Cell Profiling of Acute Kidney Injury to Chronic Kidney Disease Reveals Novel Oxidative Stress Characteristics in the Failed Repair of Proximal Tubule Cells
Abstract
1. Introduction
2. Results
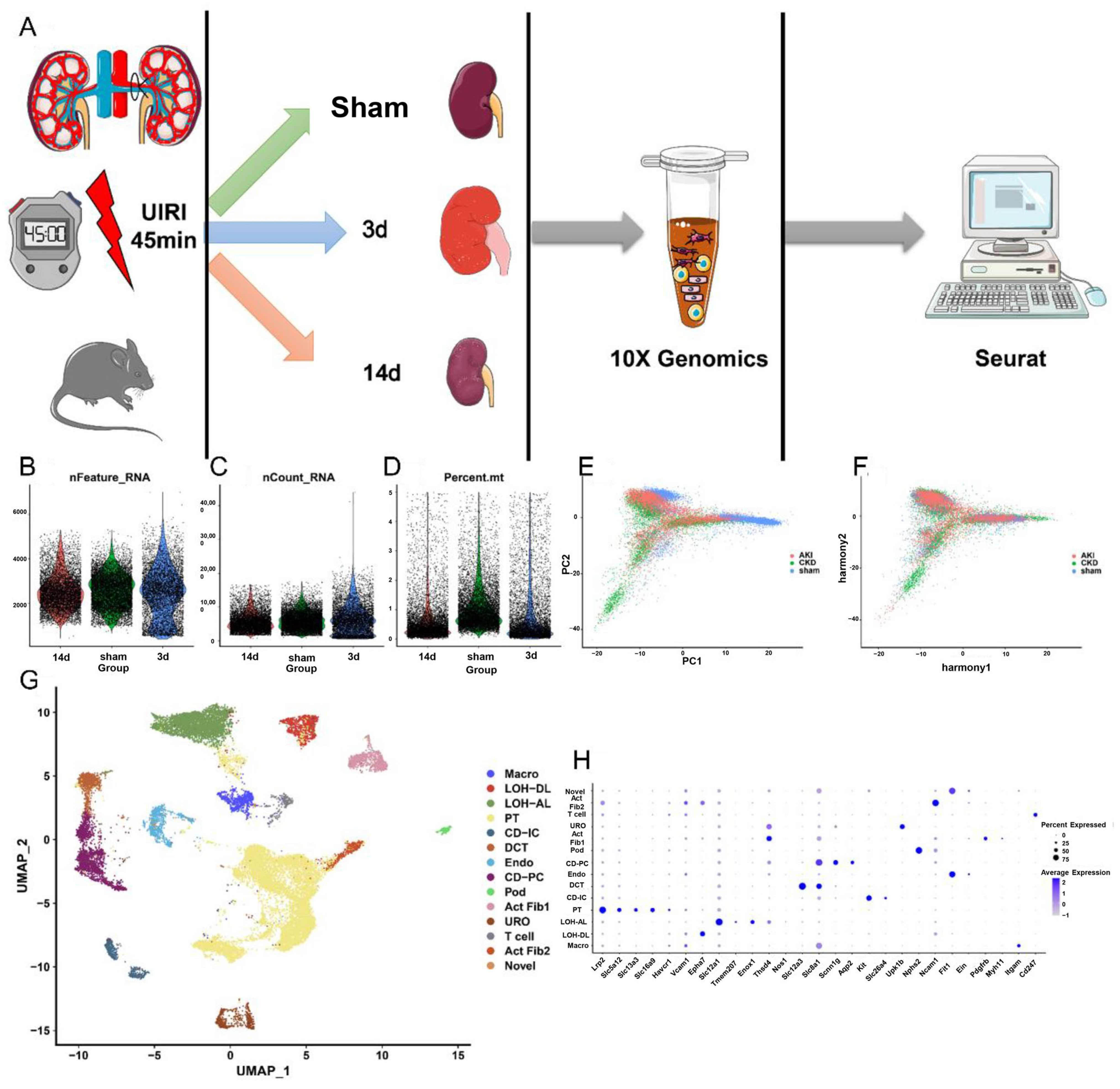
2.1. SnRNA and ScRNA Profiling of Kidney Cells from AKI to CKD
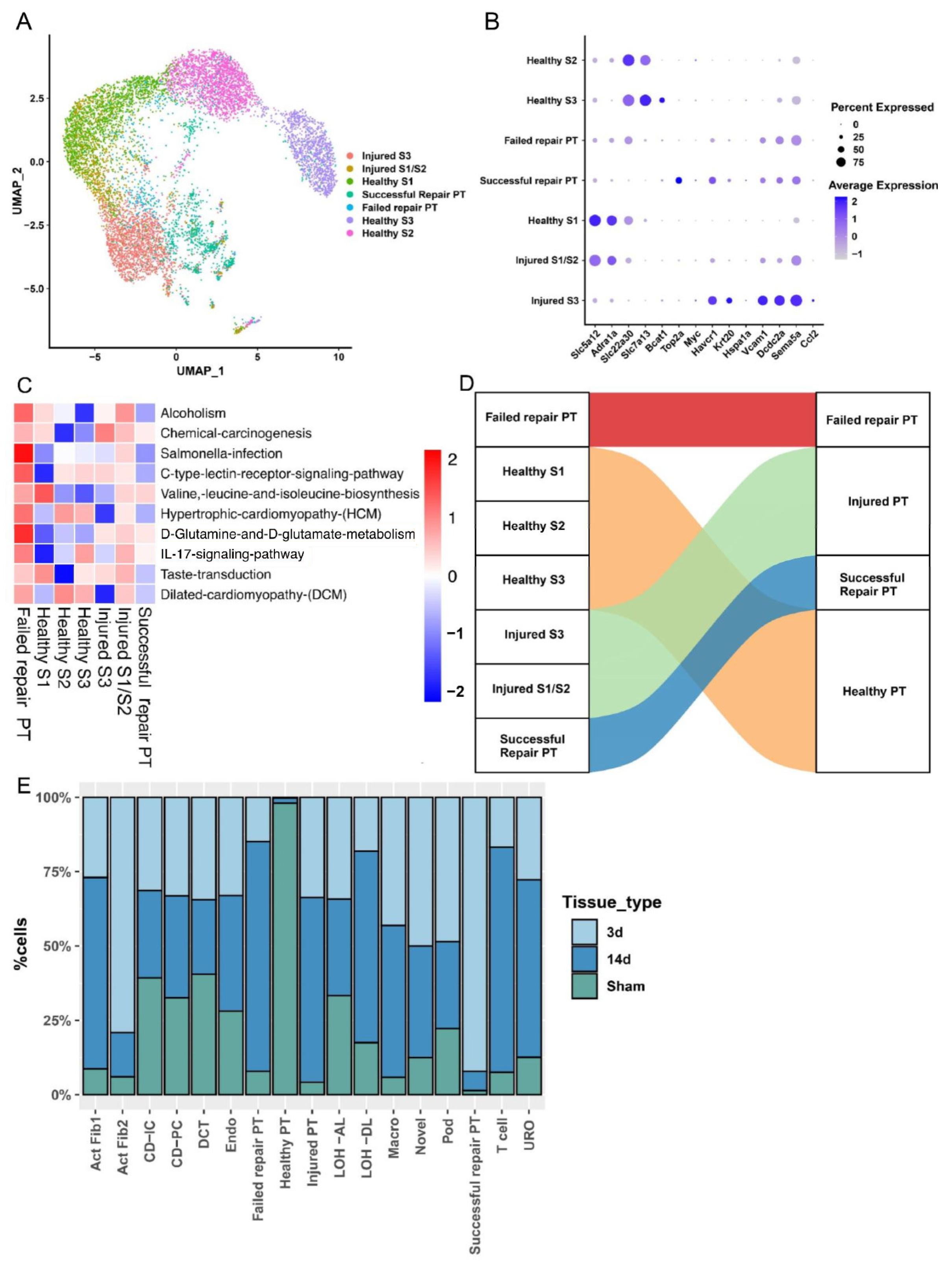
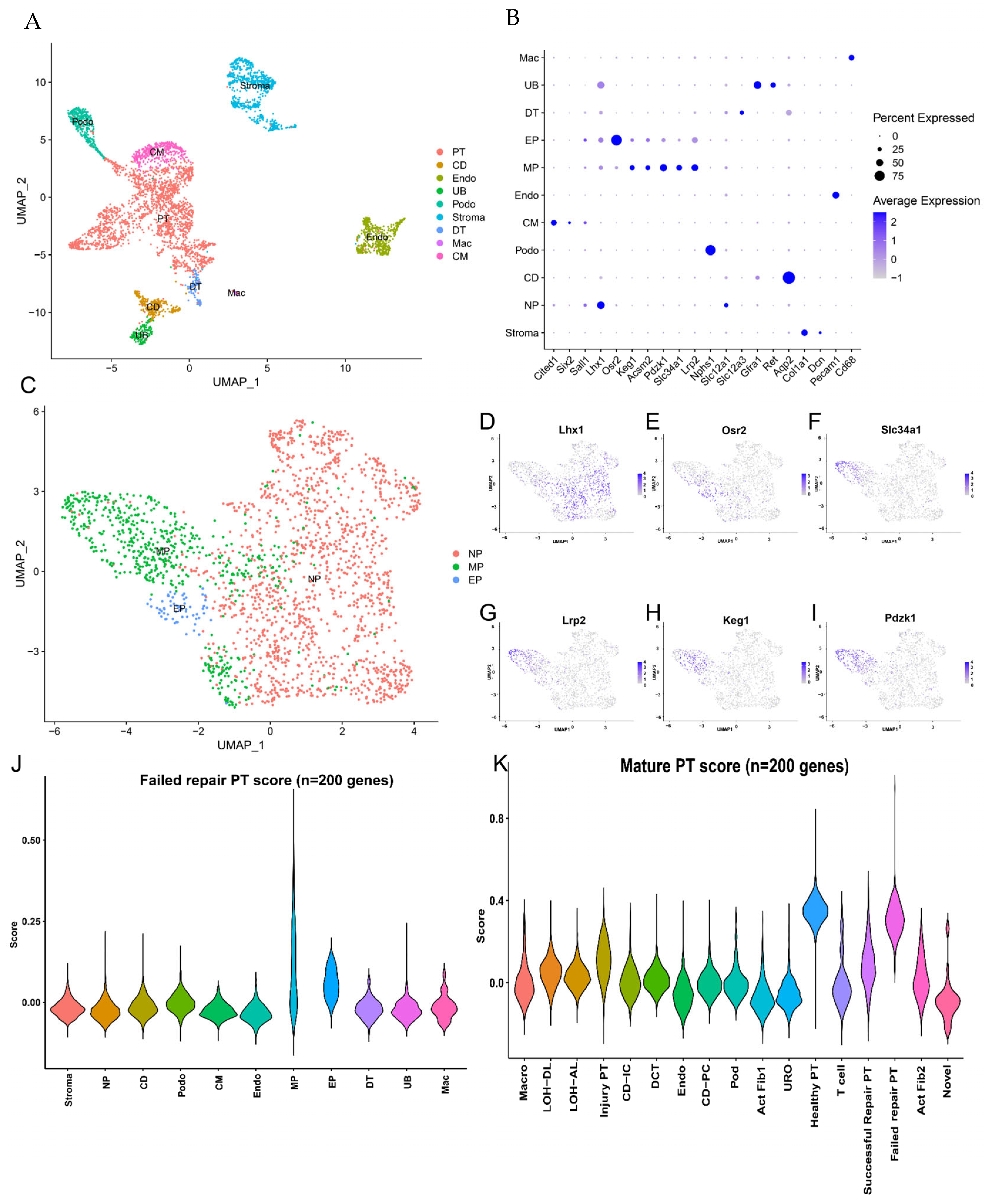
2.2. PT Cell Subsets from AKI to CKD Progression
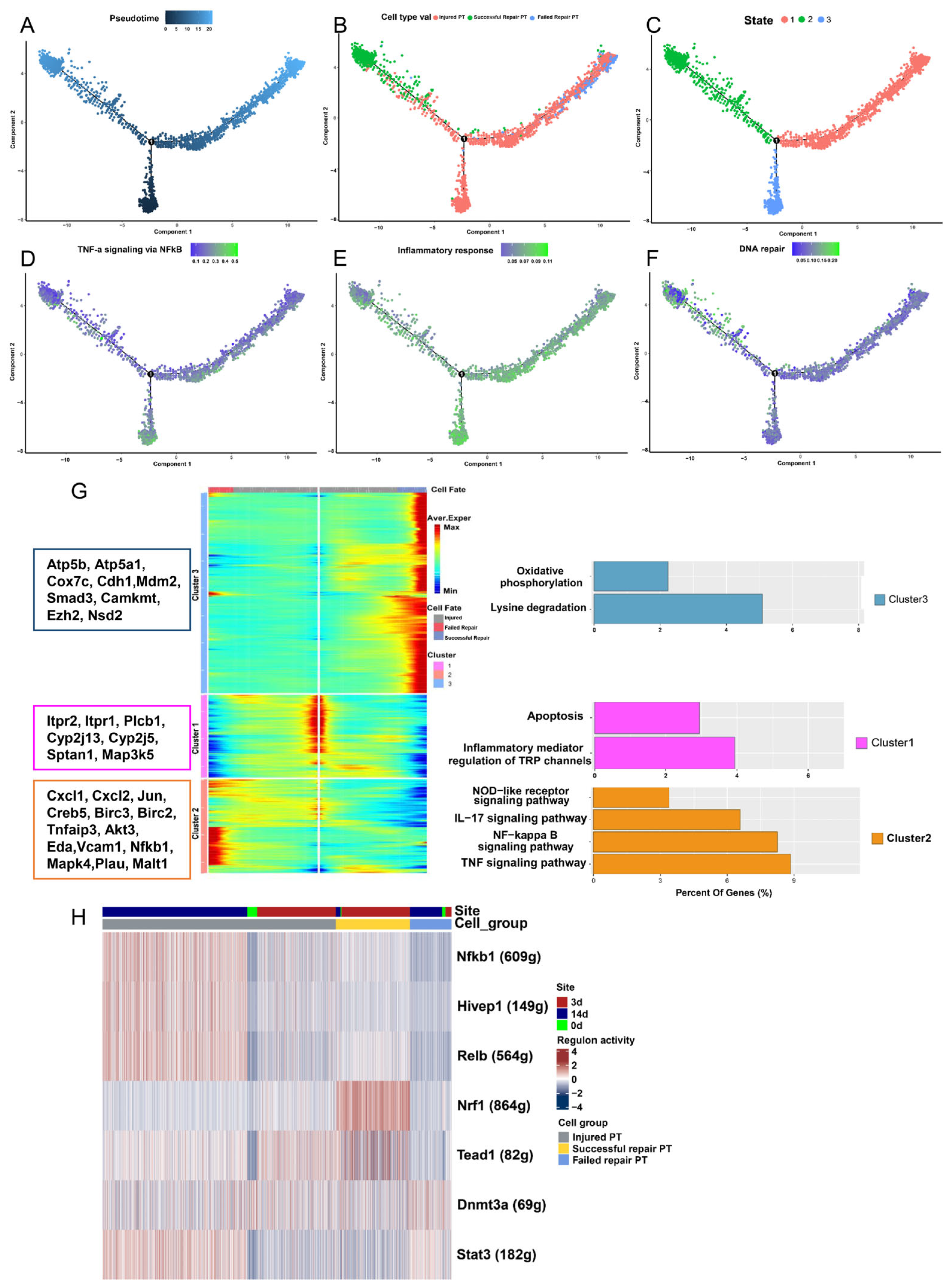
2.3. Analysis of Differentiated State Characteristics
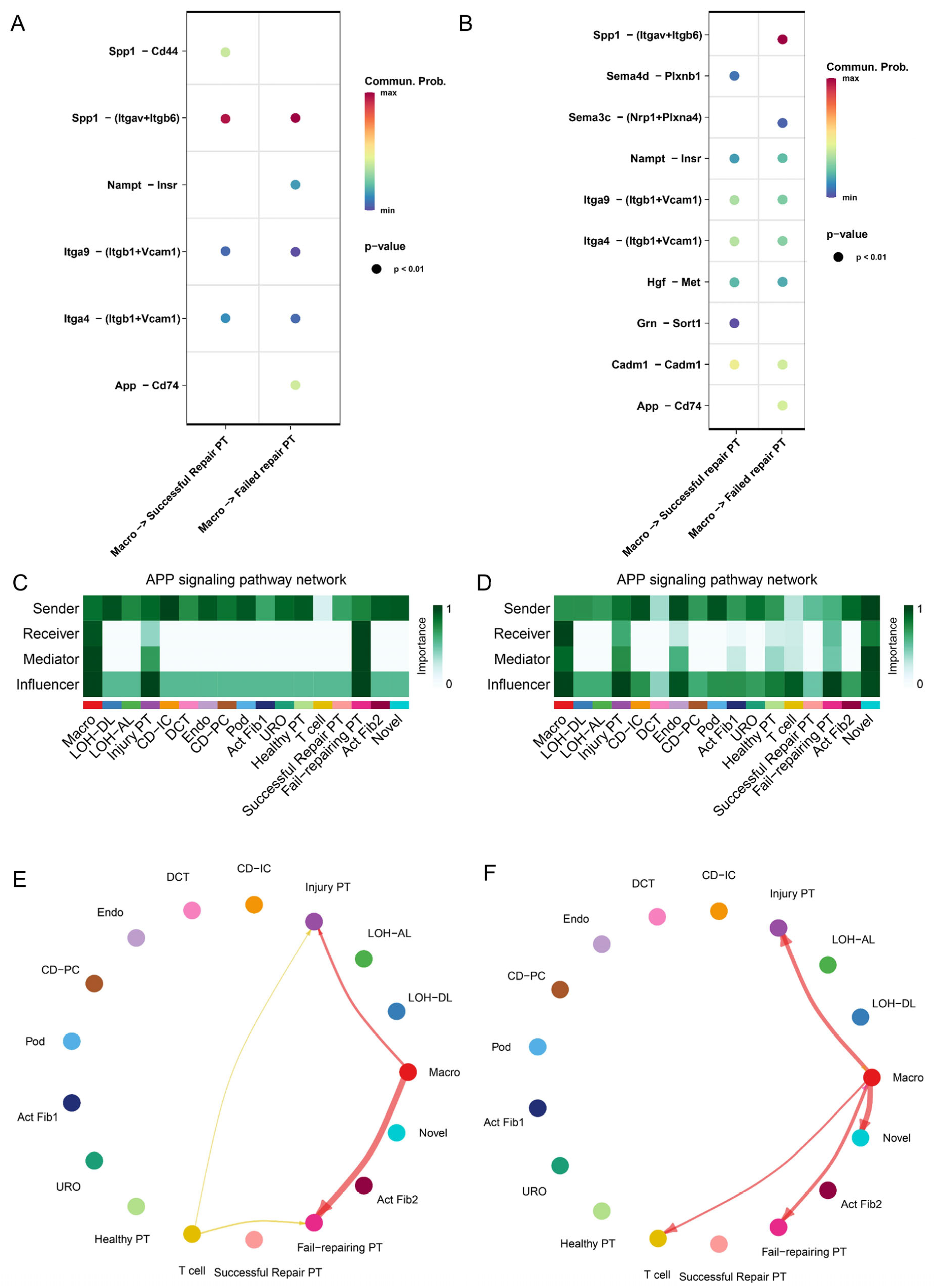
2.4. Cross-Talk between PT Cell Subsets and Immune Microenvironment from AKI to CKD
2.5. Failed Repair PT Cells Emerge with the Oxidative Stress Status after AKI
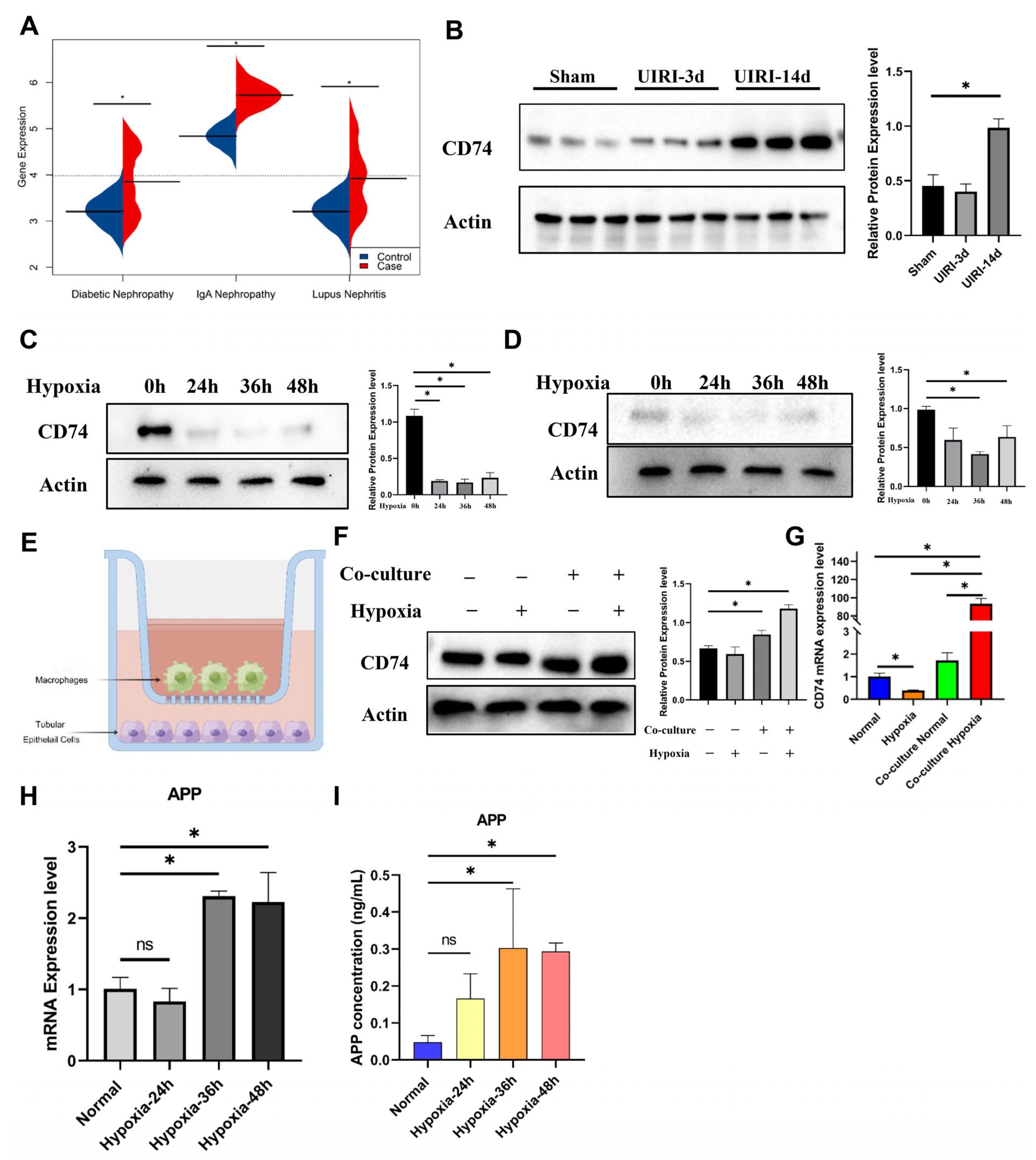
2.6. The IL-17 Pathway and CD74 Play a Key Role in the Progression of FR-PT Cells after Clinical AKI
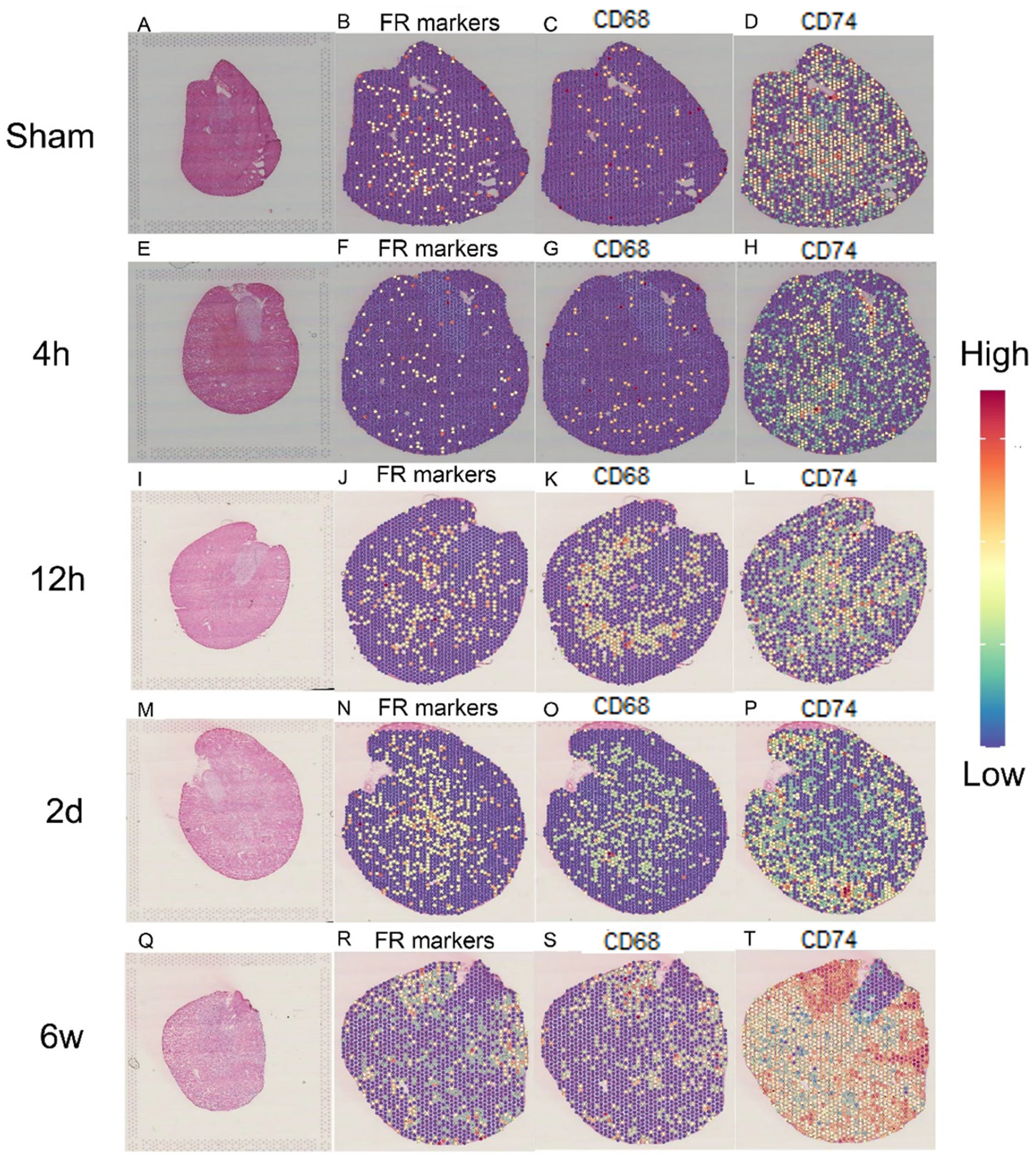
2.7. Spatial Transcriptomics Reveals the Changes after Kidney Injury
2.8. CD74 Upregulation in Tubular Epithelial Cells after Co-Culture with Macrophages
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Mouse Kidney Samples
4.3. 10× Genomics-Based snRNA-seq
4.4. snRNA-seq Data Processing
4.5. Pseudotemporal Analysis
4.6. Cell Communication Analysis
4.7. Gene Enrichment Analysis
4.8. Cell Type Correlation Analysis
4.9. Pathway Annotation Analysis
4.10. Spatial Transcriptomics
4.11. Cell Culture
4.12. Co-Culture
4.13. Quantitative Real-Time PCR (qRT-PCR)
4.14. Western Blotting
4.15. Enzyme-Linked Immunosorbent Assay (ELISA)
4.16. Immunohistochemistry (IHC)
5. Conclusions
Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kurzhagen, J.T.; Dellepiane, S.; Cantaluppi, V.; Rabb, H. AKI: An increasingly recognized risk factor for CKD development and progression. J. Nephrol. 2020, 33, 1171–1187. [Google Scholar] [CrossRef] [PubMed]
- Rewa, O.; Bagshaw, S.M. Acute kidney injury-epidemiology, outcomes and economics. Nat. Rev. Nephrol. 2014, 10, 193–207. [Google Scholar] [CrossRef]
- Hong, S.; Healy, H.; Kassianos, A.J. The Emerging Role of Renal Tubular Epithelial Cells in the Immunological Pathophysiology of Lupus Nephritis. Front. Immunol. 2020, 11, 578952. [Google Scholar] [CrossRef] [PubMed]
- Chang-Panesso, M.; Humphreys, B.D. Cellular plasticity in kidney injury and repair. Nat. Rev. Nephrol. 2017, 13, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Zuk, A.; Bonventre, J.V. Acute Kidney Injury. Annu. Rev. Med. 2016, 67, 293–307. [Google Scholar] [CrossRef]
- Bonventre, J.V. Dedifferentiation and proliferation of surviving epithelial cells in acute renal failure. J. Am. Soc. Nephrol. JASN 2003, 14 (Suppl. 1), S55–S61. [Google Scholar] [CrossRef]
- Guo, J.K.; Cantley, L.G. Cellular maintenance and repair of the kidney. Annu. Rev. Physiol. 2010, 72, 357–376. [Google Scholar] [CrossRef]
- Zheng, Z.; Li, C.; Shao, G.; Li, J.; Xu, K.; Zhao, Z.; Zhang, Z.; Liu, J.; Wu, H. Hippo-YAP/MCP-1 mediated tubular maladaptive repair promote inflammation in renal failed recovery after ischemic AKI. Cell Death Dis. 2021, 12, 754. [Google Scholar] [CrossRef]
- He, L.; Wei, Q.; Liu, J.; Yi, M.; Liu, Y.; Liu, H.; Sun, L.; Peng, Y.; Liu, F.; Venkatachalam, M.A.; et al. AKI on CKD: Heightened injury, suppressed repair, and the underlying mechanisms. Kidney Int. 2017, 92, 1071–1083. [Google Scholar] [CrossRef]
