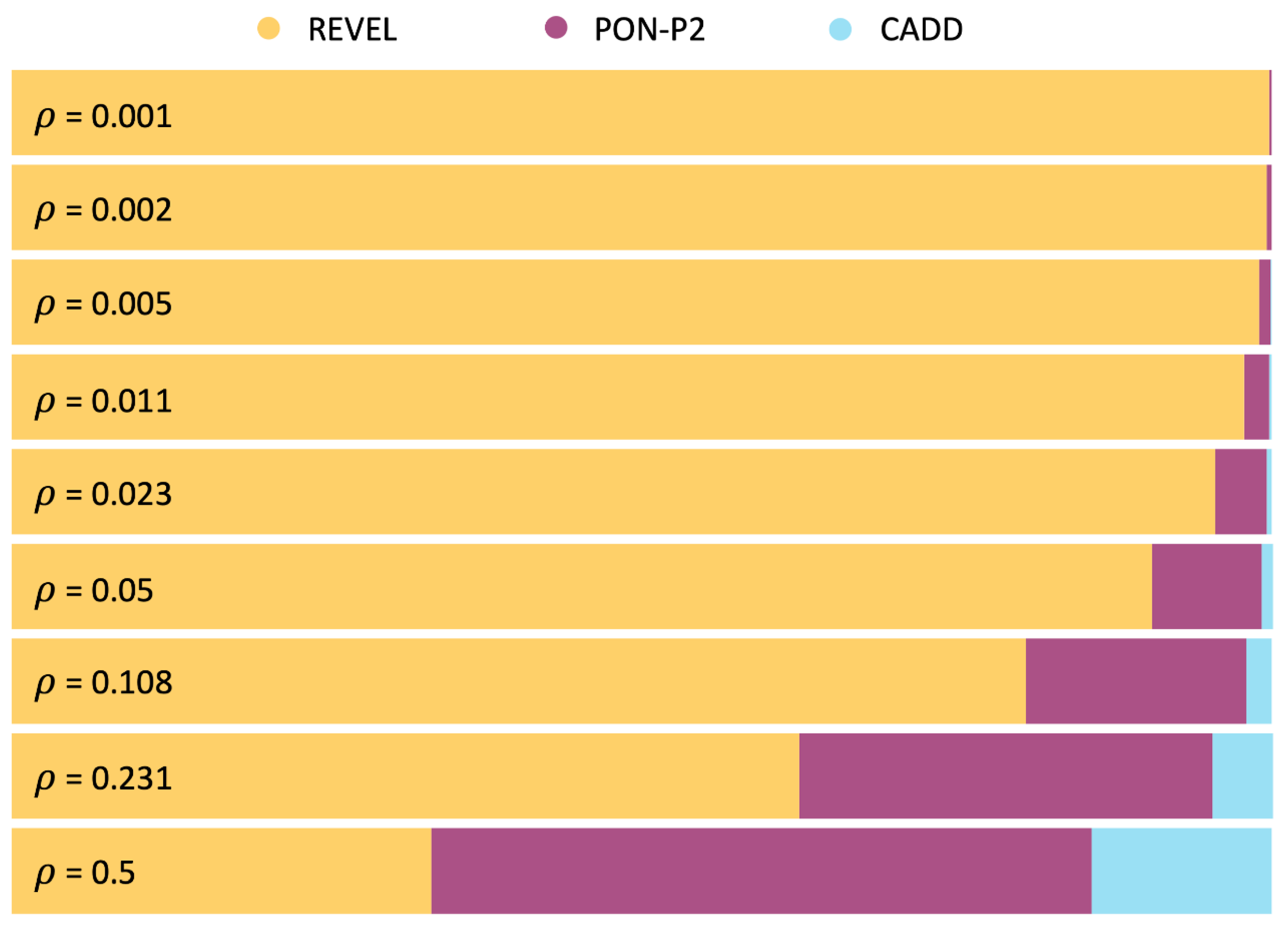
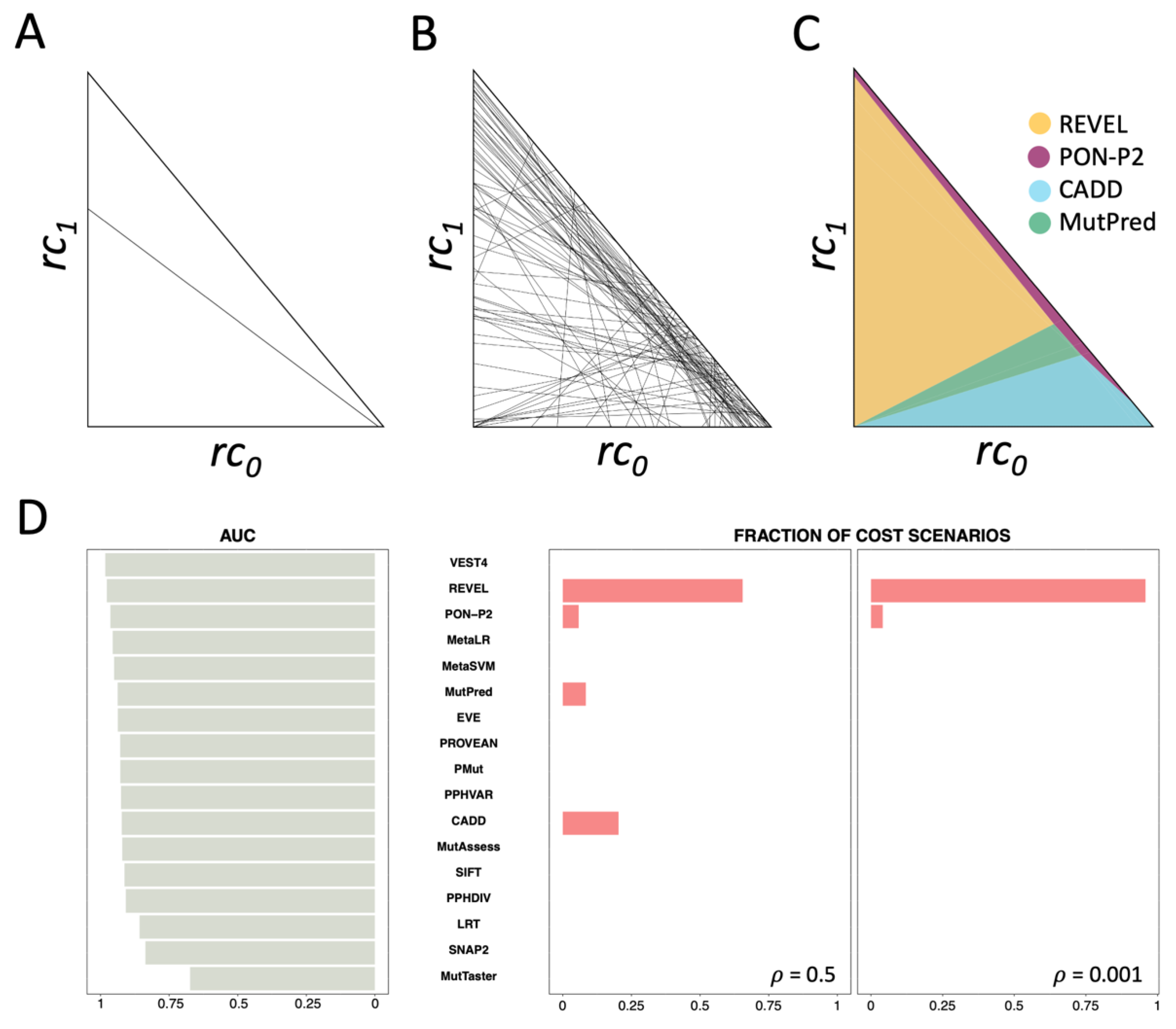
2.5. Cost Analysis of the Pathogenicity Predictors Studied by Pejaver et al. [33]
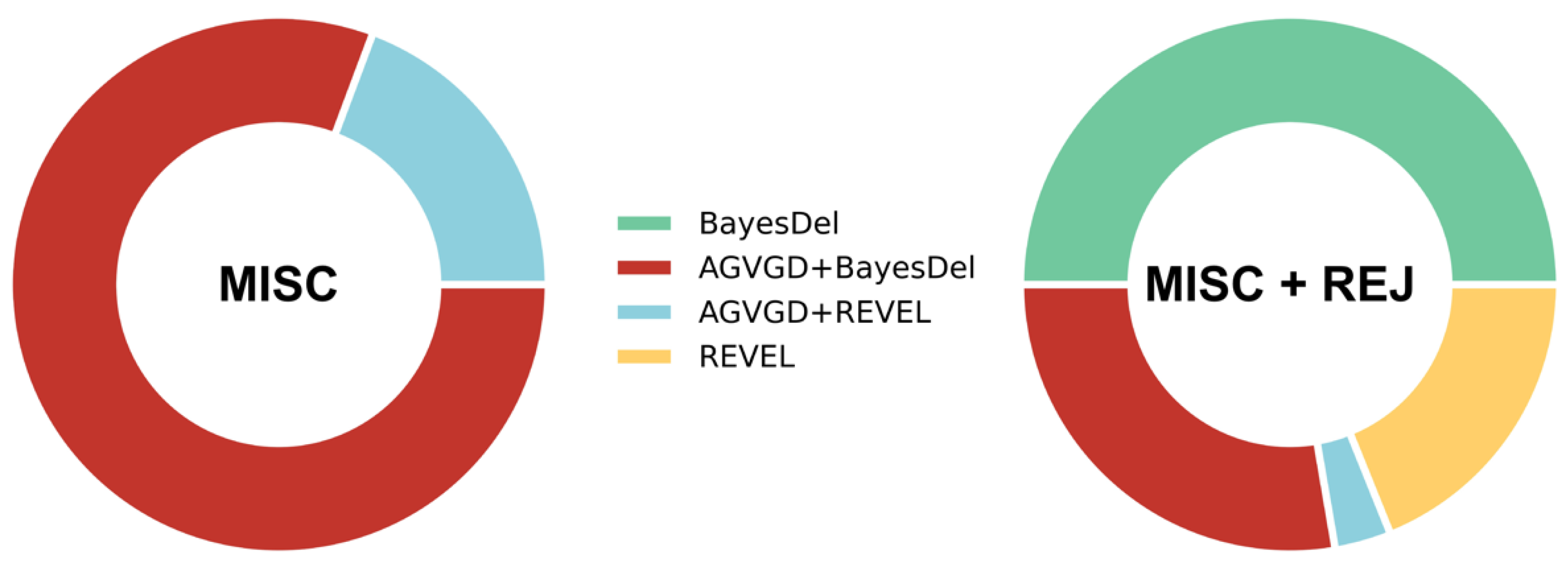
2.6. Cost-Based Comparison of the Computational Evidence Used in the Two ATM-Adapted ACMG/AMP Guidelines
3. Discussion
4. Methods and Materials
4.1. Framework for Comparing Classifiers with No Reject Option
4.1.1. Cost Model for Misclassification Errors Only
4.1.2. Predictor Comparison across Clinical Scenarios
4.2. Framework for Comparing Classifiers with Reject Option
4.2.1. Cost Model for Misclassification Errors plus Rejection
4.2.2. Predictor Comparison across Clinical Scenarios
Finding the {rk, k = 1, m} Regions from the Polygons in PN
Using an Adapted Breadth First Search (BFS) to Generate Polygons in PN
4.3. Variant Dataset
4.4. Pathogenicity Predictors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lázaro, C.; Lerner-Ellis, J.; Spurdle, A. Clinical DNA Variant Interpretation, 1st ed.; Lázaro, C., Lerner-Ellis, J., Spurdle, A., Eds.; Academic Press: London, UK, 2021. [Google Scholar]
- Shendure, J.; Findlay, G.M.; Snyder, M.W. Genomic Medicine–Progress, Pitfalls, and Promise. Cell 2019, 177, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Chapman, M.; Evans, K.; Azevedo, L.; Hayden, M.; Heywood, S.; Millar, D.S.; Phillips, A.D.; et al. The Human Gene Mutation Database (HGMD®): Optimizing Its Use in a Clinical Diagnostic or Research Setting. Hum. Genet. 2020, 139, 1197–1207. [Google Scholar] [CrossRef] [PubMed]