- Basile, D.P.; Anderson, M.D.; Sutton, T.A. Pathophysiology of acute kidney injury. Compr. Physiol. 2012, 2, 1303–1353. [Google Scholar]
- Kinsey, G.R. Macrophage dynamics in AKI to CKD progression. J. Am. Soc. Nephrol. JASN 2014, 25, 209–211. [Google Scholar] [CrossRef] [PubMed]
- Reid, S.; Scholey, J.W. Recent Approaches to Targeting Canonical NFκB Signaling in the Early Inflammatory Response to Renal IRI. J. Am. Soc. Nephrol. JASN 2021, 32, 2117–2124. [Google Scholar] [CrossRef] [PubMed]
- Kirita, Y.; Wu, H.; Uchimura, K.; Wilson, P.C.; Humphreys, B.D. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc. Natl. Acad. Sci. USA 2020, 117, 15874–15883. [Google Scholar] [CrossRef]
- Tang, R.; Meng, T.; Lin, W.; Shen, C.; Ooi, J.D.; Eggenhuizen, P.J.; Jin, P.; Ding, X.; Chen, J.; Tang, Y.; et al. A Partial Picture of the Single-Cell Transcriptomics of Human IgA Nephropathy. Front. Immunol. 2021, 12, 645988. [Google Scholar] [CrossRef] [PubMed]
- Ide, S.; Kobayashi, Y.; Ide, K.; Strausser, S.A.; Abe, K.; Herbek, S.; O’Brien, L.L.; Crowley, S.D.; Barisoni, L.; Tata, A.; et al. Ferroptotic stress promotes the accumulation of pro-inflammatory proximal tubular cells in maladaptive renal repair. eLife 2021, 10, e68603. [Google Scholar] [CrossRef] [PubMed]
- Adam, M.; Potter, A.S.; Potter, S.S. Psychrophilic proteases dramatically reduce single-cell RNA-seq artifacts: A molecular atlas of kidney development. Development 2017, 144, 3625–3632. [Google Scholar] [CrossRef]
- Puthumana, J.; Thiessen-Philbrook, H.; Xu, L.; Coca, S.G.; Garg, A.X.; Himmelfarb, J.; Bhatraju, P.K.; Ikizler, T.A.; Siew, E.D.; Ware, L.B.; et al. Biomarkers of inflammation and repair in kidney disease progression. J. Clin. Investig. 2021, 131, e139927. [Google Scholar] [CrossRef]