- Özkan, S.; Padilla, N.; Moles-Fernández, A.; Diez, O.; Gutiérrez-Enríquez, S.; de la Cruz, X. The Computational Approach to Variant Interpretation: Principles, Results, and Applicability. In Clinical DNA Variant Interpretation: Theory and Practice; Lázaro, C., Lerner-Ellis, J., Spurdle, A., Eds.; Elsevier Inc./Academic Press: London, UK, 2021; pp. 89–119. [Google Scholar]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C.; Wagstaff, K.L. Machine Learning for Science and Society. Mach. Learn. 2014, 95, 1–9. [Google Scholar] [CrossRef]
- Vihinen, M. How to Evaluate Performance of Prediction Methods? Measures and Their Interpretation in Variation Effect Analysis. BMC Genom. 2012, 13, S2. [Google Scholar] [CrossRef]
- Adams, N.M.; Hand, D.J. Comparing Classifiers When the Misallocation Costs Are Uncertain. Pattern Recognit. 1999, 32, 1139–1147. [Google Scholar] [CrossRef]
- Pepe, M.S. The Statistical Evaluation of Medical Tests for Classification and Prediction; Oxford University Press: Oxford, UK, 2003. [Google Scholar]
- OECD. Health at a Glance 2021: OECD Indicators; OECD Publishing: Paris, France, 2021; pp. 1–275. ISSN 1999-1312. [Google Scholar]
- Mulcahy, A.W.; Whaley, C.M.; Gizaw, M.; Schwam, D.; Edenfield, N.; Becerra-Ornelas, A.U. International Prescription Drug Price Comparisons: Current Empirical Estimates and Comparisons with Previous Studies; RAND Corporation: Santa Monica, CA, USA, 2021. [Google Scholar]
- Hand, D.J. Classifier Technology and the Illusion of Progress. Stat. Sci. 2006, 21, 1–15. [Google Scholar] [CrossRef]
- Herbei, R.; Wegkamp, M.H. Classification with Reject Option. Can. J. Stat. 2006, 34, 709–721. [Google Scholar] [CrossRef]
- Hanczar, B. Performance Visualization Spaces for Classification with Rejection Option. Pattern Recognit. 2019, 96, 106984. [Google Scholar] [CrossRef]
- Feliubadaló, L.; Moles-Fernández, A.; Santamariña-Pena, M.; Sánchez, A.T.; López-Novo, A.; Porras, L.-M.; Blanco, A.; Capellá, G.; de la Hoya, M.; Molina, I.J.; et al. A Collaborative Effort to Define Classification Criteria for ATM Variants in Hereditary Cancer Patients. Clin. Chem. 2021, 67, 518–533. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. PolyPhen-2: Prediction of Functional Effects of Human NsSNPs. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the Effects of Coding Non-Synonymous Variants on Protein Function Using the SIFT Algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the Deleteriousness of Variants throughout the Human Genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation Prediction for the Deep-Sequencing Age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef]
- Reva, B.; Antipin, Y.; Sander, C. Predicting the Functional Impact of Protein Mutations: Application to Cancer Genomics. Nucleic Acids Res. 2011, 39, e118. [Google Scholar] [CrossRef]