- Malone, A.F.; Wu, H.; Fronick, C.; Fulton, R.; Gaut, J.P.; Humphreys, B.D. Harnessing Expressed Single Nucleotide Variation and Single Cell RNA Sequencing To Define Immune Cell Chimerism in the Rejecting Kidney Transplant. J. Am. Soc. Nephrol. JASN 2020, 31, 1977–1986. [Google Scholar] [CrossRef]
- Dixon, E.E.; Wu, H.; Muto, Y.; Wilson, P.C.; Humphreys, B.D. Spatially Resolved Transcriptomic Analysis of Acute Kidney Injury in a Female Murine Model. J. Am. Soc. Nephrol. JASN 2022, 33, 279–289. [Google Scholar] [CrossRef]
- Wei, C.; Banu, K.; Garzon, F.; Basgen, J.M.; Philippe, N.; Yi, Z.; Liu, R.; Choudhuri, J.; Fribourg, M.; Liu, T.; et al. SHROOM3-FYN Interaction Regulates Nephrin Phosphorylation and Affects Albuminuria in Allografts. J. Am. Soc. Nephrol. JASN 2018, 29, 2641–2657. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, J.; Tian, J.; Virzì, G.M.; Digvijay, K.; Cueto, L.; Yin, Y.; Rosner, M.H.; Ronco, C. Mitochondria in Sepsis-Induced AKI. J. Am. Soc. Nephrol. JASN 2019, 30, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Miguel, V.; Tituaña, J.; Herrero, J.I.; Herrero, L.; Serra, D.; Cuevas, P.; Barbas, C.; Puyol, D.R.; Márquez-Expósito, L.; Ruiz-Ortega, M.; et al. Renal tubule Cpt1a overexpression protects from kidney fibrosis by restoring mitochondrial homeostasis. J. Clin. Investig. 2021, 131, e140695. [Google Scholar] [CrossRef]
- Zhao, Z.; Wu, J.; Xu, H.; Zhou, C.; Han, B.; Zhu, H.; Hu, Z.; Ma, Z.; Ming, Z.; Yao, Y.; et al. XJB-5-131 inhibited ferroptosis in tubular epithelial cells after ischemia-reperfusion injury. Cell Death Dis. 2020, 11, 629. [Google Scholar] [CrossRef] [PubMed]