- Ioannidis, N.M.; Rothstein, J.H.; Pejaver, V.; Middha, S.; McDonnell, S.K.; Baheti, S.; Musolf, A.; Li, Q.; Holzinger, E.; Karyadi, D.; et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016, 99, 877–885. [Google Scholar] [CrossRef]
- Chun, S.; Fay, J.C. Identification of Deleterious Mutations within Three Human Genomes. Genome Res. 2009, 19, 1553–1561. [Google Scholar] [CrossRef]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef]
- Dong, C.; Wei, P.; Jian, X.; Gibbs, R.; Boerwinkle, E.; Wang, K.; Liu, X. Comparison and Integration of Deleteriousness Prediction Methods for Nonsynonymous SNVs in Whole Exome Sequencing Studies. Hum. Mol. Genet. 2015, 24, 2125–2137. [Google Scholar] [CrossRef]
- Carter, H.; Douville, C.; Stenson, P.D.; Cooper, D.N.; Karchin, R. Identifying Mendelian Disease Genes with the Variant Effect Scoring Tool. BMC Genom. 2013, 14 (Suppl. 3), S3. [Google Scholar] [CrossRef]
- Pejaver, V.; Urresti, J.; Lugo-Martinez, J.; Pagel, K.A.; Lin, G.N.; Nam, H.J.; Mort, M.; Cooper, D.N.; Sebat, J.; Iakoucheva, L.M.; et al. Inferring the Molecular and Phenotypic Impact of Amino Acid Variants with MutPred2. Nat. Commun. 2020, 11, 5918. [Google Scholar] [CrossRef] [PubMed]
- Niroula, A.; Urolagin, S.; Vihinen, M. PON-P2: Prediction Method for Fast and Reliable Identification of Harmful Variants. PLoS ONE 2015, 10, e0117380. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, Y.; Yachdav, G.; Rost, B. SNAP Predicts Effect of Mutations on Protein Function. Bioinformatics 2008, 24, 2397–2398. [Google Scholar] [CrossRef] [PubMed]