- Xu, L. The Role of Myeloid Cells in Acute Kidney Injury and Kidney Repair. Kidney360 2021, 2, 1852–1864. [Google Scholar] [CrossRef] [PubMed]
- Chou, Y.N.; Lee, M.M.; Deng, J.S.; Jiang, W.P.; Lin, J.G.; Huang, G.J. Water Extract from Brown Strain of Flammulina velutipes Alleviates Cisplatin-Induced Acute Kidney Injury by Attenuating Oxidative Stress, Inflammation, and Autophagy via PI3K/AKT Pathway Regulation. Int. J. Mol. Sci. 2023, 24, 9448. [Google Scholar] [CrossRef]
- Wang, W.; Sai, W.L.; Yang, B. The role of macrophage polarization and interaction with renal tubular epithelial cells in ischemia-reperfusion induced acute kidney injury. Sheng Li Xue Bao 2022, 74, 28–38. [Google Scholar]
- Loverre, A.; Tataranni, T.; Castellano, G.; Divella, C.; Battaglia, M.; Ditonno, P.; Corcelli, M.; Mangino, M.; Gesualdo, L.; Schena, F.P.; et al. IL-17 expression by tubular epithelial cells in renal transplant recipients with acute antibody-mediated rejection. Am. J. Transplant. 2011, 11, 1248–1259. [Google Scholar] [CrossRef]
- Cao, Y.; Chen, X.; Sun, H. Silencing of O-linked N-acetylglucosamine transferase ameliorates hypercalcemia-induced neurotoxicity in renal failure by regulating EZH2/KLF2/CXCL1 axis. Cell Death Dis. 2021, 12, 819. [Google Scholar] [CrossRef]
- Andrade-Oliveira, V.; Foresto-Neto, O.; Watanabe, I.K.M.; Zatz, R.; Câmara, N.O.S. Inflammation in Renal Diseases: New and Old Players. Front. Pharmacol. 2019, 10, 1192. [Google Scholar] [CrossRef]
- Lu, Y.; Li, K.; Hu, Y.; Wang, X. Expression of Immune Related Genes and Possible Regulatory Mechanisms in Alzheimer’s Disease. Front. Immunol. 2021, 12, 768966. [Google Scholar] [CrossRef]
- Fletcher, J.S.; Springer, M.G.; Choi, K.; Jousma, E.; Rizvi, T.A.; Dombi, E.; Kim, M.O.; Wu, J.; Ratner, N. STAT3 inhibition reduces macrophage number and tumor growth in neurofibroma. Oncogene 2019, 38, 2876–2884. [Google Scholar] [CrossRef] [PubMed]
- Celada, L.J.; Kropski, J.A.; Herazo-Maya, J.D.; Luo, W.; Creecy, A.; Abad, A.T.; Chioma, O.S.; Lee, G.; Hassell, N.E.; Shaginurova, G.I.; et al. PD-1 up-regulation on CD4(+) T cells promotes pulmonary fibrosis through STAT3-mediated IL-17A and TGF-β1 production. Sci. Transl. Med. 2018, 10, eaar8356. [Google Scholar] [CrossRef] [PubMed]
- Abplanalp, W.T.; Cremer, S.; John, D.; Hoffmann, J.; Schuhmacher, B.; Merten, M.; Rieger, M.A.; Vasa-Nicotera, M.; Zeiher, A.M.; Dimmeler, S. Clonal Hematopoiesis-Driver DNMT3A Mutations Alter Immune Cells in Heart Failure. Circ. Res. 2021, 128, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Pei, W.; Zhang, M.; Yang, H.; Zhong, M.; Kong, X.; Xu, Y.; Zhu, X.; Chen, T.; et al. LncRNA Dnmt3aos regulates Dnmt3a expression leading to aberrant DNA methylation in macrophage polarization. FASEB J. 2020, 34, 5077–5091. [Google Scholar] [CrossRef]
- Wilkins, H.M.; Swerdlow, R.H. Amyloid precursor protein processing and bioenergetics. Brain Res. Bull. 2017, 133, 71–79. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, Q.; Chen, S.; Xu, C. Functions of amyloid precursor protein in metabolic diseases. Metab. Clin. Exp. 2021, 115, 154454. [Google Scholar] [CrossRef]
- Valiño-Rivas, L.; Baeza-Bermejillo, C.; Gonzalez-Lafuente, L.; Sanz, A.B.; Ortiz, A.; Sanchez-Niño, M.D. CD74 in Kidney Disease. Front. Immunol. 2015, 6, 483. [Google Scholar] [CrossRef]
- He, S.; Wang, L.H.; Liu, Y.; Li, Y.Q.; Chen, H.T.; Xu, J.H.; Peng, W.; Lin, G.W.; Wei, P.P.; Li, B.; et al. Single-cell transcriptome profiling of an adult human cell atlas of 15 major organs. Genome Biol. 2020, 21, 294. [Google Scholar] [CrossRef]