- Frazer, J.; Notin, P.; Dias, M.; Gomez, A.; Min, J.K.; Brock, K.; Gal, Y.; Marks, D.S. Disease Variant Prediction with Deep Generative Models of Evolutionary Data. Nature 2021, 599, 91–95. [Google Scholar] [CrossRef]
- López-Ferrando, V.; Gazzo, A.; De La Cruz, X.; Orozco, M.; Gelpí, J.L. PMut: A Web-Based Tool for the Annotation of Pathological Variants on Proteins, 2017 Update. Nucleic Acids Res. 2017, 45, W222–W228. [Google Scholar] [CrossRef]
- Ernst, C.; Hahnen, E.; Engel, C.; Nothnagel, M.; Weber, J.; Schmutzler, R.K.; Hauke, J. Performance of in Silico Prediction Tools for the Classification of Rare BRCA1/2 Missense Variants in Clinical Diagnostics. BMC Med. Genom. 2018, 11, 35. [Google Scholar] [CrossRef]
- Hereditary Breast, Ovarian and Pancreatic Cancer Variant Curation Expert Panel. ClinGen Hereditary Breast, Ovarian and Pancreatic Cancer Expert Panel Specifications to the ACMG/AMP Variant Interpretation Guidelines for ATM Version 1.1; ClinGen: Bethesda, MD, USA, 2022. [Google Scholar]
- Pejaver, V.; Byrne, A.B.; Feng, B.; Radivojac, P.; Brenner, S.E.; Pejaver, V.; Byrne, A.B.; Feng, B.; Pagel, K.A.; Mooney, S.D.; et al. Calibration of Computational Tools for Missense Variant Pathogenicity Classification and ClinGen Recommendations for PP3/BP4 Criteria. Am. J. Hum. Genet. 2022, 109, 2163–2177. [Google Scholar] [CrossRef]
- ClinGen TP53 Expert Panel Specifications to the ACMG/AMP Variant Interpretation Guidelines Version 1. Available online: https://www.clinicalgenome.org/affiliation/50013/ (accessed on 1 June 2023).