- Zhou, Y.; Yu, Z.; Liu, L.; Wei, L.; Zhao, L.; Huang, L.; Wang, L.; Sun, S. Construction and evaluation of an integrated predictive model for chronic kidney disease based on the random forest and artificial neural network approaches. Biochem. Biophys. Res. Commun. 2022, 603, 21–28. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Qiu, X.; Mao, Q.; Tang, Y.; Wang, L.; Chawla, R.; Pliner, H.A.; Trapnell, C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 2017, 14, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Ji, A.L.; Rubin, A.J.; Thrane, K.; Jiang, S.; Reynolds, D.L.; Meyers, R.M.; Guo, M.G.; George, B.M.; Mollbrink, A.; Bergenstråhle, J.; et al. Multimodal Analysis of Composition and Spatial Architecture in Human Squamous Cell Carcinoma. Cell 2020, 182, 497–514.e22. [Google Scholar] [CrossRef] [PubMed]
- Berglund, E.; Maaskola, J.; Schultz, N.; Friedrich, S.; Marklund, M.; Bergenstråhle, J.; Tarish, F.; Tanoglidi, A.; Vickovic, S.; Larsson, L.; et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 2018, 9, 2419. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, D.; Yang, Q.; Lv, X.; Huang, W.; Zhou, Z.; Wang, Y.; Zhang, Z.; Yuan, T.; Ding, X.; et al. Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat. Commun. 2020, 11, 6322. [Google Scholar] [CrossRef]
- Poorghobadi, S.; Agharezaei, M.; Ghanbari, M.; Bahramali, G.; Abbasian, L.; Sajadipour, M.; Baesi, K. Discordant immune response among treatment experienced patients infected with HIV-1: Crosstalk between MiRNAs expression and CD4+ T cells count. Int. Immunopharmacol. 2023, 114, 109533. [Google Scholar] [CrossRef]
- Zhang, M.; Wu, L.; Deng, Y.; Peng, F.; Wang, T.; Zhao, Y.; Chen, P.; Liu, J.; Cai, G.; Wang, L.; et al. Single Cell Dissection of Epithelial-Immune Cellular Interplay in Acute Kidney Injury Microenvironment. Front. Immunol. 2022, 13, 857025. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Z.; Zhou, Y.; Zhang, Y.; Ning, X.; Li, T.; Wei, L.; Wang, Y.; Bai, X.; Sun, S. Cell Profiling of Acute Kidney Injury to Chronic Kidney Disease Reveals Novel Oxidative Stress Characteristics in the Failed Repair of Proximal Tubule Cells. Int. J. Mol. Sci. 2023, 24, 11617. https://doi.org/10.3390/ijms241411617
Yu Z, Zhou Y, Zhang Y, Ning X, Li T, Wei L, Wang Y, Bai X, Sun S. Cell Profiling of Acute Kidney Injury to Chronic Kidney Disease Reveals Novel Oxidative Stress Characteristics in the Failed Repair of Proximal Tubule Cells. International Journal of Molecular Sciences. 2023; 24(14):11617. https://doi.org/10.3390/ijms241411617
Chicago/Turabian StyleYu, Zhixiang, Ying Zhou, Yuzhan Zhang, Xiaoxuan Ning, Tian Li, Lei Wei, Yingxue Wang, Xiao Bai, and Shiren Sun. 2023. "Cell Profiling of Acute Kidney Injury to Chronic Kidney Disease Reveals Novel Oxidative Stress Characteristics in the Failed Repair of Proximal Tubule Cells" International Journal of Molecular Sciences 24, no. 14: 11617. https://doi.org/10.3390/ijms241411617
APA StyleYu, Z., Zhou, Y., Zhang, Y., Ning, X., Li, T., Wei, L., Wang, Y., Bai, X., & Sun, S. (2023). Cell Profiling of Acute Kidney Injury to Chronic Kidney Disease Reveals Novel Oxidative Stress Characteristics in the Failed Repair of Proximal Tubule Cells. International Journal of Molecular Sciences, 24(14), 11617. https://doi.org/10.3390/ijms241411617