- Rehm, H.L.; Berg, J.S.; Brooks, L.D.; Bustamante, C.D.; Evans, J.P.; Landrum, M.J.; Ledbetter, D.H.; Maglott, D.R.; Martin, C.L.; Nussbaum, R.L.; et al. ClinGen—The Clinical Genome Resource. N. Engl. J. Med. 2015, 372, 2235–2242. [Google Scholar] [CrossRef]
- Fortuno, C.; James, P.A.; Young, E.L.; Feng, B.; Olivier, M.; Pesaran, T.; Tavtigian, S.V.; Spurdle, A.B. Improved, ACMG-Compliant, in Silico Prediction of Pathogenicity for Missense Substitutions Encoded by TP53 Variants. Hum. Mutat. 2018, 39, 1061–1069. [Google Scholar] [CrossRef]
- Drummond, C.; Holte, R.C. Cost Curves: An Improved Method for Visualizing Classifier Performance. Mach. Learn. 2006, 65, 95–130. [Google Scholar] [CrossRef]
- Baldi, P.; Brunak, S.; Chauvin, Y.; Andersen, C.A.F.; Nielsen, H. Assessing the Accuracy of Prediction Algorithms for Classification: An Overview. Bioinformatics 2000, 6, 412–424. [Google Scholar] [CrossRef]
- Hernández-Orallo, J.; Flach, P.; Ferri, C. A Unified View of Performance Metrics: Translating Threshold Choice into Expected Classification Loss. J. Mach. Learn. Res. 2012, 13, 2813–2869. [Google Scholar]
- De Berg, M.; Cheong, O.; van Kreveld, M.; Overmars, M. Computational Geometry: Algorithms and Applications, 3rd ed.; Springer: New York, NY, USA, 2008. [Google Scholar]
- Liu, X.; Li, C.; Mou, C.; Dong, Y.; Tu, Y. DbNSFP v4: A Comprehensive Database of Transcript-Specific Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Genome Med. 2020, 12, 103. [Google Scholar] [CrossRef]
- Grimm, D.G.; Azencott, C.-A.; Aicheler, F.; Gieraths, U.; MacArthur, D.G.; Samocha, K.E.; Cooper, D.N.; Stenson, P.D.; Daly, M.J.; Smoller, J.W.; et al. The Evaluation of Tools Used to Predict the Impact of Missense Variants Is Hindered by Two Types of Circularity. Hum. Mutat. 2015, 36, 513–523. [Google Scholar] [CrossRef]
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to Accessing Data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef]
- Katsonis, P.; Wilhelm, K.; Williams, A.; Lichtarge, O. Genome Interpretation Using in Silico Predictors of Variant Impact. Hum. Genet. 2022, 141, 1549–1577. [Google Scholar] [CrossRef]
- Liu, X.; Wu, C.; Li, C.; Boerwinkle, E. DbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum. Mutat. 2016, 37, 235–241. [Google Scholar] [CrossRef]
- Vihinen, M. Problems in Variation Interpretation Guidelines and in Their Implementation in Computational Tools. Mol. Genet. Genomic Med. 2020, 8, e1206. [Google Scholar] [CrossRef]
- Chow, C.K. On Optimum Recognition Error and Reject Tradeoff. IEEE Trans. Inf. Theory 1970, 6, 41–46. [Google Scholar] [CrossRef]
- Lee, J.M. Axiomatic Geometry; American Mathematical Society: Providence, RI, USA, 2012. [Google Scholar]
- Yaglom, I.M.; Boltyanskii, V.G. Convex Figures; Holt, Rinehart and Winston: New York, NY, USA, 1961. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguirre, J.; Padilla, N.; Özkan, S.; Riera, C.; Feliubadaló, L.; de la Cruz, X. Choosing Variant Interpretation Tools for Clinical Applications: Context Matters. Int. J. Mol. Sci. 2023, 24, 11872. https://doi.org/10.3390/ijms241411872
Aguirre J, Padilla N, Özkan S, Riera C, Feliubadaló L, de la Cruz X. Choosing Variant Interpretation Tools for Clinical Applications: Context Matters. International Journal of Molecular Sciences. 2023; 24(14):11872. https://doi.org/10.3390/ijms241411872
Chicago/Turabian StyleAguirre, Josu, Natàlia Padilla, Selen Özkan, Casandra Riera, Lídia Feliubadaló, and Xavier de la Cruz. 2023. "Choosing Variant Interpretation Tools for Clinical Applications: Context Matters" International Journal of Molecular Sciences 24, no. 14: 11872. https://doi.org/10.3390/ijms241411872
APA StyleAguirre, J., Padilla, N., Özkan, S., Riera, C., Feliubadaló, L., & de la Cruz, X. (2023). Choosing Variant Interpretation Tools for Clinical Applications: Context Matters. International Journal of Molecular Sciences, 24(14), 11872. https://doi.org/10.3390/ijms241411872






